Abstract
The southwestern highlands forests of Ethiopia are the origin of the coffee plant Coffea arabica. The production of coffee in this area is affected by tracheomycosis caused by a soil-born fungus Gibberella xylarioides. The use of endemic antagonistic strains of mycoparasitic Trichoderma species would be a nature conserving means to combat this disease. We have used molecular methods to reveal that the community of Trichoderma in the rhizosphere of C. arabica in its native forests is highly diverse and includes many putatively endemic species. Among others, the putative new species were particularly efficient to inhibit growth of G. xylarioides.
Keywords:
biocontrol; coffee; diversity; DNA barcode; Ethiopia; Hypocrea; molecular phylogeny; rhizosphere 1. Introduction
Coffee (Coffea arabica L.) is a tropical crop, which today contributes 70% of the world’s commercial coffee market, and is currently grown in 75 countries with a total production close to 108 million ton per year [1,2]. For the African country of Ethiopia, coffee is the major export crop, accounting for over 60% of total value of exports [3]. It originated in the former Province of Kaffa in the southwest of the country [4,5,6] rendering Ethiopia the origin and center of infraspecific genetic diversity for C. arabica.
The production of coffee is today severely affected by fungal wilt diseases including tracheomycosis (coffee wilt disease) caused by Fusarium xylarioides Steyaert (teleomorph: Gibberella xylarioides Heim and Saccas) [7,8,9,10]. The disease is responsible for a reduction in the production of coffee beans and is also accompanied by severe damage and death of millions of coffee bushes [7].
Currently, attempts to control coffee wilt disease are fundamentally based on the breeding of resistant plants, plant and environmental management, and synthetic fungicides [11]. The high cost of pesticides, the emergence of fungicide-resistant pathogen biotypes and other social and health-related impacts of conventional agriculture on the environment have however recently led to an increased interest in agricultural sustainability and biodiversity conservation [12]. Thus, there is a need for new solutions of plant disease problems that provide effective control while minimizing cost and negative consequences for human health and the environment [13].
Biological control—i.e., the antagonism and eventual killing of plant pathogens by other living organisms, which are themselves not harmful to the plants—could present an attractive alternative for combating wilt disease. Species of the anamorphic genus Trichoderma (teleomorph: Hypocrea, Ascomycota) have been proven as effective biocontrol agents of soil-borne plant diseases [14,15]. Trichoderma would be especially suitable for combating coffee wilt disease because many of its species are rhizosphere competent [16,17], and the coffee roots are the first target for the attack by pathogens [18,19]. In support of this hypothesis, Trichoderma spp. have already been applied successfully to suppress Fusarium spp. causing Asparagus root rot [20], bean root rot [21], and carnation wilt [22].
Yet the antagonistic efficacy of a biocontrol agent in the field will also depend on the environment where it will be applied [23], and therefore—ideally—screening for appropriate biocontrol agents should be done in the same environment. This would also avoid the introduction of invasive microorganisms and conserve the microbial composition of the respective biotopes. To date, information about the diversity and abundance of Trichoderma in natural for coffee forest of Ethiopia is unknown.
The objective of this study was to survey the diversity of Trichoderma inhabiting the rhizosphere of C. arabica; and to compare two different coffee habitats, native populations and plantations in major coffee growing regions of Ethiopia with respect to their abundance and distribution of Trichoderma species.
2. Results
2.1. Habitats of C. arabica and the Sampling Strategy
C. arabica is a relatively small evergreen shrub (normally up to 5 m tall), which grows in shady places in subtropical forests. It usually inhabits the highland areas around 1500 m above the mean sea level, but can be found up to 2800 m. C. arabica can grow best on deep, free-draining, loamy soils, with a good water holding capacity and a slightly acid soil.
The four major coffee growing regions of Ethiopia lie in the south, south-western and south-eastern parts of the country corresponding to Wellega, Jimma, Hararghe and SNNP (Southern Nations, Nationalities and Peoples) regions, respectively (Figure 1). The samples were taken during the rainy season of 2006 (August) on the basis of a road survey. More than 160 soil samples were collected along the main roads every several hundred meters in a way that (with the exception of Hararghe) both undisturbed native forests and disturbed semi-forests were equally represented. The latter ecosystem is common in Ethiopia. It is formed by coffee growers, when they intentionally thin out the forest in a way that the coffee plants get more sunlight but at the same time still have enough of large trees to provide adequate shade. In addition in semi-forests farmers also regularly slash the weeds to facilitate harvesting of the coffee beans. In extreme cases the artificial semi-forest may form after the primary burning of the native forest and subsequent introduction of fast growing large trees, which provide shade to coffee shrubs. The native forest is not disturbed by any anthropogenic pressure. Neither coffee plantations nor gardens were sampled.
As wild coffee inhabits a specific ecological niche it is not surprising that the soils sampled from the rhizosphere of C. arabica are similar in their physicochemical properties (Table 1). The representative samples from four studied regions were similar in carbon (approximately 2.3% in average) and nitrogen (approximately 0.3%) content. The pH of soil solutions was around 5 reaching a maximum of 6.5 in samples from the native forest in Hararghe.

Table 1.
Description of sampled areas and physical and chemical properties of soils.
| Region | Ecosystem | Code | Alt. range (m) | Soil properties | No. of samples | No. of Trichoderma isolates | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Color | Texture | C% | N% | pH | ||||||
| Jimma | native forest | JF | 1320–2150 | Reddish brown | Sandy loam | 2.7 | 0.27 | 4.80 | 30 | 26 |
| semi-forest | JC | 1370–2400 | Reddish brown | Silt loam | 2.7 | 0.6 | 4.72 | 29 | 27 | |
| Wellega | native forest | WF | 1500–2300 | Reddish brown | Silt loam | 2.4 | 0.34 | 4.97 | 15 | 13 |
| semi-forest | WC | 1570–2400 | Reddish brown | Sandy loam | 1.6 | 0.24 | 4.67 | 53 | 11 | |
| SNNP | native forest | SF | 1650–2050 | Reddish brown | Sandy loam | 2.3 | 0.26 | 6.01 | 13 | 14 |
| semi-forest | SC | 1670–2080 | Reddish brown | Silt loam | 2.3 | 0.32 | 4.81 | 12 | 37 | |
| Hararghe | native forest | HF | 2150 | Reddish brown | Sandy loam | 2.5 | 0.23 | 6.53 | 2 | 2 |
| semi-forest | HC | 1580–2350 | Reddish brown | Sandy loam | 2 | 0.19 | 4.01 | 6 | 4 | |
2.2. Occurrence of Known Trichoderma Species
There were 134 isolates of Trichoderma recovered from the four major coffee growing regions of Ethiopia (Appendix 1; Figure 1). Among them, 54 came from the rhizosphere of coffee plants in forest soil, whereas 80 were obtained from C. arabica rhizosphere from semi-forests. Among these, 11 and 15 of the native (forest) and disturbed (semi-forest) soil samples respectively, were from plants infected with wilt disease (the presence of the disease was not visible at the time of sampling but later discovered in the laboratory).
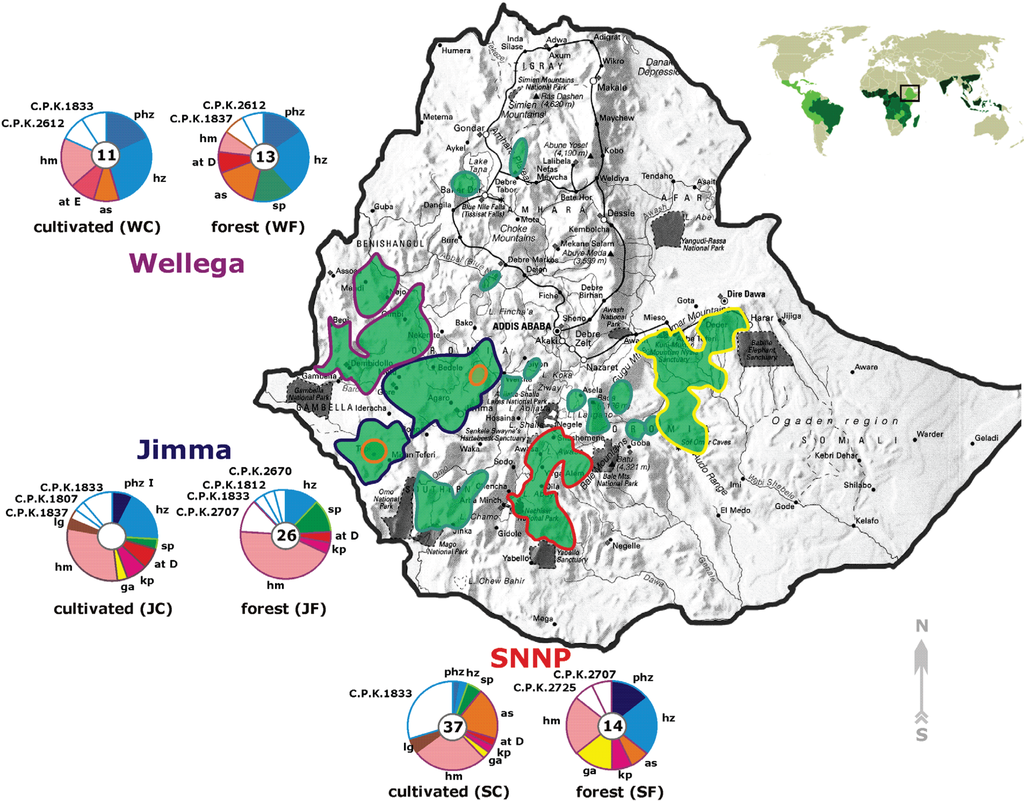
Figure 1.
Location of the main coffee growing areas (green areas) and the corresponding diversity of Hypocrea/Trichoderma in the rhizosphere of C. arabica. The purple line corresponds to Wellega, blue to Jimma, red for the SNNP region and yellow for the Hararghe region. The orange circles indicate the main commercial coffee plantation sites in Ethiopia (Jimma). The diversity of Hypocrea/Trichoderma is shown by separate pie plots (disturbed semi-forest and native forest, respectively) for all coffee growing areas except Hararghe, which was undersampled. The number in the center of each plot indicates the total number of Trichoderma strains isolated from each area. Species are abbreviated as T. harzianum sensu stricto (hz), ‘pseudoharzianum matrix’ (phz), T. hamatum (hm), Hypocrea atroviridis clade E (at E), H. atroviridis clade D (at D), T. asperelloides (as), T. spirale (sp), T. longibrachiatum (lg), T. gamsii (ga), and H. koningiopsis (kp), all potentially new species are numbered as “T. sp.” followed by the corresponding strain number. The world map inset on the upper right side of the image shows the location of Ethiopia in the “coffee belt” where the light green indicates C. arabica production areas, and the dark green shows C. robusta.
The ITS1 and 2 oligonucleotide barcode program TrichOKey [24] identified 104 of the 134 isolates at the species level. Eight known species were found: T. harzianum sensu lato and T. hamatum were most abundant (30 and 35 isolates respectively), followed by T. asperelloides (11 isolates), T. spirale (eight isolates), H. atroviridis/T. atroviride (six isolates) and H. koningiopsis/T. koningiopsis (five isolates). T. gamsii and T. longibrachiatum, were represented by four and three specimens respectively. While ITS1 and 2 sequences are sufficient for distinguishing the majority of Hypocrea/Trichoderma species [24], they have only limited power to differentiate at the infraspecific level. As all identified species are very common and known to be cosmopolitan, it is interesting to learn whether the isolates from Ethiopia represent also common haplotypes of these species, which are already known from other geographic regions. In order to learn this, we sequenced the hypervariable 4th long intron of the elongation factor 1-alpha (tef1) of the corresponding strains. Comparison of the resulting sequences with those of the Database of Industrially Important Fungi of Vienna University of Technology and the NCBI GenBank databases showed that the tef1 sequences of T. longibrachiatum, H. koningiopsis/T. koningiopsis and T. gamsii strains were identical to those of strains known from Europe or South America (data not shown). Isolates of T. harzianum sensu lato also covered a large part of the genetic diversity known for this complex taxon, although most strains were identical or highly similar to strains from Africa (Cameroon, Egypt). The majority of strains belonged to the network of recombining holomorphic strains known as the ‘pseudoharzianum matrix’ [25], while at least four isolates were attributed to the new putative agamospecies T. sp. ‘afroharzianum’ [25] and a single isolate C.P.K. 1818 resembled the ex-type strain of T. harzianum CBS 226.95 (i.e., T. harzianum sensu stricto), which is usually found in a temperate climate [26].
Eleven isolates were initially identified as T. asperellum. However this species has been recently reconsidered as an aggregate of two morphological cryptic species T. asperellum s. s. and T. asperelloides [27]. Phylogenetic analysis of tef1 intron and the detection of the diagnostic SNP of ITS2 of the rRNA gene cluster resulted in the attribution of Ethiopian isolates to T. asperelloides (Figure 2), which is frequent in Africa.
A different situation was encountered with the other three species: strains of T. hamatum exhibited three tef1 alleles, which formed a clade separate from all other known T. hamatum strains in the phylogenetic analysis (data not shown). Most of the strains identified as H. atroviridis/T. atroviride clustered in H. atroviridis clade D [28], forming a terminal branch shared by an isolate from Nepal (C.P.K. 300). One isolate (C.P.K. 2622), although belonging to the H. atroviridis/T. atroviride clade, formed a basal single branch, which cannot be attributed to any clades specified Dodd et al. [28] (Figure 2).
A high, as yet unnoticed genetic diversity was found for T. spirale: a phylogenetic tree of tef1 sequences resulted in the formation of several statistically supported clades with the ex-type strain CBS 346.93 forming a separate lineage (Figure 3). Six of the eight Ethiopian T. spirale isolates formed a so far unique terminal clade remote from the ex-type strain, whereas one (C.P.K. 2606) was located within a closely related subclade and one formed a single lineage which occupies an unresolved position on the T. spirale s. l. cladogram.
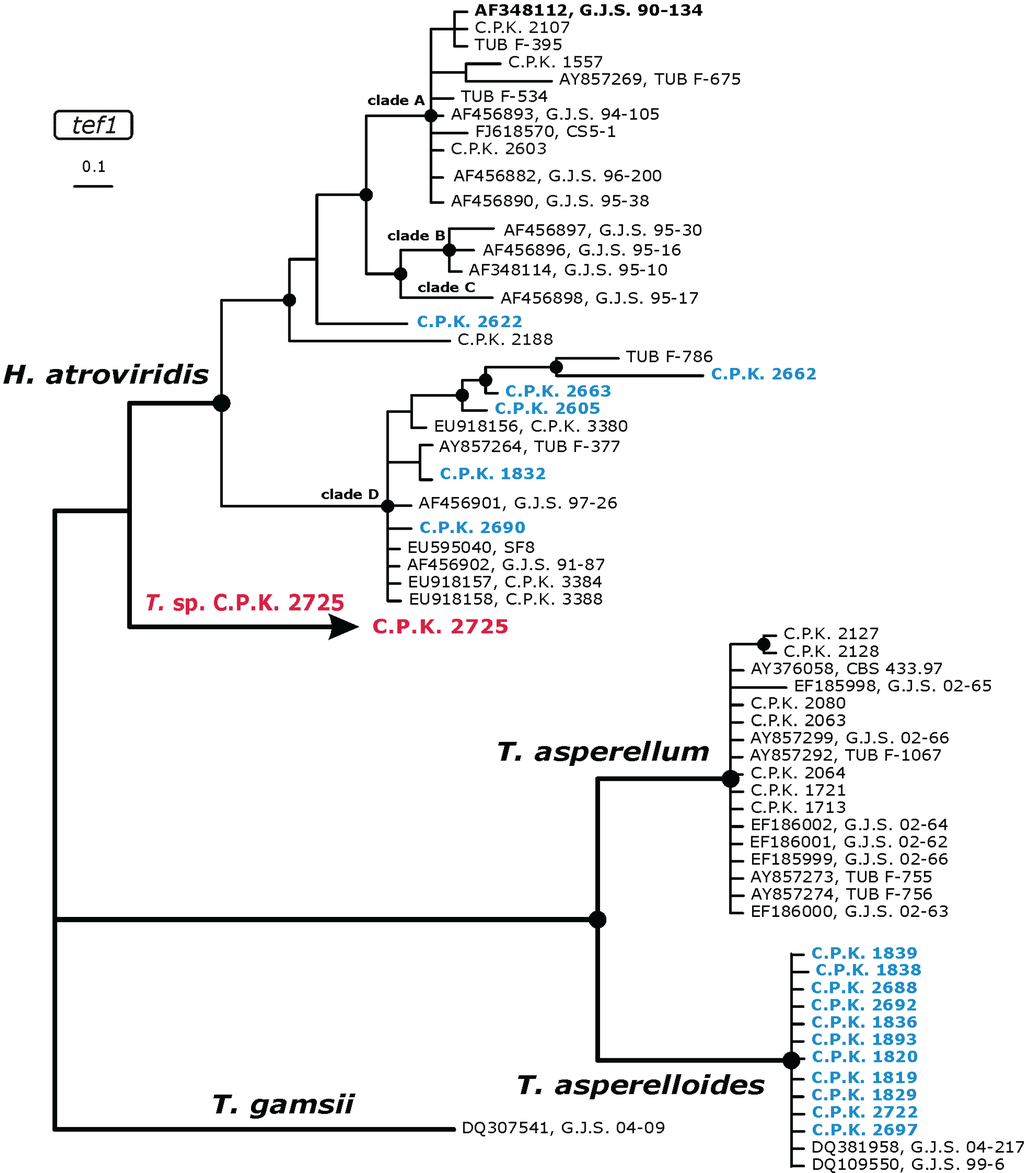
Figure 2.
Identification phylogram of Trichoderma diversity from rhizosphere of C. arabica in a vicinity of H. atroviridis/T. atroviride and T. asperellum inferred from a Bayesian analysis of tef1 intron alignments. The position of the potentially new species is shown in red, while hypothetically endemic populations of known species are marked by blue. Nodes with black circles indicate posterior probabilities >0.94. All reference strains are given by the accession number of the corresponding tef1 sequence in GenBank and/or strain number. Infraspecific groups and species names are given above branches leading to the corresponding node.
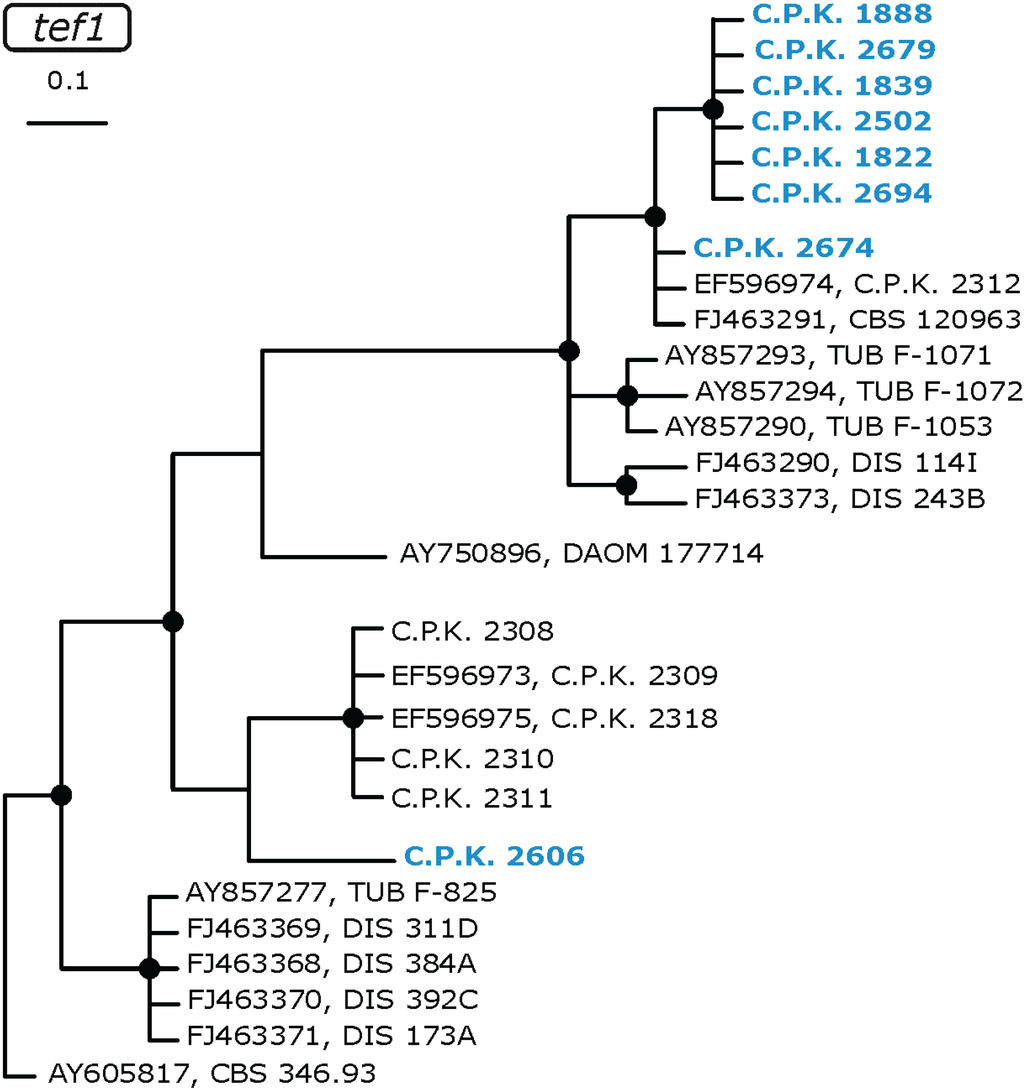
Figure 3.
Identification phylogram of Ethiopian tef1 alleles (in blue) of T. spirale based on a Bayesian analysis. Nodes marked by black circles indicate posterior probabilities >0.94. All reference strains are given by the accession number of the corresponding tef1 sequence in GenBank and the strain number.
2.3. Occurrence of Potentially Endemic Trichoderma Taxa
Thirty (22.6%) of the isolates were identified as eight new alleles of ITS1 and 2 by TrichOKey and thus may constitute new, undescribed species. One of them (T. sp. C.P.K. 1833) accounted for the majority of isolates (16), whereas five of the others putative taxa were encountered only as single specimens. Five, three and two isolates were found for T. sp. C.P.K. 2707, T. sp. C.P.K. 1837 [29] and T. sp. C.P.K. 2612, respectively. Trichoderma sp. C.P.K. 1837 was recently shown to be a new phylogenetic species closely related to H. orientalis and T. longibrachiatum [29] of Trichoderma section Longibrachiatum. For the other putative new species hallmark sequences suggested that they are new members of Trichoderma section Pachybasium and section Trichoderma.
In order to test the hypothesis that these are new phylogenetic species, we also sequenced the tef1 fragment from these 30 isolates and used them for an analysis together with sequences of the most closely related reference species. Figure 4A shows that T. sp. C.P.K. 1828 is a member of the Brevicompactum clade [30], forming a separate branch between T. arundinaceum and H. rodmanii, and the four isolates of T. sp. C.P.K. 2707 form a sister branch to T. helicum (Figure 4B).
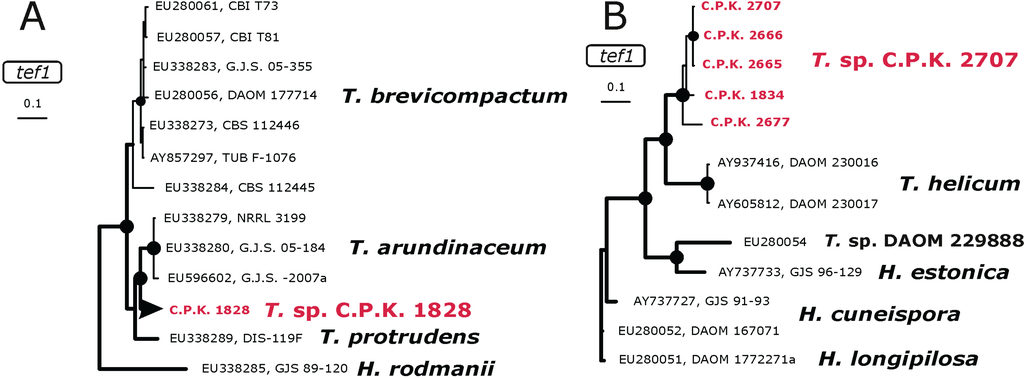
Figure 4.
Identification phylogram of a potentially new species within the Brevicompactum clade (a) and a potentially new species related to T. helicum (b) based on the Bayesian analysis of tef1 intron. Nodes marked by black circles indicate posterior probabilities > 0.94. All reference strains are given by the accession number of the corresponding tef1 sequence in GenBank and strain number.
The remaining five putative new species were located within the Harzianum clade (Figure 5): one branch leading to the most abundant T. sp. C.P.K. 1833 and the single lineage corresponding to T. sp. C.P.K. 2670 formed a sister clade to the T. cerinum–T. tomentosum species pair. The two isolates of T. sp. C.P.K. 2612 formed an isolated branch basal to all other species. And the remaining two hypothetical new species—T. sp. C.P.K. 1812 and C.P.K. 1807 were located on basal branches to the clades containing e.g., Trichoderma species causing green mould disease of Pleurotus (T. pleuroticola and T. pleurotum) and Agaricus (T. aggressivum) respectively (Figure 5). The single putative new species of section Trichoderma (T. sp. C.P.K. 2725) formed a basal branch to H. atroviridis as shown in Figure 2.
2.4. Habitat Preference of Trichoderma
In order to assess the richness of Trichoderma in the rhizosphere of C. arabica, we calculated Shannon’s biodiversity index H and evenness E, as well as the Simpson’s biodiversity index (Table 2). Simpson’s index and the evenness index were close to 1, indicating very high diversity and lack of dominance of any species. When the indexes are calculated separately for cultivated and uncultivated areas it becomes apparent that the diversity and evenness is equal in both ecosystems.
The high diversity was also reflected in the distribution of Trichoderma species. With the exception of species which were only obtained as single isolates, all other species were encountered from more than one area indicating that they may be evenly distributed within the coffee rhizosphere. Yet some remarkable exceptions could be seen: T. sp. C.P.K. 1833 was mostly isolated from semi-forest soils and T. sp. C.P.K. 2707 was only observed in the native forest. We also note that T. hamatum was particularly abundant in Jimma province, both in semi-forest as well as native forest rhizosphere (Figure 1).
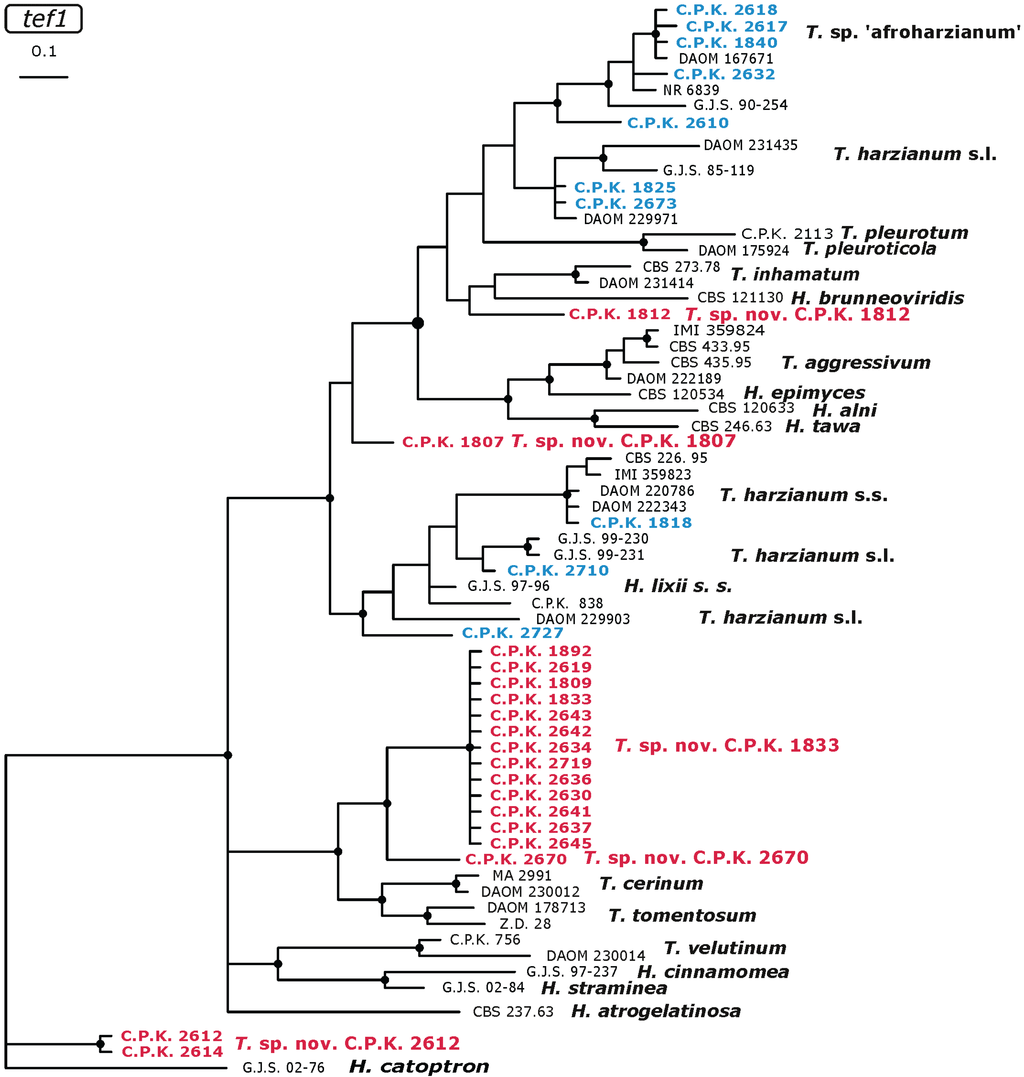
Figure 5.
Distribution of Ethiopian Trichoderma strains within Harzianum clade as inferred from a Bayesian analysis of tef1 intron showing the position of potentially new species (red) and hypothetically endemic populations (blue). Nodes marked by black circles indicate posterior probabilities >0.94. GenBank accession numbers of the corresponding tef1 sequence of the reference strains are given in [25].
In order to learn whether there would be any ecological preference of individual species, we performed a correlation analysis of the occurrence of strains with habitat specific properties such as altitude, pH, and the carbon and nitrogen content of the soils. Undisturbed forest soils in SNNP and Hararghe areas had a higher pH values (6.0–6.5) compared to other locations. Nevertheless, results from these analyses showed that there was no correlation between occurrence of any Trichoderma species and any of factors monitored in this study. Only exception was the observation that cosmopolitan species which were also known from other geographic regions and continents appeared as better adapted to grow on soils poor in nitrogen and carbon. For example, occurrence of T. asperelloides was significantly higher on such soils (ANOVA, P < 0.05).

Table 2.
Diversity of Trichoderma in major Ethiopian coffee growing regions.
| Index | Average | Ecosystem | Sampled regions | |||||
|---|---|---|---|---|---|---|---|---|
| Forest | Semi-forest | Wellega | Jimma | SNNP | Hararghe | |||
| Shannon diversity | H | 2.47 | 2.34 | 2.29 | 2.14 | 2.19 | 2.15 | 1.56 |
| Simpson | D | 0.88 | 0.87 | 0.87 | 0.9 | 0.84 | 0.86 | 0.93 |
2.5. Antagonistic Activity of Trichoderma Isolates against Gibberella xylarioides
In order to test whether the isolates of this study would be applicable as agents of biological control against plant pathogenic fungi (biocontrol agents), we tested a subset of randomly selected strains against an indigenous strain of G. xylarioides causing the coffee wilt in Ethiopia. Table 3 shows that all 10 isolates of Trichoderma tested were in fact able to inhibit the growth of G. xylarioides in vitro (PIRG between 55% and 76%). The hypothetically new taxon T sp. C.P.K. 2612 showed the highest antagonistic activity in this test. Interestingly, three strains (T. sp. C.P.K. 1817, H. atroviridis/T. atroviride C.P.K. 2622, and T. sp. C.P.K. 1834) also produced a zone of inhibition indicative of the formation of secondary metabolite(s) inhibiting G. xylarioides.
3. Discussion
Rhizosphere competence—i.e., the ability to competitively colonize plant roots—has often been emphasized as a prerequisite for strains of genus Trichoderma to act as biocontrol agents (cf. [14]). However, to the best of our knowledge, the issue whether Trichoderma spp. prefer the rhizosphere as an ecological niche in soil has never been systematically addressed. In this study, we show that the rhizosphere of a C. arabica exhibits a notably high diversity of Trichoderma taxa. In fact, a correlation analysis failed to detect any other factor which appeared to influence species richness and distribution, and we thus conclude that the ecological-physiological constraint put by C. arabica ecological niche is the main parameter determining the presence of Trichoderma.
The major coffee growing regions of Ethiopia constitute one of the 34 biodiversity hotspot areas of the world [31]. In total they comprise 80% of the landmass of the Afromontane biodiversity hotspot that is inhabited by more than 700 plant species. The rhizosphere of C. arabica exhibited one of the highest richness of Trichoderma species detected so far (cf. [32,33,34]). The only study which may be comparable in sampling size relative to the studied area was performed in neotropical forests of South America, mainly in Colombia [34]. In that study the exclusively high diversity of Trichoderma (29 species detected among 183 isolates) was also accompanied by a high fraction of putatively new species (11 species corresponding to 6% of the sample). However the study of Hoyos-Carvajal et al. [34] included strains isolated from a great diversity of ecological niches and substrata and therefore the observed species richness likely reflects the overall microbial biodiversity potential for neotropical regions. The principal difference of our findings is that we studied a limited and well defined microecological niche of rhizosphere of C. arabica. In another investigation of Trichoderma diversity based on a cultivation-dependent approach on Sardinia [32], around 500 Trichoderma strains have been isolated from different non-rhizosphere soil samples. The Shannon index calculated for all samples from the rhizosphere of C. arabica in Ethiopia is nearly two times higher compared to the same statistics calculated for soils in Sardinia (2.47 versus 1.59 respectively), although that sample was three times larger as compared to the present study. As Sardinian ecosystems were found to be seriously disturbed by various human activities and while in Ethiopia we sampled the largely undisturbed areas, our current results demonstrate that Trichoderma diversity may be used as an ecological indicator of ‘soil health’.

Table 3.
Ability of selected Trichoderma strains isolated from rhizosphere of C. arabica to inhibit G. xylarioides.
| Isolate No. | Species | PIRG1 | Clear Zone |
|---|---|---|---|
| C.P.K. 2612 | T. sp. C.P.K.2612 | 76a | - |
| C.P.K. 2614 | T. sp. C.P.K.2612 | 72b | - |
| C.P.K. 1808 | T. sp. C.P.K.1833 | 64c | - |
| C.P.K. 2619 | T. sp. C.P.K.1833 | 62c | - |
| C.P.K. 1817 | T. sp. C.P.K.1837 | 62c | 1.5 |
| C.P.K. 2698 | T. hamatum | 60c | - |
| C.P.K. 2622 | H. atroviride | 56d | 2 |
| C.P.K. 1819 | T. asperelloides | 56d | - |
| C.P.K. 1888 | T. spirale | 55d | - |
| C.P.K. 1834 | T .sp. C.P.K.2707 | 55d | 1.5 |
| F. xylarioides (Control) | 0 | - | |
1 See the explanations in the Experimental Section.
The samples isolated from the rhizosphere of coffee plants also contain more of putatively new taxa than any other previous study (28% of all isolates; eight known species versus nine potentially new taxa; compare with Migheli et al. [32], Zachow et al. [33] Hoyos-Carvajala [34] or Kullnig et al. [35]). No new species have been detected among 500 isolates from soils in Sardinia. Although our identification of new taxa was based only on ITS1 and 2 sequence analysis and tef1 phylogeny, and this claim still needs to be verified by analysis of additional loci, we consider this procedure reliable. In all our previous studies, prediction of new taxa by a combination of ITS1 and 2 and tef1 polymorphisms has later on always been confirmed by multiloci phylogeny (e.g., [35,36]).
One of these potentially new species (T. sp. C.P.K. 1833) was remarkable as it was the third most frequent taxon sampled in this study (16 isolates), while at the same time mainly being recovered from C. arabica rhizosphere growing in semi-forests. Comparison with our database revealed that strains of this putative species have been isolated previously from soil in Siberia ([35], erroneously identified as T. oblongisporum), Guatemala [24], Slovakia and Kenya (unpublished data), and thus it is likely cosmopolitan. Phylogenetically, this species appears to be closely related to T. tomentosum and T. cerinum. The fact that it was so abundant in Ethiopia and that its phylogenetic clade was even accompanied by a basal branch to a further potentially new taxon (T. sp. C.P.K. 2670) may suggest that T. sp. C.P.K. 1833 has an East African origin. Its high antagonistic ability renders it a potential candidate for biological control trials, because it has been isolated from disturbed forests.
The diversity of known species was dominated by T. harzianum, T. hamatum, T. spirale and T. asperelloides. All of these species are regarded as the most frequent representatives of the genus in soil and known to be cosmopolitan. They all have been also abundantly detected in Sardinian soils. We therefore consider them as ecological opportunists, which have very efficient abilities in conidial dispersal and germination. Their presence could thus be interpreted to be due to invasion. However, T. spirale is more frequently isolated from tropical environments [24,34,35,36], and also T. asperelloides has been reported to be abundant in South America and Cameroon [27,37]. In addition, most strains of T. harzianum sensu lato, for instance, T. sp. ‘afroharzianum’ [25] also belonged to tef1 phylogenetic clades that were either rich in tropical isolates or being exclusively African.
In this study, we also tested the hypothesis that Trichoderma taxa hypothetically endemic to the rhizosphere of C. arabica would exhibit antagonistic activity against the coffee pathogen G. xylarioides. Although we randomly selected only a small subsample of our isolates for these tests and made only in vitro dual confrontation assays, the results performed verify this hypothesis. Moreover, in planta experiments, performed with different varieties of C. arabica, gave similar results (T. B. Mulaw, unpublished data). The fact, that the putative new taxa were particularly efficient antagonists, supports the above hypothesis for enrichment of these taxa in the coffee rhizosphere. Selecting biocontrol strains by looking for isolates associated with the respective plant may therefore be a method best suited for ecosystems with a largely preserved microbial flora.
4. Experimental Section
4.1. Soil Sampling and Characterization
At each sampling site, a visually healthy adult C. arabica plant was located and a sample was taken within its rhizosphere area. The top layer of a soil litter and the upper soil horizon (4–6 cm) was discarded, and 100 g of soil from approximately 10 cm depth was collected, placed in polyethylene bags and labeled. Afterwards, similar samples from one subarea (geographic region, type of growth of C. arabica, latitude and tracheomycosis state of the forest) were merged in several larger samples, which were subsequently sieved and dried on sterile paper for 2–3 days. Thereafter, they were stored at 4 °C until isolation of Trichoderma strains occurred.
The soil color was defined using a standard color scale for soil science (Munsell Soil Color Charts, U.S. Dept. of Agriculture). All chemical analyses were performed using the fine earth fraction. To measure pH, 1 g of soil was suspended in 100 mL of 1M KCl, and after shaking for one hour pH was determined with a glass electrode. The total nitrogen content was determined according to the Kjeldahl method [38] on a Vapodest 30 (Gerhardt, Germany). The total organic carbon content was measured using the Liechtenfelder method [38], which oxidizes carbon with potassium dichromate, and quantifies the generated Cr3+ photometrically (DIN 19684).
4.2. Strain Isolation and Purification
10 g of soil samples (if necessary pulverized by means of a mortar and pestle, and passed through a 0.5 mm soil screen mesh to remove large debris and root fragments; [39] ) were suspended in 90 mL sterile distilled water and thoroughly mixed. A 10 mL aliquot was then used to prepare a series of dilutions in the range of 10–1 to 10–3, and inoculated on to potato dextrose agar (PDA), malt extract agar (MEA) and synthetic low nutrient agar (SNA) [40], each supplemented with streptomycin (50 mg/L) to prevent bacterial growth. Three replicates were done for each medium and soil sample. Plates were incubated at 25 °C for a period of 10 days, and examined daily for colony development. Putative Trichoderma colonies, indicated by their extensive fluffy white mycelium and distinctively green sporulation, were collected, and used to prepare single spore cultures. They were maintained in 50% (w/v) glycerol at −80 °C in the Collection of Industrially Important Microorganisms of Vienna University of Technology, Austria.
4.3. DNA Extraction, PCR Amplification and Sequencing
Genomic DNA was extracted from mycelia grown on MEA with the Plant DNeasy Minikit (QIAgen GmbH, Hilden, Germany) according to the manufacturer’s instructions. A region of nuclear DNA containing the ITS1 and two regions of the rRNA gene cluster, was amplified by PCR with the primer combinations SR6R and LR1 [40], and an approximately 1-kb portion of the gene encoding translation elongation factor 1-alpha (tef1) was amplified and sequenced using the primers EF1 (5’-ATGGGTAAGGA(A/G)GACAAGAC-3’) and EF2 (GGA(G/A)GTACCA GT(G/C)ATCATGTT-3) as described by Jaklitsch et al. [40]. PCR products were purified with the QIAquick PCR Purification Kit (QIAgen) and were subjected to automated sequencing at MWG (Martinsried, Germany). The sequences obtained in this study have been deposited at GenBank; their accession numbers are given in Appendix 1.
4.4. Identification of Trichoderma
For species identification, ITS 1 and 2 sequences were subjected to analysis by TrichOKey (http://www.isth.info/tools/molkey/index.php; [24]). In ambiguous cases, the result was re-checked by sequence analysis of the large intron of tef1 using a sequence similarity search against a database of type sequences implemented in TrichoBLAST (www.isth.info/tools/blast, [41]). For analysis of unusual ITS1 and 2 or tef1 alleles, sequences were automatically aligned with ClustalX [42] and visually checked using Genedoc 2.6.002 [43]. Potentially unique alleles were then confirmed by BLAST analysis against NCBI GenBank, and the database of Collection of Industrially Important Microorganisms of Vienna University of Technology, Austria which currently contains more than 3500 Hypocrea/Trichoderma strains with more than 5800 sequences of phylogenetic marker loci.
4.5. Molecular Phylogenetic Analysis
A multiple sequence alignment file was formatted with PAUP*4.0b10, and manually adapted for the MrBayes v 3.0B4 program [44,45,46]. The model of evolution and prior settings for individual loci were used as estimated for different taxa of Hypocrea/Trichoderma [47]. Metropolis-coupled Markov chain Monte Carlo (MCMCMC) sampling was performed with four incrementally heated chains that were simultaneously run for 1,000,000 and 3,000,000 generations. To check for potentially poor mixing of MCMCMC, each analysis was repeated several times. The convergence of MCMCMC was monitored by examining the value of the marginal likelihood through generations. Convergence of substitution rate and rate heterogeneity model parameters were also checked. Bayesian posterior probabilities (PP) were obtained from the 50% majority rule consensus of trees sampled every 100 generations after removing the 500 first trees.
4.6. Antagonism of Trichoderma Isolates against Gibberella xylarioides
Ten different isolates of Trichoderma were individually tested for their antagonistic property against G. xylarioides using the dual culture technique. Agar pieces of 6 mm diameter of G. xylarioides and Trichoderma isolates were taken from pure cultures 7 days old, and placed on plates of potato dextrose agar (3%, Merck, Germany) in a distance 6 cm between G. xylarioides and the Trichoderma strain. Plates were incubated at room temperature for 5 days. Five plates were prepared for each isolate. Plates inoculated with G. xylarioides alone served as control. Clear zone of inhibition (CZI) was also determined by measuring the clearance between the colony margins of the G. xylarioides and Trichoderma. Radial growth of both G. xylarioides and Trichoderma were measured after 5 days after inoculation.
The percentage of inhibition of radial growth (PIRG) was calculated as ([R1-R2]x100)/R1, where R1 is the colony diameter of the pathogen in the control and R2 is the diameter of the pathogen during antagonistic interaction. Antagonism was assessed in semi-quantitative means [48]: >75 PIRG indicating very high antagonistic activity, 61–75 PIRG indicating high antagonistic activity, 51–61 PIRG, indicating moderate antagonistic activity, <50 PIRG, indicating low antagonistic activity, and 0 indicating no activity.
The data were analyzed using SPSS version 17.0 software.
4.7. Statistical Analysis
The Shannon biodiversity index H was used to evaluate the species diversity, which appears as the product of evenness E and the number of species [49]. It measures the likelihood that the next individual will be the same species as the previous sample. Given a sample size with many (more than 5) species, a value near 0 would indicate that every species in the sample is the same, whereas a value near or 4.6 would indicate that the number of individuals is evenly distributed between the 5 species. Dominance of individual species was also measured by Simpson's diversity index [50], using formula D= 1 − ∑ini (ni − 1)i / N(N − 1), where ni represents specimens of a species and N represents the total number of species. This index reflects the probability that two individuals randomly selected from a sample will belong to different species.
5. Conclusions
Our data strongly support the speculation that the C. arabica rhizosphere in Ethiopia is a hotspot for speciation of several Trichoderma spp. The putative Ethiopian endemic nature of several of the new taxa and populations encountered in this study would be consistent with an allopatric speciation scenario. Yet this usually occurs by geographic isolation [51], and although it is possible that the East African highland could present a barrier causing such isolation, the cosmopolitan nature of many of the taxa found in this study argues against this possibility. Therefore, we rather suppose that the association with rhizosphere of C. arabica, which is native for the region, considerably contributed to this speciation. C. arabica is known to display a high genetic diversity in Ethiopia [52] and Tanzania [53]. We consider it possible that this genetic diversity has given rise to new Trichoderma populations and taxa capable of establishing themselves in the rhizosphere of a genetically variable plant. It will thus be interesting to look for Trichoderma populations outside the rhizosphere of C. arabica or/and to compare rhizosphere samples from Tanzania, as well as in areas growing other coffee species (for instance, C. robusta) and/or genetically less variable cultivars of Arabica coffee such as Indonesia or Central America [54].
Given that this investigation is the first of its kind in coffee growing areas of Ethiopia, and that studies on the wild C. arabica-associated microbial community are generally lacking, there is high demand for further research in this field. It is also recommended that further studies be conducted to determine microbial communities using both culture-dependent and culture-independent techniques to reveal the true picture of Trichoderma diversity in such specific microbial communities.
Acknowledgements
This work was partly supported by the Austrian Science Fund grants FWF P-17859 to I.S.D and by OeAD 894/07 North-South Dialogue Program, doctorate scholarship to T.B. Mulaw. The authors are grateful to Monika Komon-Zelazowska (Vienna University of Technology, Austria) for her laboratory assistance and to Vera Terekhova (Moscow State University, Soil Science Faculty, Russia) for her help with the soil analyses.
References
- Coffee. 2009. Available online: http://www.coffeeresearch.org/ (accessed on 5 May 2009).
- Davids, K. Alternative Africas: Rwanda, Tanzania, Zimbabwe. Available online: http://www.coffeereview.com/ (accessed on 5 May 2009).
- Fair Trade Farmers in Ethiopia. 2004. Available online: http://www.globalexchange.org/campaigns/fairtrade/coffee/EthiopiaFlyer.pdf (accessed on 5 May 2009).
- Wellman, F.L. Coffee: Botany, Cultivation and Utilization. World Crop Series; Inter Science: New York, NY, USA, 1961. [Google Scholar]
- Meyer, F. Notes on wild Coffea arabica from southwestern Ethiopia, with some historical considerations. Econ. Bot. 1965, 19, 136–151. [Google Scholar] [CrossRef]
- Friis, I. The wild populations of Coffea arabica L., and cultivated coffee. In Taxonomic Aspects of African Economic Botany, Proceedings of the IX plenary meeting of AETFAT, Las Palmas de Gran Canaria, 18–23 March, 1978; Kunkel, G., Ed.; Bentham-Moxon Trust: Les Palmas de Gran Canaria, Spain, 1979. [Google Scholar]
- Adugna, G.; Hulluka, M.; Hindorf, H. Incidence of tracheomycosis, Gibberella xylarioides (Fusarium xylarioides), on Arabica coffee in Ethiopia. Ztschr. Pflanzenkrankh. Pflanzenschutz 2001, 108, 136–142. [Google Scholar]
- Geiser, D.M.; Ivey, M.L.L.; Hakiza, G.; Juba, J.H.; Miller, S.A. Gibberella xylarioides (anamorph: Fusarium xylarioides), a causative agent of coffee wilt disease in Africa, is a previously unrecognized member of the G. fujikuroi species complex. Mycologia 2005, 97, 191–201. [Google Scholar] [CrossRef]
- Serani, S.; Taligoola, H.K.; Hakiza, G.J. An investigation into Fusarium spp. associated with coffee and banana plants as potential pathogens of robusta coffee. Afric. J. Ecol. 2007, 45, 91–95. [Google Scholar]
- Silva, M.C.; Várzea, V.; Guerra-Guimarães, L.; Azinheira, H.G.; Fernandez, D.; Petitot, A.-S.; Bertrand Lashermes, B.P.; Nicole, M. Coffee resistance to the main diseases: Leaf rust and coffee berry disease. Braz. J. Plant Physiol. 2006, 18, 119–147. [Google Scholar] [CrossRef]
- Strange, R.N. Plant Disease Control: Towards Environmentally Acceptable Methods; Chapman and Hall: New York, NY, USA, 1993. [Google Scholar]
- Van der Vossen, H.A.M. A critical analysis of the agronomic and economic sustainability of organic coffee production. Exper. Agricult. 2005, 41, 449–473. [Google Scholar] [CrossRef]
- Cook, R.J.; Bruckart, W.L.; Coulson, J.R.; Goettel, M.S.; Humber, R.A.; Lumsden, R.D.; Maddox, J.V.; McManus, M.L.; Moore, L.; Meyer, S.F.; Quimby, P.C.; Stack, J.P.; Vaughn, J.L. Safety of microorganisms intended for pest and plant disease control: A framework for scientific evaluation. Biol. Control 1996, 7, 333–351. [Google Scholar] [CrossRef]
- Harman, G.E.; Howell, C.R.; Viterbo, A.; Chet, I.; Lorito, M. Trichoderma species—opportunistic, avirulent plant symbionts. Nature Rev. Microbiol. 2004, 2, 43–56. [Google Scholar]
- Jensen, D.F.; Wolffhechel, H. The use of fungi, particularly Trichoderma spp. and Gliocladium spp to control root rot and damping-off diseases. In Biological Control: Benefits and Risks; Hokkanen, H.M.T., Lynch, J.M., Eds.; Cambridge University Press: Cambridge, UK, 1993; pp. 177–189. [Google Scholar]
- Bailey, B.A.; Lumsden, R.D. Direct effects of Trichoderma and Gliocladium on plant growth and resistance to pathogens. In Trichoderma and Gliocladium Vol. 2; Harman, G.E., Kubicek, C.P., Eds.; Taylor and Francis: London, UK, 1998; pp. 185–204. [Google Scholar]
- Whipps, J.M. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 2001, 52, 487–511. [Google Scholar] [CrossRef]
- Bowen, G.D.; Rovira, A.D. Microbial colonization of plant roots. Annu. Rev. Phytopathol. 1976, 14, 121–144. [Google Scholar] [CrossRef]
- Weller, D.M. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu. Rev. Phytopathol. 1988, 26, 379–407. [Google Scholar] [CrossRef]
- Rubio-Pérez, E.; Molinero-Ruiz, M.L.; Melero-Vara, J.M.; Basallote-Ureba, M.J. Selection of potential antagonists against asparagus crown and root rot caused by Fusarium spp. Commun. Agric. Appl. Biol. Sci. 2008, 73, 203–206. [Google Scholar]
- Gilardi, G.; Baudino, M.; Gullino, M.L.; Garibaldi, A. Attempts to control Fusarium root rot of bean by seed dressing. Commun. Agric. Appl. Biol. Sci. 2008, 73, 75–80. [Google Scholar]
- Shanmugam, V.; Sharma, V.; Ananthapadmanaban, B. Genetic relatedness of Trichoderma isolates antagonistic against Fusarium oxysporum f.sp. dianthi inflicting carnation wilt. Folia. Microbiol. 2002, 53, 130–138. [Google Scholar]
- Ojiambo, P.S.; Scherm, H. Biological and application-oriented factors influencing plant disease suppression by biological control: a meta-analytical review. Phytopathology 2006, 96, 1168–1174. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Kopchinskiy, A.G.; Komon, M.; Bissett, J.; Szakacs, G.; Kubicek, C.P. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet. Biol. 2005, 42, 813–828. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Kubicek, C.P.; Komon-Zelazowska, M.; Mulaw, T.B.; Bissett, J. The Trichoderma harzianum demon: complex speciation history resulting in coexistence of hypothetical biological species, recent agamospecies and numerous relict lineages. BMC Evol. Biol. 2010, in press. [Google Scholar]
- Nagy, V.; Seidl, V.; Szakacs, G.; Komon-Zelazowska, M.; Kubicek, C.P.; Druzhinina, I.S. Application of DNA BarCodes for screening of industrially important fungi: the haplotype of Trichoderma harzianum sensu stricto indicates superior chitinase formation. Appl. Environ. Microbiol. 2007, 73, 7048–7058. [Google Scholar] [CrossRef]
- Samuels, G.J.; Ismaiel, A.; Bon, M.-C.; De Respinis, S.; Petrini, O. Trichoderma asperellum sensu lato consists of two cryptic species. Mycologia 2009. [Google Scholar] [CrossRef]
- Dodd, S.L.; Lieckfeldt, E.; Samuels, G.J. Hypocrea atroviridis sp. nov., the teleomorph of Trichoderma atroviride. Mycologia 2003, 95, 27–40. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Komoń-Zelazowska, M.; Kredics, L.; Hatvani, L.; Antal, Z.; Belayneh, T.; Kubicek, C.P. Alternative reproductive strategies of Hypocrea orientalis and genetically close but clonal Trichoderma longibrachiatum, both capable to cause invasive mycoses of humans. Microbiology 2008, 154, 3447–3459. [Google Scholar] [CrossRef]
- Degenkolb, T.; Dieckmann, R.; Nielsen, K.F.; Grafenhan, T.; Theis, C.; Zafari, D.; Chaverri, P.; Ismaiel, A.; Bruckner, H.; Von Döhren, H.; Thrane, U.; Petrini, O.; Samuels, G.J. The Trichoderma brevicompactum clade: a separate lineage with new species, new peptaibiotics, and mycotoxins. Mycol. Prog. 2008, 7, 177–219. [Google Scholar] [CrossRef]
- Mittermeier, R.A.; Robles, G.P.; Hoffman, M.; Pilgrim, J.; Brooks, T.; Mittermeier, C.G.; Lamourex, J.; da Fonseca, G.A.B. Hotspots Revisited: Earth’s Biologically Richest and Most Endangered Terrestrial Ecoregions; CEMEX: Mexico City, Mexico, 2004. [Google Scholar]
- Migheli, Q.; Balmas, V.; Komoń-Zelazowska, M.; Scherm, B.; Caria, R.; Kopchinskiy, A.; Kubicek, C.P.; Druzhinina, I.S. Soils of a Mediterranean hotspot of biodiversity and endemism (Sardinia, Tyrrhenian Islands) are inhabited by pan-European and likely invasive species of Hypocrea/Trichoderma. Environ. Microbiol. 2009, 11, 35–46. [Google Scholar] [CrossRef]
- Zachow, C.; Berg, C.; Müller, H.; Meincke, R.; Komon-Zelazowska, M.; Druzhinina, I.S.; Kubicek, C.P.; Berg, G. Fungal biodiversity in the soils/rhizospheres of Tenerife (Canary Islands): relationship to vegetation zones and environmental factors. ISME J. 2009, 3, 79–92. [Google Scholar] [CrossRef]
- Hoyos-Carvajala, L.; Orduz, S.; Bissett, J. Genetic and metabolic biodiversity of Trichoderma from Colombia and adjacent neotropic regions. Fungal Genet. Biol. 2009, 46, 615–631. [Google Scholar] [CrossRef]
- Kullnig, C.; Szakacs, G.; Kubicek, C.P. Molecular identification of Trichoderma species from Russia, Siberia and the Himalaya. Mycol. Res. 2000, 104, 1117–1125. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Bissett, J.; Kullnig-Gradinger, C.M.; Druzhinina, I.S.; Szakacs, G. Genetic and metabolic diversity of Trichoderma: a case study on South-East Asian isolates. Fungal Genet. Biol. 2003, 38, 310–317. [Google Scholar] [CrossRef]
- Zhang, C.L.; Druzhinina, I.S.; Kubicek, C.P.; Xu, T. Biodiversity of Trichoderma in China: evidence for a North to South difference of species distribution in East Asia. FEMS Microbiol. Letts. 2005, 251, 251–257. [Google Scholar] [CrossRef]
- Batjes, N.H. Total carbon and nitrogen in the soils of the world. Eur. J. Soil Sci. 1996, 47, 151. [Google Scholar] [CrossRef]
- Tondje, P.R.; Roberts, D.P.; Bon, M.C.; Widmer, T.; Samuels, G.J.; Ismaiel, A.; Begoude, A..; Tchana, T.; Nyemb-Tshomb, E.; Ndoumbe-Nkeng, M.; Bateman, R.; Fontem, D.; Hebbar, K.P. Isolation and identification of mycoparasitic isolates of Trichoderma asperellum with potential for suppression of black pod disease of cacao in Cameroon. Biol. Control 2007, 43, 202–212. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Komon, M.; Kubicek, C.P.; Druzhinina, I.S. Hypocrea crystalligena sp. nov., a common European species with a white spored Trichoderma anamorph. Mycologia 2006, 98, 500–514. [Google Scholar]
- Kopchinskiy, A.; Komon, M.; Kubicek, C.P.; Druzhinina, I.S. TRICHOBLAST: a mulilocus database for Trichoderma and Hypocrea identifications. Mycol. Res. 2005, 109, 658–660. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acid. Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Nicholas, K.B.; Nicholas, H.B. Genedoc, a Tool for Editing and Annotating Multiple Sequence Alignments. 1996. Available online: http://www.cris.com/ketchup/genedoc.html (accessed on 1 January 2008).
- Rannala, B.; Yang, Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996, 43, 304–311. [Google Scholar] [CrossRef]
- Yang, Z.; Rannala, B. Bayesian phylogenetic inference using DNA sequences: a Markov chain Monte Carlo method. Mol. Biol. Evol. 1997, 14, 717–724. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Chaverri, P.; Fallah, P.; Kubicek, C.P.; Samuels, G.J. Hypocrea flaviconidia, a new species from Costa Rica with yellow conidia. Stud. Mycol. 2004, 50, 401–407. [Google Scholar]
- Rita, N.; Tricita, H.Q. Soil mycoflora of black pepper rhizosphere in the Philippines and their In-vitro antagonism against phytophthora capsici L. Indo. J. Agric. Sci. 2004, 5, 1–10. [Google Scholar]
- Shannon, C.E.; Weiner, W. The Mathematical Theory of Communication; University of Illionis Press: Urbana, IL, USA, 1963. [Google Scholar]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Giraud, T.; Refrégier, G.; de Vienne, D.; Le Gac, M. Speciation in fungi. Fungal Genet. Biol. 2008, 45, 791–802. [Google Scholar] [CrossRef]
- Aga, E.; Bryngelsson, T.; Bekele, E.; Salomon, B. Genetic diversity of forest arabica coffee (Coffea arabica L.) in Ethiopia as revealed by random amplified polymorphic DNA (RAPD) analysis. Hereditas 2003, 138, 36–46. [Google Scholar] [CrossRef]
- Masumbuko, L.I.; Bryngelsson, T.; Mneney, E.E.; Salomon, B. Genetic diversity in Tanzanian Arabica coffee using random amplified polymorphic DNA (RAPD) markers. Hereditas 2003, 139, 56–63. [Google Scholar] [CrossRef]
- Anthony, F.; Combes, C.; Astorga, C.; Bertrand, B.; Graziosi, G.; Lashermes, P. The origin of cultivated Coffea arabica L. varieties revealed by AFLP and SSR markers. Theor. Appl. Genet. 2002, 104, 894–900. [Google Scholar] [CrossRef]
Appendix

Appendix 1.
Strains of Trichoderma isolated in this study and NCBI GenBank accession numbers of their sequences.
| Region | Ecosystem | Alt. (m) | Species | No. of Isolates | Strain No. | GenBank accession No. | ||
|---|---|---|---|---|---|---|---|---|
| C.P.K. | ITS 1 and 2 | tef1 | ||||||
| Wellega | disturbed semi-forest | 2500 | T. sp. ‘afroharzianum’ | 2 | 2618 | FJ412028 | FJ436158 | |
| 2400 | 2617 | FJ412059 | FJ436157 | |||||
| 2250 | T. harzianum sensu lato | 3 | 2620 | FJ412029 | ||||
| 2500 | 2616 | FJ412026 | ||||||
| 2800 | 2615 | FJ412025 | ||||||
| 1900 | T. asperelloides | 1 | 1839 | FJ412074 | FJ436141 | |||
| 2800 | H. atroviridis/T. atroviride clade E | 1 | 2622 | FJ412094 | FJ436160 | |||
| 1570 | T. hamatum | 2 | 1826 | FJ411979 | FJ436130 | |||
| 2400 | 2613 | FJ411986 | ||||||
| 2500 | T. sp. C.P.K.2612 | 1 | 2614 | FJ412013 | FJ436156 | |||
| 2400 | T. sp. C.P.K.1833 | 1 | 2619 | FJ412060 | FJ436159 | |||
| native forest | 1750 | T. harzianum sensu lato | 5 | 2610 | FJ412024 | FJ436154 | ||
| 1900 | 1825 | FJ412017 | FJ436129 | |||||
| 1750 | 3 | 2611 | FJ412027 | |||||
| 1750 | 2608 | FJ412023 | ||||||
| 1750 | 2609 | FJ412032 | ||||||
| 1750 | T. spirale | 2 | 2606 | FJ412078 | FJ436153 | |||
| 1750 | 2607 | FJ412079 | ||||||
| 1560 | T. asperelloides | 2 | 1893 | FJ412049 | FJ436148 | |||
| 1500 | 1838 | FJ412047 | FJ436140 | |||||
| 1950 | H. atroviridis/T. atroviride clade D | 1 | 2605 | FJ412093 | FJ436152 | |||
| 2300 | T. hamatum | 1 | 1827 | FJ411980 | ||||
| 1750 | T. sp. C.P.K.2612 | 1 | 2612 | FJ412011 | FJ436155 | |||
| 1740 | T. sp. C.P.K. 1837 | 1 | 1837 | FJ412090 | FJ436139 | |||
| Hararghe | disturbed semi-forest | 2300 | T. sp. ‘afroharzianum’ | 1 | 1840 | FJ412018 | FJ436142 | |
| 2100 | T. asperelloides | 1 | 1829 | FJ412048 | FJ436132 | |||
| 2080 | T. sp. C.P.K.1828 | 1 | 1828 | FJ412085 | FJ436131 | |||
| 1580 | T. sp. C.P.K. 1837 | 1 | 1841 | FJ436143 | ||||
| native forest | 2150 | T. harzianum sensu lato | 2 | 2623 | FJ412030 | |||
| 2150 | 2624 | FJ412031 | ||||||
| Jimma | disturbed semi-forest | 1950 | T. harzianum sensu lato | 1 | 2673 | FJ412038 | FJ436175 | |
| 2200 | T. harzianum sensu stricto | 1 | 1818 | FJ412016 | FJ436123 | |||
| 1750 | T. harzianum sensu lato | 5 | 2664 | FJ412034 | ||||
| 1800 | 2506 | FJ412020 | ||||||
| 1850 | 2507 | FJ412021 | ||||||
| 1950 | 2672 | FJ412037 | ||||||
| 1590 | 2684 | FJ412036 | ||||||
| 1660 | T. spirale | 1 | 1888 | FJ412075 | FJ436144 | |||
| 1650 | H. atroviridis/T. atroviride clade D | 2 | 2662 | FJ412095 | FJ436170 | |||
| 1700 | 2663 | FJ412096 | FJ436171 | |||||
| 1360 | H. koningiopsis/T. koningiopsis | 2 | 1816 | FJ412100 | FJ436121 | |||
| 1850 | T. gamsii | 1 | 2508 | FJ412103 | FJ436151 | |||
| 1600 | T. hamatum | 8 | 2682 | FJ411994 | ||||
| 1500 | 2686 | FJ411996 | ||||||
| 1500 | 2687 | FJ411997 | FJ436180 | |||||
| 2400 | 1810 | FJ411977 | FJ436116 | |||||
| 1770 | 1814 | FJ411981 | FJ436119 | |||||
| 1500 | 2685 | FJ411995 | FJ436179 | |||||
| 1680 | 1811 | FJ411978 | ||||||
| 2000 | 2505 | FJ411985 | FJ436150 | |||||
| 1330 | H. koningiopsis/T. koningiopsis | 1 | 1831 | FJ412091 | FJ436133 | |||
| 1950 | T. longibrachiatum | 1 | 1815 | FJ412086 | FJ436120 | |||
| 2300 | T. sp. C.P.K. 1837 | 1 | 1817 | FJ412089 | FJ436122 | |||
| 2150 | T. sp. C.P.K.1807 | 1 | 1807 | FJ412014 | FJ436113 | |||
| 2250 | T. sp. C.P.K.1833 | 3 | 1808 | FJ412055 | FJ436114 | |||
| 2300 | 1809 | FJ412056 | FJ436115 | |||||
| 1800 | 2671 | FJ412069 | ||||||
| native forest | 2100 | T. harzianum sensu lato | 3 | 2510 | FJ412022 | |||
| 1750 | 2504 | FJ412019 | ||||||
| 1700 | 2668 | FJ412035 | ||||||
| 1670 | T. spirale | 3 | 2674 | FJ412071 | FJ436176 | |||
| 1880 | 2679 | FJ412072 | FJ436178 | |||||
| 1800 | 2502 | FJ412076 | FJ436149 | |||||
| 1800 | H. atroviridis/T. atroviride clade D | 1 | 1832 | FJ412092 | FJ436134 | |||
| 1950 | H. koningiopsis/T. koningiopsis | 1 | 1813 | FJ412099 | FJ436118 | |||
| 1750 | T. hamatum | 11 | 2499 | FJ411982 | ||||
| 1880 | 2678 | FJ411991 | ||||||
| 1900 | 2680 | FJ411992 | ||||||
| 1780 | 2675 | FJ411989 | ||||||
| 1600 | 2500 | FJ411983 | ||||||
| 1700 | 2501 | FJ411984 | ||||||
| 1800 | 2667 | FJ411987 | ||||||
| 1640 | 2669 | FJ411988 | ||||||
| 1780 | 2676 | FJ411990 | ||||||
| 1900 | 2681 | FJ411993 | ||||||
| 1950 | 1835 | FJ436137 | ||||||
| 1960 | T. sp. C.P.K.1833 | 1 | 1833 | FJ412058 | FJ436135 | |||
| 2150 | T. sp. C.P.K.1812 | 1 | 1812 | FJ412015 | FJ436117 | |||
| 1720 | T. sp. C.P.K.2670 | 1 | 2670 | FJ412107 | FJ436174 | |||
| 1830 | T. sp. C.P.K.2707 | 3 | 2677 | FJ412083 | FJ436177 | |||
| 1600 | 2666 | FJ412082 | FJ436173 | |||||
| 1800 | 1834 | FJ412080 | FJ436136 | |||||
| SNNP | disturbed semi-forest | 2070 | T. sp. ‘afroharzianum’ | 1 | 2632 | FJ412033 | FJ436162 | |
| 2000 | T. harzianum sensu lato | 1 | 2718 | FJ412039 | ||||
| 1750 | T. spirale | 2 | 2694 | FJ412077 | FJ436184 | |||
| 1670 | 1822 | FJ412073 | FJ436127 | |||||
| 1670 | T. asperelloides | 7 | 1836 | FJ412046 | FJ436138 | |||
| 1750 | 2692 | FJ412051 | FJ436183 | |||||
| 2000 | 2722 | FJ412053 | FJ436191 | |||||
| 1750 | 2697 | FJ412052 | FJ436185 | |||||
| 1750 | 1819 | FJ412044 | FJ436124 | |||||
| 1700 | 1820 | FJ412045 | FJ436125 | |||||
| 1750 | 2688 | FJ412050 | FJ436181 | |||||
| 1750 | H. atroviridis/T. atroviride clade D | 1 | 2690 | FJ412097 | FJ436182 | |||
| 1780 | H. koningiopsis/T. koningiopsis | 1 | 1823 | FJ412102 | FJ436128 | |||
| 2000 | T. gamsii | 1 | 2723 | FJ412106 | FJ436192 | |||
| 1750 | T. hamatum | 10 | 2689 | FJ411998 | ||||
| 1750 | 2700 | FJ412004 | ||||||
| 1750 | 2703 | FJ412007 | ||||||
| 1750 | 2691 | FJ411999 | ||||||
| 1750 | 2695 | FJ412000 | ||||||
| 1750 | 2696 | FJ412001 | ||||||
| 1750 | 2698 | FJ412002 | ||||||
| 1750 | 2699 | FJ412003 | ||||||
| 1750 | 2702 | FJ412005 | ||||||
| 1750 | 2704 | FJ412006 | ||||||
| 1800 | T. longibrachiatum | 2 | 1889 | FJ412087 | FJ436145 | |||
| 1770 | 1890 | FJ412088 | FJ436146 | |||||
| 1690 | T. sp. C.P.K.1833 | 11 | 2645 | FJ412068 | FJ436169 | |||
| 2000 | 2719 | FJ412070 | FJ436190 | |||||
| 2200 | 2634 | FJ412063 | FJ436163 | |||||
| 1750 | 1892 | FJ412057 | FJ436147 | |||||
| 1850 | 2626 | FJ412061 | ||||||
| 1970 | 2630 | FJ412062 | FJ436161 | |||||
| 2200 | 2636 | FJ412064 | FJ436164 | |||||
| 2260 | 2637 | FJ412065 | FJ436165 | |||||
| 1790 | 2641 | FJ412066 | FJ436166 | |||||
| 1690 | 2642 | FJ412067 | FJ436167 | |||||
| 1700 | 2643 | FJ436168 | ||||||
| native forest | 1850 | T. harzianum sensu lato | 5 | 2710 | FJ412040 | FJ436189 | ||
| 1900 | 2727 | FJ412012 | FJ436194 | |||||
| 1850 | 2712 | FJ412042 | ||||||
| 1850 | 2711 | FJ412041 | ||||||
| 1900 | 2728 | FJ412043 | ||||||
| 2000 | T. asperelloides | 1 | 2724 | FJ412054 | ||||
| 1750 | H. koningiopsis/T. koningiopsis | 1 | 1821 | FJ412101 | FJ436126 | |||
| 1650 | T. gamsii | 2 | 2706 | FJ412104 | FJ436186 | |||
| 2000 | 2709 | FJ412105 | FJ436188 | |||||
| 1650 | T. hamatum | 3 | 2705 | FJ412008 | ||||
| 2050 | 2715 | FJ412009 | ||||||
| 2050 | 2717 | FJ412010 | ||||||
| 2050 | T. sp. C.P.K. 2725 | 1 | 2725 | FJ412098 | FJ436193 | |||
| 1950 | T. sp. C.P.K.2707 | 1 | 2707 | FJ412084 | FJ436187 | |||
Bold font indicates potentially new species.
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).