Abstract
Molecular analysis of the bacterial community structure associated with sludge processed by autothermal thermophilic aerobic digestion (ATAD), was performed using a number of extraction and amplification procedures which differed in yield, integrity, ability to amplify extracted templates and specificity in recovering species present. Interference to PCR and qPCR amplification was observed due to chelation, nuclease activity and the presence of thermolabile components derived from the ATAD sludge. Addition of selected adjuvant restored the ability to amplify community DNA, derived from the thermophilic sludge, via a number of primer sets of ecological importance and various DNA polymerases. Resolution of community profiles by molecular techniques was also influenced by the ATAD sludge extraction procedure as demonstrated by PCR-DGGE profiling and comparison of taxonomic affiliations of the most predominant members within 16S rRNA gene libraries constructed from ATAD DNA extracted by different methods. Several modifications have been shown to be necessary to optimize the molecular analysis of the ATAD thermal niche which may have general applicability to diversity recovery from similar environments.
1. Introduction
The development of DNA-based techniques has revolutionized the ability to characterise and identify the diversity and taxonomy of environmental organisms in a wide variety of niches [1,2], such as food [3], soil [4], water [5] and the human body [6]. A major advantage of this approach is that it allows monitoring, detection and analysis of the genetic targets of interest directly from environmental samples, without the additional steps of cultivation and recovery [7,8], which are known to be inefficient in recovering symbiotic, facultative, stationary, slow growing, pH sensitive and various other fastidious microorganisms [9,10]. In spite of its attractiveness, many molecular studies applied to soil and water have indicated that the choice of processing method and the design of extraction protocols may affect the degree of lysis of the microorganisms present in the sample (and hence the recovery of their template DNAs) [11], the integrity and size of DNA obtained [12] and the extent of co-extraction of both organic and inorganic impurities which may interfere with PCR amplification [13]. These factors may also affect the usefulness and applicability of the DNA for further molecular analysis [14] and drastically effect the recovery of molecular diversity, leading to mistakes in the interpretation of the true diversity and taxa present [15,16].
It is thus essential, when examining any new ecological niche to carefully apply methodology to evaluate the efficiency of sample extraction and determine the heterogeneity of the amplified DNA target obtained [17]. Among the various methods applied, denaturing gradient gel electrophoresis (DGGE) has been shown to be effective in rapid screening of diversity within the pool of amplified PCR fragments from taxonomic targets like 16S rRNA [18,19,20]. To provide precise identification and quantification of the phylotypes present in different DNA samples, amplification of a near full-length taxonomic marker, such as 16S rRNA, is necessary, followed by clone library generation and sequencing [21,22]. However, a key initial factor of both methodologies, is successful template amplification and here multiple strategies have been used in an attempt to overcome inhibition of amplification of problematic DNA samples. These strategies include serial dilution of the DNA [23] and titration with MgCl2 [24]. However this can reduce the resolution power of DNA based techniques and lead to loss of amplification of rare genetic targets (particularly by dilution) while giving rise to low reproducibility and reduced diversity [25,26]. A range of chemical additives (adjuvants) have been shown to be beneficial to Taq-polymerase performance in PCR reactions but their applicability and effectiveness depends on the nature and origin of the DNA samples, as well as the types of inhibitor potentially present in the samples [27,28,29]. The choice of adjuvant can be made by empirically screening numerous compounds in the PCR reaction until the desired effects are achieved, which in this case would be successful amplification of diverse targets. Thus for each new environmental niche being analysed, a number of suitable adjuvants could be screened based on previous experience with particular samples or based on the nature of impurities expected or found associated with that niche. Such an analysis should be a key component in the optimisation of molecular examination for any environmental niche. A range of commercial extraction kits are now available and many are attractive in terms of short operation times, ease of performance, lack of refrigeration necessary in the field, and the minimal set of chemicals and equipment required. Indeed there have been numerous reports of their successful application in a variety of niches [30,31]. However the composition of ingredients is often proprietary, a full understanding of the protocol and what is being achieved is often absent and most importantly the extent to which the full biodiversity of the niche under examination is being explored is rarely determined or examined.
Bioengineered ecosystems are artificially created habitats, designed to utilise the metabolic and genetic potential of living organisms [32] and such systems often vary in design, size, operational parameters and purpose [33,34]. Autothermal thermophilic aerobic digestion (ATAD) is such a system, which utilises the metabolic activity of predominately aerobic microorganisms to generate a thermophilic degradative environment for sludge treatment, producing a stabilized class A biosolids product capable of land spread without site restriction. Such an engineered environment can be expected to select novel communities of microorganisms adapted to the physicochemical environment pertaining within and adapting to changes in conditions of feed, oxygen, temperature, pH and products formed. The community structure operating within ATAD systems treating domestic sludge has been little studied. There has been limited culture based examination of ATAD systems [35,36], and limited DNA-based examination of ATAD systems treating swine-waste [37], pharmaceutical waste [38] and secondary sludge [39]. In all cases DNA extraction tools were used with no comparison or optimisation of the techniques used to examine the diversity obtained. Here we have examined an ATAD system treating domestic sludge operating up to 65 oC to determine optimal strategies to maximise recovery of molecular targets of taxonomical, biotechnological and ecological interest.
2. Experimental Section
2.1. Sampling, ATAD Location, Feed Characteristics and Sampling Procedure
Sludges were collected from the Killarney Sewage Treatment Works, which processes the majority of the domestic wastewater for a population of 20,000–51,000 people (mean solids production of 500 tones per annum). The ATAD feed consists of a combination of primary and secondary treated sludges, thickened to 4–6 % TS on a belt filter and is processed in a semi-batch process via two reactor systems with aeration, one operating at mesophilic temperatures (Reactor 1A) and the other at thermophilic temperatures (Reactor 2A), details of the process have been described [34,39]. Reactors 1A and 2A (110 m3) are operated in series, with partially digested sludge being fed from the first mesophilic ATAD reactor (operational temperature range 35–49 °C) to the second thermophilic ATAD reactor (operation temperature range 58–65 °C). Temperatures were monitored via in line thermocouples during each ATAD cycle and while sampling, process variations have been described [34,39,40]. Samples were taken aseptically from ATAD reactors during or following thermal treatment. For sampling of reactor 2A, samples of sludge were taken after 23 hours of digestion following the sludge feeding procedure at the later stages of processing.
2.2. DNA Extraction
2.2.1. Solvent-based method
One mL of ATAD sludge was centrifuged at 30,000 x g to maximise collection of microcolloidal particles, flocs and bacterial cells. Extracellular DNA was removed by washing pelleted sludge twice with TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0) and collected by centrifugation at 16000 x g at 16 °C for 20 min and the supernatant discarded. The pellet was resuspended in 100 µL of pre-treatment buffer I (TE buffer + 20 mg/mL of lysozyme), and 0.03 g of 0.1 mm, 0.5 mm or 1.2 mm acid-washed beads (G8772, G1145, G152, Sigma-Aldrich) were added to the mixture [41]. After incubation on ice for 15 min and at 37 °C for 10 min three short pulses (5 s each) of high speed vortexing was performed to physically disintegrate sludge flocs, microparticles and cells. Following this step, 0.5 μL of lysis solution II was added, containing 0.05 M Tris–HCl pH 7.6, 0.1 M NaCl, 0.05 M EDTA, 2% SDS (to dissolve hydrophobic material or cell membranes), 0.2% PVP (facilitating penetration of extracelluar materials and removal of phenolics) and 0.1% β-mercaptoethanol (to inactivate proteases and nucleases) [42]. The mixture was incubated at room temperature for 15–30 min, followed by incubation at 65 °C for 30 min after which the cell debris was pelleted by centrifugation at 4,000 x g for 5 min at room temperature. Removal of protein was performed by the addition of 0.4 mL of Tris-buffered phenol (pH 8.0). The phases were mixed again gently for 5 min at room temperature, 0.3 mL of chloroform was then added, the phases mixed and then separated by centrifugation at 13,000 x g for 10 min. The aqueous phase was re-extracted with a phenol: chloroform mixture (1:1 ratio) [43]. Traces of phenol were removed via a single chloroform extraction (0.8 mL). Total nucleic acid was precipitated by adding 0.6 volumes of isopropanol for 30 min and incubated at room temperature to minimise co-precipitation of inorganic, organic or humic components, which can occur during incubation at lower temperatures and in the presence of ethanol. The resulting precipitate was collected by centrifugation at 13,000 x g, at room temperature for 30 min and washed twice with 70% ethanol to remove traces of isopropanol. DNA pellets were resuspended in 100 µL of 10 mM Tris-HCl (pH 8.0) [43] for immediate analysis. For long term storage, DNA pellets were kept in 80% ethanol, and stored at −80 °C until further analysis. Before further analysis, DNA sample were defrosted on ice and then the DNA pellets were air dried and dissolved in 100 µl of 10 mM Tris (pH 8.0) [43].
2.2.2. Extraction of genomic DNA via the MoBIO commercial kit
Total genomic DNA was isolated from ATAD sludge using the Power Soil DNA isolation kit (MoBIO, UK) according to the manufacturer’s instructions. DNA was eluted in 100 µl of 10 mM Tris-HCl (pH 8.0) [43] for immediate analysis. For long term storage, DNA was precipitated by addition of 3 volumes of 96 % ethanol, 0.3M sodium acetate (pH 5.5) and 20 ng of glycogen (Roche, UK) [43] or stored as in 2.2.1.
2.3. Analysis of Integrity and Size of Extracted Genomic DNA
Agarose gels (0.8% w/v) were prepared in 1xTAE buffer and 1 µL of each DNA extract was electrophoresed at 80 V in 1xTAE buffer and post-stained in 1x TAE buffer containing 0.1% Ethidium Bromide [43].
2.4. Amplification of Extracted DNA and Detection of Co-Extracted Contaminants
2.4.1. PCR conditions
A list of oligonucleotides used is presented in Table 1. Conditions for amplification of the 16S rRNA, rpoB and integrase genes were applied as described elsewhere [44,45,46,47,48,49].

Table 1.
Oligonucleotides used in this study.
| Primer name | Nucleotide sequence (5’–3’) | Target sequences | Techniques | Reference |
|---|---|---|---|---|
| 27f | AGAGTTTGATCCTGGCTCAG | V1, 16S rRNA gene | PCR | [44] |
| 1492r | GGTTACCTTGTTACGACTT | V 9 , 16S rRNA gene | PCR | [44] |
| 518r | ATTACCGCGGCTGCTGG | V5, 16S rRNA gene | PCR, DGGE, | [45] |
| 338fGC a | ACTCCTACGGGAGGCAGCAG | V3, 16S rRNA gene | DGGE | [45] |
| 968f | AACGCGGAAGAACCTTAC | V6, 16S rRNA gene | PCR | [46] |
| 1401r | CGGTGTGTACAAGAAGACCC | V8 , 16S rRNA gene | PCR | [44] |
| 1698f | AACATCGGTTTGATCAAC | rpoB | PCR | [47] |
| 2041r | CGTTGCATGTTGGTACCCAT | rpoB | PCR | [47] |
| Int R391f | AACTAGGGCTGGGCTTATAACATGGCC | Integrase, R391 | PCR | [48,49] |
| Int R391r | AAAGATGGCAGCTTGCCGCAACCTC | Integrase, R391 | PCR | [48,49] |
| Kanf | tatcgattgtatgggaagcc | aph, R391 [46] | PCR, cloning, qRT-PCR | [This study] |
| Kanr | cagcgcatcaacaatattttca | aph, R391 [46] | PCR, cloning, qRT-PCR | [This study] |
| T7f | ATTTAGGTGACACTATAG | pGEM vector | sequencing | Promega, UK |
| SP6r | TAATACGACTCACTATAGGG | pGEM vector | sequencing | Promega, UK |
a -GC clamp added to the 5' end of the primer.338F, 5'-CGCCCGCCG CGCGCGGCGGGCGGGGCGGGGGCACGGGGGG-3'.
2.4.2. Addition of additives to improve PCR amplification and relieve inhibition of ATAD sludge derived components
A number of adjuvants, shown to be effective in relieving the inhibitory effect of various inhibitory compounds derived from soil, meat, blood, organic polluted samples and stool samples [50,51,52,53,54,55,56,57,58,59,60], were added to the reaction mix to evaluate their ability to facilitate amplification, reverse or prevent inhibitory effect. The additives used were 50 ng non-acetylated Bovine Serum Albumin (naBSA), 0.1% (w/v) PVPP, 10% (v/v) glycerol and DMSO.
2.5. The Effect of Inhibiting Co-Contaminants on the Activity of Taq-Polymerases of Different Origin
Nine different DNA polymerases, differing in origin, specificity, polymerisation and proof-reading capacity, were analyzed for their ability to amplify ATAD DNA samples obtained following solvent-based extraction to determine the sensitivity of the DNA polymerases to inhibitory compounds and unknown impurities which may be present in the thermophilic sludge. Primer set 27f and 1492r was used for PCR amplification of DNA obtained by the two extraction methods following serial dilution of the extracts with and without addition of adjuvants. The following DNA polymerases were analysed: AmpliTaq Gold DNA polymerase (Thermus flavus), Expand High Fidelity (Expand HF) PCR system (a mixture of two DNA polymerases, Taq (Thermus aquaticus) and Pwo (Pyrococcus woese)), Pwo DNA polymerases (Pyrococcus woesei), rTth DNA polymerase (Thermus thermophilus), Taq DNA polymerase (Thermus aquaticus) (Bio-Line), Tfl DNA polymerase (Thermus flavus) (Promega,UK), RedTaq Genomic DNA polymerase (Sigma) and Long-target genomic DNA Taq DNA polymerase (TaKaRa), HotTub DNA polymerase (Thermus ubiquatous). Conditions were as recommended by the manufacturer.
2.6. Development of an Internal Standard for the qRT-PCR Assay and Inhibition Assessment
The presence of PCR inhibitory substances in ATAD DNA extracts obtained by the solvent-based method, was analysed by the method described by Kleiboeker [61]. PCR performance, target enumeration, and degree of relief of inhibition due to addition of additives were determined by monitoring the change in detection of the arbitrary fluorescence threshold Ct value (Δ Ct) for a number of target molecules added to the Real-Time PCR reaction (exogeneous genomic target) with and without the addition of various amounts of ATAD DNA extract.
Primer set, Kanf and Kanr MWG Biotech (Germany) (Table 1), annealing to the aminoglycoside phosphotransferase gene [orf 20 of the Providencia rettgeri ICE R391 (NCBI accession number AY090559 [48])], were designed for this study to amplify a 250 bp DNA fragment as a model target. DNA was extracted from Escherichia coli R391 and PCR performed using primers Kanr and Kanf (Table 1) with an initial denaturation at 95 °C for 5 min, followed by 35 cycles of 95 °C for 20 s, 53 °C for 10 s, and 72 °C for 15 s, followed by a final extension at 72 °C for 9 min. The resulting PCR product was purified using a Wizard SV gel and PCR clean-up system (Promega), ligated into the pGEM-TA vector and trasformed into Escherichia coli JM109 by electroporation [43]. Sequence identity was confirmed by sequencing and the construct termed pGEM-TA Kan.
ATAD DNA samples extracted by the solvent-based protocol were diluted 10 and 100 fold and samples without pGEM-TA-Kan were assessed by qPCR for the presence of the kanamycin resistance determinant and shown to be negative. To ‘spike’ the ATAD DNA, 103 copies of the target gene (1 µL pGEM-TA-Kan plasmid construct) were mixed with diluted and non-diluted ATAD DNA extracts and examined by qRT-PCR to determine potential inhibition and interference of potential co-contaminants. Real-time fluorescence measurements were recorded via a Light Cycler II instrument (Roche). Cycle thresholds (Ct) values were recorded for plasmid standards, and plasmid standards spiked with ATAD DNA extracts at various dilutions for three independent experiments. The ΔCt values were calculated by subtracting the mean control Ct value for the uninhibited control reactions from the sample containing plasmid control Ct.
2.7. Analysis of the Total Bacterial Community Profile and Predominant and Rare Species as a Function of Extraction Procedures
ATAD DNA extracts were amplified with primer pair 338f GC and 518r [45] annealing to the V3-V5 region of 16S rRNA gene. 3−5 µg of PCR product was analysed via a DGGE gel system (Shaw, USA). DGGE gels (dimension 1.5 mm/22 cm/22 cm) contained 4% (w/v) PAAG, 0.5 × TAE buffer) Gradients were formed between 30% and 80% denaturant. Electrophoresis was performed at constant temperature of 60 °C, and 25 V (15 min) followed by 75 V for 18 hours. Gels were stained with EtBr, digitally imaged to allow band detection and DGGE profile comparison.
PCR amplification for each ATAD DNA extract was also carried out using universal bacterial 1492r and 27f primers (Table 1) via touchdown PCR, and low cycle number [62,63,64] to avoid PCR biases. PCR products were ligated into the pGEM-TA vector (Promega), transformed into electrocomptent JM109 cells and positive clones with vector insert of correct size confirmed by amplification with vector specific primers T7f and SP6r (Table 1). These PCR amplicons were subjected for ARDRA analysis [65] with CfoI, RsaI, MspI and AluI restriction enzymes. Plasmids which were shown to posess a unique ARDRA pattern were used as templates for amplification of the V3-V5 region of the 16S rRNA gene followed by DGGE analysis. Clones with unique ARDRA and DGGE patterns were sequenced using the SequiTherm Excel II DNA sequencing KiT-LC (Epicentre Technologies, Madison, Wisconsin, USA), with fluorescence DNA primers (MWG-Biotech, Germany), labelled at the 5’end with the dye IRD-800 (Li-COR Inc.,USA) and analysed on a LI-COR Inc (Lincoln, Nebraska, USA) LONGREADER 4200 DNA sequencer as recommended by the manufacturer. The newly obtained sequencing reads were aligned and assembled to a reference sequence within CLC Biosoftware [66]. Sequence trimming and removal of vector contamination was performed using the on-line bioinformatic tool VecScreen at the NCBI [67]. The directionality of all sequences was synchronised using “Sequence manipulation suite” [68]. “CHECK_CHIMERA” from the Ribosomal Database Project (RDP) [69] was applied to all sequences to examine possible chimeric artifacts. Nucleotide sequences were compared with GeneBank entries using BlastN [70] and SEQUENCE_MATCH from the RDP [71].
The 16S rRNA gene sequences obtained in this study have been deposited in the GenBank database with accession numbers GU320654, GU320658, GU320663, GU325832, GU325838, GU320661, GU320662, GU325830, GU325829 and GU325836.
3. Results and Discussion
The suitability of genomic DNA extracted from ATAD sludge for further molecular analysis can be determined by three important characteristics, its molecular weight, the absence of PCR amplification inhibitors and extracts that recover all possible genomes present. Two DNA extraction protocols were compared for their ability to produce ATAD derived genomic DNA which fulfills these requirements.
3.1. Integrity, Size and Stability of the Extracted Nucleic Acid
The two methods applied were the commercial MoBIO Power DNA extraction kit and a modified laboratory method based on a solvent-extraction technique (solvent-based method). The color of the DNA extracts varied from a slightly-yellowish (MoBIO kit) to a slightly-brown color (the solvent method) which is reflective of humic substances [72,73], known to co-purify because of their chemical characteristics. Such substances can be the major source of inhibitors of molecular manipulations with microbial DNA [74] with as little [75] as 1 μL of undiluted humic-acid-like substance completely inhibiting DNA polymerases activity.
The MoBIO commercial kit produced DNA of greater than 15 kb (Figure 1a) while DNA extracted by the solvent-based method, incorporating a glass beating technique, resulted in some shearing. (Figure 1b) possibly as a consequence of the physical disruption step. DNA yield was shown to decrease as a function of ATAD operation temperature and may reflect a decrease in sludge biomass. Typical yields of 34 ± 2.3 μg of DNA from 1 ml of the sludge from ATAD Reactor 1A and 11.6 ± 1.9 μg of DNA from 1 ml of the sludge from ATAD Reactor 2A were obtained. Replicate extractions of DNA from ATAD sludge obtained early from the thermophilic reactor (Figure 1b, Lanes 1−3) and sludge at a later stage (Figure 1b, Lanes 4−6) indicated that the procedure was reproducible and yielded DNA suitable for further analysis.
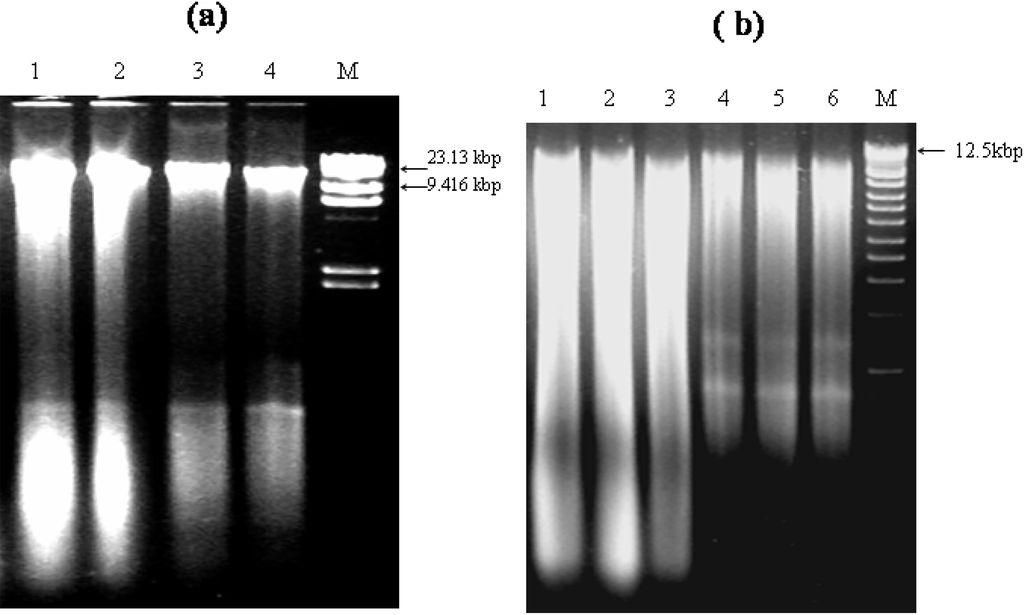
Figure 1.
Integrity and molecular size of extracted ATAD DNA by two extraction methods as visualized by agarose gel electrophoresis: (a) commercial MoBIO kit from ATAD Reactor 1A (a, Lanes 1−2) and ATAD Reactor 2A (a, Lanes, 3−4) and (b) by solvent-based method. ATAD Reactor 1A (b, Lanes 1−3) and ATAD Reactor 2A (b, lanes 4−6). (a) M, λ DNA/Hind III molecular weight marker (Invitrogen) (a) and (b) M, 1 kb DNA Ladder (Invitrogen) were used as recommended by manufacturer.
A near-full length 16S rRNA gene fragment (1450 bp) was successfully amplified from samples of ATAD DNA extracted by the MoBIO extraction kit (Figure 2, Lane 1−3), whereas amplification failed on ATAD DNA extracted by the solvent-based protocol (Figure 2, Lane 4–6) showing inhibition with no visible products, excess primers or primer-dimers. Replicate DNA extracts from thermophilic sludge in Reactor 2A at 10 different sampling points all showed signs of PCR inhibition and amplification with 35 cycles was unsuccessful.

Figure 2.
The amplifiability of ATAD DNA extracts from Reactor 2A with two extraction protocols using primers 27f−1491r (Table 1) (with expected amplicon size of 1450 bp) analysed via 1% agarose TBE gel electrophoresis, visualised by EtBr. Lane M, 1kb DNA Marker (Invitrogen) (5 μL per lane), Lanes 1−3, MoBIO extracted ATAD DNA. Lanes 4−6, Solvent extracted ATAD DNA.
To evaluate the nature of this inhibition five different primers sets for various DNA target sequences (R391 integrase gene, the rpoB gene and the V3-V5, V6-V8 and V1-V9 regions of the bacterial 16S rRNA gene) and nine different DNA polymerases were used for PCR amplification [76]. No visible PCR product was detected for any set of primers and the activity of seven of the nine polymerases tested were inhibited by co-extracted impurities from the ATAD DNA samples obtained by the solvent-based method. Such inhibition of PCR amplification could compromise attempts to apply DNA-based molecular analysis of ATAD DNA samples. When the ATAD DNA extract was added to the control plasmid, used as an internal control, the Ct value of the qRT-PCR reaction lowered to a non-detectable level after 45 cycles of amplification. This data confirmed that poor amplification results from exogenous inhibitory components which lower the detection sensitivity and may thus lead to underestimation of the amount of genetic target under investigation.

Figure 3.
Nuclease activity in ATAD DNA extracted from ATAD sludge following solvent extraction and the ability of various chemical and physical factors to inhibit these co-extracted nucleases. M, GeneRuler 1kb DNA ladder (Fermentas) (5 μL per lane). Lane C, Genomic bacterial DNA (200 ng) and pGEM-TA plasmid DNA (100 ng) were run alone untreated. Lane 1, 1 μL of ATAD DNA (20 ng) were added to substrate mixture of genomic bacterial DNA (200 ng) and pGEM-TA plasmid DNA (100 ng) and incubated at 37 ºC. Lane 2, 1 μL of ATAD DNA (20 ng) were added to substrate mixture of genomic bacterial DNA (200 ng) and pGEM-TA plasmid DNA (100ng) and incubated at 65 ºC. Lane 3, 1 μL of ATAD DNA (20ng) was added to a substrate mixture of genomic bacterial DNA (200 ng) and pGEM-TA plasmid DNA (100 ng) with addition of 5 mM EDTA, and incubated at 55 ºC for 30 min. Lane 4, 1 μL of ATAD DNA heat treated 95 ºC for 20 min (20 ng) with substrate mixture of genomic bacterial DNA (200 ng) and pGEM-TA plasmid DNA (100 ng) with incubation at 55 ºC for 30 min. Lane 5, 1 μL of ATAD DNA (20 ng) was added to a substrate mixture of genomic bacterial DNA (200 ng) and pGEM-TA plasmid DNA (100 ng) with addition of 1% formamide and incubated at 55 ºC for 30 min. Lane 6, Control sample without addition of ATAD DNA, incubated at 55 ºC for 30 min. After incubation all samples were extracted with phenol/chloroform solution and 10 μL separated by agarose gel electrophoresis and visualized by EtBr.
Degradation of target DNA or short oligonucleotides (primers) by nuclease action may be a cause of poor amplification and may be a particular issue with ATAD sludge. These nucleases result from thermal lysis, may be thermostable and survive the thermal cycling reactions during PCR. ATAD DNA samples occasionally showed evidence of degradation particularly following storage and hence the presence of nucleases was investigated by addition of exogenous plasmid or genomic DNA to ATAD extracts. These added DNA substrates were found to be rapidly degraded within 30 min after addition of ATAD DNA extracts at 37 ºC (Figure 3, Lane 1) and 65 ºC (Figure 3, Lane 2). The loss of integrity of the substrates supports the hypothesis, that active nucleases are present within the ATAD DNA extracts in spite of using EDTA as a nuclease inhibitor. Approaches to inhibit nuclease activity included the addition of formamide, EDTA, and temperature treatment followed by assessment of changes in added substrate integrity during subsequent incubation with ATAD DNA. Addition of up to 5mM EDTA (Figure 3, Lane 3) to the incubation mix, did not inhibit nuclease activity and heating up to 95 ºC for 20 min before addition to the substrate mixture had only a slight effect on nuclease activity (Figure 3, Lane 4). These data may only be explained by the presence of thermostable nucleases originating from thermophilic microorganisms in the ATAD sludge. The addition of formamide however (Figure 3, Lane 5) was found to be a major aid in reducing nuclease action and maintaining the DNA extracts on ice during manipulation appeared to be a key factor in stabilising ATAD DNA integrity. Nuclease action was only manifest at higher incubation temperatures as might be expected for thermostable nucleases with high optimum operating temperatures. Given the reaction conditions operating during PCR there activity at elevated temperatures would be a major cause of reduced diversity recovery not only from ATAD sludge but potentially for any thermal source being investigated. Due to these observations DNA extraction from ATAD sludge was carried out immediately on sampling, longer-term storage of ATAD DNA samples was achieved as a pellet in 80% ethanol while addition of 1% formamide with storage at -80 ºC was found to be an effective method of maintaining ATAD DNA integrity.
Other factors associated with the high organics content of ATAD sludge may also contribute to poor efficiency of genetic target recovery by PCR amplification through binding to DNA or inhibiting polymerase action. Apparently inert components such as cellulose and cellulose derivatives, which are present in the ATAD sludge matrix samples [39] are known to adsorb to DNA molecules and may be highly inhibitory. Many DNA purification protocols applied to ATAD sludge extracts such as precipitation of DNA with 5% polyethylene glycol 8000 (PEG)/0.6 M NaCl [77] and filtration with a combined Sepharose 4B/polyvinyl polypyrrolidone (PVPP) spin column [78] were shown to have little effect on amplification but did reduce DNA yields. Thus, while it may not be possible to remove all impurities from ATAD DNA samples, a more satisfactory approach to resolve problem of PCR inhibition would be to relieve interference of co-extracted compounds, rather than attempt to remove all offending substances.
3.3. Other Strategies to Overcome Inhibition of PCR Amplification
Mg2+ ions are a vital cofactor for DNA polymerase and their concentration will affect the success and specificity of amplification [79] while the sequestration of Mg2+ ions by various compounds and interference by Ca2+ ions may inhibit amplification [80]. Addition of Mg2+ in the range of 2.5–8 mM in PCR mixture had a slight effect on the amplification efficacy indicating that the presence of chelating agents in the non-diluted ATAD DNA extracts may also contribute to the PCR inhibition. Dilution of DNA extracts was examined as a means of alleviating inhibition using 10- and 100-fold dilutions of ATAD DNA samples obtained by the solvent–based method. Partial relief of inhibition occurred on dilution suggesting that other factors which potentially inhibit DNA-polymerase action may also be present. Different primer sets were effected, the least effected primer set 25f–1497r, amplified near-full length 16S rRNA gene amplicons (Figure 4b, Lane 2). Given that dilution should equally dilute co-extracted materials this observation may indicate that the affinity of this primer set may be greater for this DNA target than some of the other primer sets used. The efficiency of PCR amplification did not correlate with the length of amplicon, and primers sets for the V3-V5 and V1-V9 regions of 16S rRNA gene appeared to be more resistant to the inhibitory effects of co-extracted compounds (data not shown). Although the dilution scheme (1/10 and 1/100) was not sufficient to totally eliminate inhibition of amplification of the internal standard (Δ Ct = 6.5 and 5.5), the effect of inhibiting compounds were shown to be lowered by this approach.

Figure 4.
Effect of different adjuvants, polymerases and primer sets on PCR amplification of the 16S rRNA gene from ATAD DNA extracts. (a) 200 ng of ATAD DNA was added to reaction mixes containing adjuvant. Lane 1, No adjuvant, Lane 2, BSA (50ng), Lane 3, PVPP (0.1% w/v). Lane 4, glycerol (10% v/v), Lane 5, DMSO (5%), Lane 6, BSA (50 ng) + formamide (1%). Lane 7, formamide (1%), M, 1 kb Plus Molecular weight DNA marker (Invitrogen) (5 μL per lane). (b) PCR amplification efficacy of 16S rRNA with primers 27f−1491r on 10-fold diluted of ATAD DNA amplified with 9 different DNA polymerases. M, GeneRuler 1kb DNA ladder (5 μL per lane). Lane1, AmpliTaq Gold DNA polymerase, Lane 2, Expand High Fidelity (Expand HF), Lane 3, Tfl DNA polymerase, Lane 4, Red Taq genomic DNA polymerase (Sigma), Lane 5, Pwo DNA polymerase, Lane 6, Taq DNA polymerase (BioLine), Lane 7, HotTub DNA polymerase, Lane 8, rTth DNA polymerases, Lane 9, TaKaRA Long-target DNA polymerase. (c) Five different sets of primers (Table 1) were screened in PCR amplification using Bio-Line Taq-DNA polymerase and PCR mix optimised with addition of adjuvants (BSA and formamide). Lane 1, V3-V5 region 16S rRNA, Lane 2, V1-V9 regions 16S rRNA, Lane 3, V6-V8 regions 16 rRNA, Lane 4, ICE R391 integrase gene, Lane 5, rpo B and M, Low Range DNA ladder (Invitrogen) (5 μL per lane).
Since many ATAD sludge factors such as nucleases, carbohydrates, humic substances, and synthetic fiber material may result in inhibition of PCR amplification, several adjuvants were added in an attempt to improve amplification and recovery of the 16S rRNA gene by various DNA polymerases (Figure 4b). With the exception of BSA and BSA with formamide (Figure 4a, Lane 2 and 9), none of the other adjuvants had positive effects. Synergistic or additive effects were observed when BSA (Figure 4a, Lane 2) was combined with formamide (Figure 4a, Lane 2) and resulted in PCR products suitable for further molecular biological analysis. Enhanced efficiency of PCR was observed for polymerase used with the addition of adjuvant (Figure 4a) and was shown to enhance detection sensitivity and recovery of amplicons from multiple primers targeting different genetic loci (Figure 4c). These targets are the most commonly utilised for investigation of molecular diversity and its dynamics. The addition of adjuvant worked well to eliminate inhibition and enhanced the efficiency of PCR in the spiked internal standard assay and restored Δ Ct (Δ Ct = 0.4) compared to mixtures without adjuvants (Δ Ct =7.5) clearly demonstrating the positive effect of adjuvants and indeed modification of different amplification parameters in optimizing recovery from ATAD sludges.
Optimising the PCR mixture allowed amplification of the 16S rRNA gene with as little as 2 pg of ATAD DNA obtained by the solvent-based method and the sensitivity was comparable with titration assays obtained on ATAD DNA extracts prepared by the commercial MoBIO kit (data not shown). The use of touchdown PCR in conjunction with optimisation of PCR mix composition did not lead to any non specific products in this study. The ability of the PCR reaction to recover rare genetic targets within ATAD samples is extremely important for diversity studies, for pathogen detection and for detection of rare species and it is clear that adjuvant addition may play a key role in this regard. BSA is thought to bind polyphenolic substances, humic substances, and anions by virtue of its high lysine content and bind lipids via hydrophobic interactions [81], all of which are present in ATAD sludge [40]. In addition BSA may provide an alternate substrate for exogenous proteases and hence protect the DNA polymerase somewhat from proteolysis. BSA in the presence of formamide was more effective (Figure 4a, Lane 6) than BSA on it own (Figure 4a, Lane 2). Formamide is known to affect the efficacy of PCR reactions via weakening of the hydrogen bonds between nucleotides, reducing the formation of complexes and secondary structures in the targeted DNA molecule [82]. It enhances specificity by lowering the effective annealing temperature of the primers and simultaneously provides effective denaturation of the DNA template during amplification [83]. Formamide has also been shown to possess DNase inhibitory activity on DNA templates isolated from human and animal saliva [84] and pathogenic biofilms [85,86] and this may be of key importance in the case of ATAD sludge.
3.4. Capacity of DNA Extraction Methods to Reproduce a Total Community Profile
The effect of the extraction procedure on the detection of predominant and rare members of bacterial community was assessed by examining amplified pools of the 16S rRNA gene from ATAD DNA extracts The migration behavior of recovered V3-V5 region amplicons of the 16S rRNA gene via DGGE and detailed taxonomical analysis based on clone library construction and nucleotide sequencing were applied. DGGE fingerprinting of the ATAD bacterial community DNA, amplified by primer pair 338fGC−518r (Table 1) was used to assess the ability of different methods to recover community DNA from predominant and rare bacterial members. DGGE profiles differed for the two extraction methods used (Figure 5b), which were stable and reproducible in replicates (Figure 5b, Lanes 1 and 3, Lane 2 and 4). Indexes of bacterial diversity were calculated from DGGE profiles, such as total band number (richness of diversity) and the number of unique bands within each profile. The MoBIO extraction method recovered a higher number of bands (N = 21) (Figure 5b, Lane 2 and 4) and had a higher mean diversity compared to the DGGE profile obtained from the DNA extracted by the solvent-based method (19 band) (Figure 5b, Lanes 1 and 3). Comparison of the two profiles revealed that each profile contained 3 unique bands, those originating from the solvent extraction method had a longer migration distance corresponding to a higher denaturant percentage within the gel suggesting a higher GC content, often associated with thermophilic organisms [87]. The two DGGE profiles shared 16 common bands, with different migration behavior and nucleotide sequences which may originate from 16 different bacterial species or indeed from a lower number of species with multiple copies of the 16S rRNA gene [88]. Taxonomical identification of dominant and rare sequences in a constructed clone library of near full length 16S rRNA gene sequences (~1450 bp) obtained by amplification of ATAD DNA was performed. The resolution power and the distribution of the predominant and rare bacterial members for each DNA extraction method is presented (Table 2).
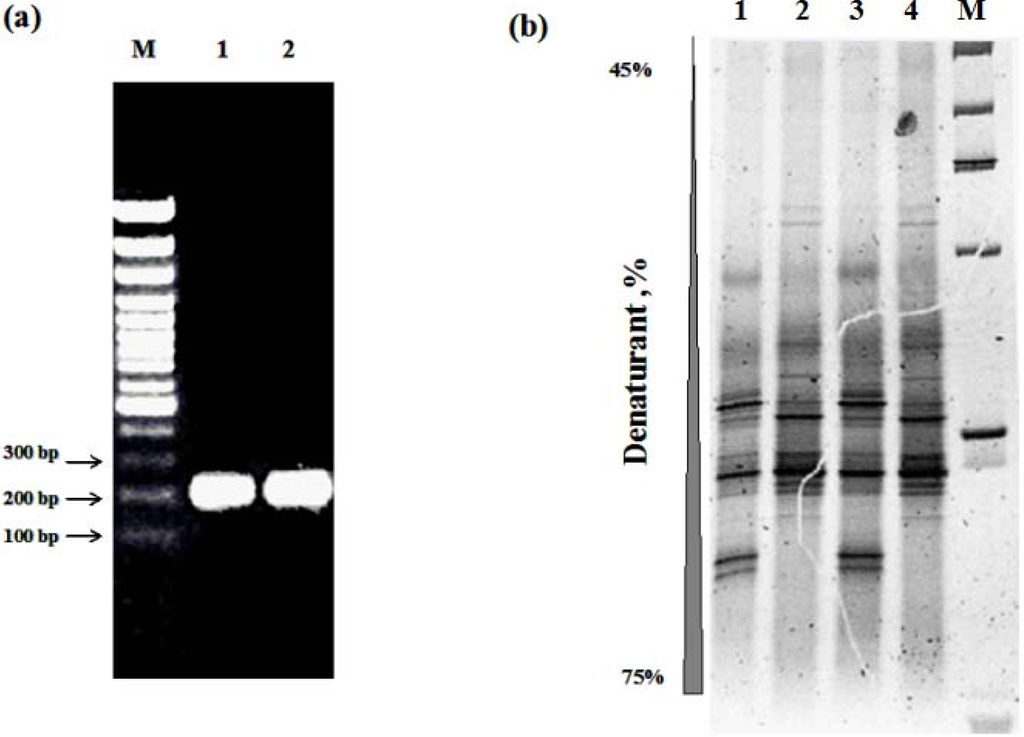
Figure 5.
DGGE analysis of ATAD bacterial community DNA extracted by the two extraction procedures with amplification of the V3-V5 hypervariable region of 16S rRNA gene. (a) Non-denaturing gel electrophoresis of PCR products of the V3-V5 region amplified from ATAD DNA extracted by MoBIO commercial kit Lane (1) and solvent-based method Lane (2). M, 100 bp DNA ladder (5 μl per lane). (b) DGGE (10% acrylamide, 30 to 80% denaturant) profile of the V3-V5 region amplified from MoBIO extracts (Lane 1 and 3), solvent based DNA extract (Lane 2 and 4), M, pBR 322/Alu I (1 μg per lane).

Table 2.
Taxonomic affiliation of predominant and unique clones within the clone libraries constructed with ATAD DNA extracts obtained by two different DNA extraction protocols.
| OTU from ATAD library | Accesssion number | Closest affiliation | % | Closest affiliation to cultivated strains | % | % of total clones in the library | |
|---|---|---|---|---|---|---|---|
| Library type | |||||||
| MoBIO kit | Solvent- based method | ||||||
| CK5 | GU320654 | Uncultured bacterium, clone H1-814, EF174267 | 99 | Clostridium ultunense Z69293 | 94 | 17.5 | 8 |
| ER78 | GU325838 | Uncultured compost bacterium, clone 4B18, DQ346582 | 99 | Bacillus sp. MSP06G, AB084065 Bacillus thermocloaceae; DSM 5250, Z26939 | 98 98 | 0 | 14.5 |
| CK27 | GU320661 | Uncultured bacterium, clone SMQ30, AM930327 | 99 | Bacillus sp. MSP06G ; AB084065 Bacillus thermocloaceae; DSM 5250; Z26939 | 98 97.5 | 4 | 17.5 |
| CK28 | GU320662 | Uncultured bacterium clone F24; AM500822 | 99 | Bacillus sp. 50LAy-1, AB375754 Ureibacillus thermosphaericus; S7, AF403019 | 99 99 | 3 | 9.5 |
| ER32 | GU325832 | Symbiobacterium sp. KY38, AB361629 | 99 | Symbiobacterium thermophilum IAM 14863 DNA AP006840 | 97 | 1 | 30.5 |
| ER 9 | GU325829 | Uncultured compost bacterium clone 1B07, DQ346486 | 98 | Moorella glycerini SQL, GQ872425 | 97 | 0 | 2.5 |
Both libraries shared most of the sequence types, but the identity of the predominant members and the presence of unique sequences differed within each library. The predominant members of the clone library (Table 2) obtained from DNA extracted by the MoBIO method were bacterial species whose nearest taxonomic identity were Clostridium ultunense (NCBI accession number EF174267), isolate Clostridial sp. PO (NCBI AN AJ002593) and Clostridium sp. PML3-1 (NCBI accession number EF165015). The presence of anaerobes in an aerobic treatment process may be reflective of poor oxygen solubility, poor mixing, insulating effects of flocs, and incomplete aeration in large scale ATAD systems. Analysis of predominant members in the clone library obtained via the solvent-based method revealed a predominance of the species with identity to Ureibacillus thermosphaericus (AB101594), Bacillus thermocloaceae (Z26939) and Symbiobacterium thermophilum (AP006840), which are known to be aerobic thermophilic bacteria with high metabolic and enzymatic activity. The presence of several clones, for example, ER 9 (closest match Moorella glycerini SQL, GQ872425), ER 17 (closest match Anoxybacillus toebii NS1-1, AY466700), ER 59 (closest match Sphingosinicella microcystinivorans, EU337119) (Table 2), were unique to the clone library constructed with DNA extracted by the solvent-based method and provides information on the presence of a more physiologically diverse group. It also indicates for the first time the presence of symbionts and ammonia reducing organisms within the ATAD sludge matrix.
The application of two DNA extraction methods to molecular analysis of ATAD sludge offers useful insights and attributes. The MoBIO method is rapid, easy and affordable, resulting in the recovery of high molecular weight ATAD DNA with little inhibition of PCR. The solvent-based method recovered other species not detected in the MoBIO extracts which leads to the conclusion that combining extracts from several procedures could aid maximal recovery of diversity. Extraction of DNA from tertiary sludge and its subsequent molecular analysis is problematic and thus far no method gives a perfect picture of the bacterial diversity. With optimisation, sample modification, use and pooling of extracts generated by different procedures, richness and diversity recovery can be improved. ATAD sludge contributes significant nuclease activity which must be addressed while other co-extracted ATAD materials influence primer specificity, PCR performance and yield. Specific modifications to extraction methods, addition of adjuvant and use and pooling of multiple extraction methodologies have been shown to be useful in diversity recovery from ATAD samples but may also have more widespread application in the analysis of other thermal environmental niches.
Acknowledgements
This work was financially supported by a collaborative research grant from the Higher Education Authority, PRTLI 4, Ireland. Authors acknowledge the assistance of staff at the Killarney WasteWater treatment facilities during sampling at the ATAD plant.
References and Notes
- Nocker, A.; Burr, M.; Camper, A.K. Genotypic microbial community profiling: a critical technical review. Microb. Ecol. 2007, 54, 276–893. [Google Scholar] [CrossRef]
- Holben, W.E.; Harris, D. DNA-based monitoring of total bacterial community structure in environmental samples. Mol. Ecol. 1995, 4, 627–631. [Google Scholar] [CrossRef]
- Giraffa, G.; Neviani, E. DNA-based, culture-independent strategies for evaluating microbial communities in food-associated ecosystems. Int. J. Food Microbiol. 2001, 20, 19–34. [Google Scholar] [CrossRef]
- Ranjard, L.; Poly, F.; Nazaret, S. Monitoring complex bacterial communities using culture-independent molecular techniques: application to soil environment. Res. Microbiol. 2000, 151, 167–177. [Google Scholar] [CrossRef]
- Kawai, M.; Matsutera, E. 16S ribosomal DNA-based analysis of bacterial diversity in purified water used in pharmaceutical manufacturing processes by PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2000, 68, 699–704. [Google Scholar] [CrossRef]
- Zhou, X.; Bent, S.J.; Schneider, M.G.; Davis, C.C.; Islam, M.R.; Forney, L.J. Characterizationof vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 2004, 150, 2565–2573. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–146. [Google Scholar]
- Dahllöf, I. Molecular community analysis of microbial diversity. Curr. Opin. Microbiol. 2002, 13, 213–217. [Google Scholar]
- Colwell, R.; Grimes, J. Semantics and strategies. In Non-culturable Microorganisms in the Environment; Colwell, R., Grimes, J., Eds.; ASM: Washington. DC, USA, 2000; pp. 1–5. [Google Scholar]
- Kazuhiro, K. Nonculturable bacterial populations that control environmental processes. Biosci. Ind. 1999, 57, 731–736. [Google Scholar]
- Frostegard, A.; Courtois, S.; Ramisse, V.; Clerc, S.; Bernillon, D.; Le Gall, F.; Jeannin, P.; Nesme, X.; Simonet, P. Quantification of bias related to the extraction of DNA directly from soils. Appl. Environ. Microbiol. 1999, 65, 5409–5420. [Google Scholar]
- Wu, L.; Li, F.; Deng, C.; Xu, D.; Jiang, S; Xiong, Y. A method for obtaining DNA from compost. Appl. Biochem. Biotechnol. 2009, 84, 389–395. [Google Scholar]
- Al-Soud, W.A.; Radstrom, P. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl. Environ. Microbiol. 1998, 64, 3748–3753. [Google Scholar]
- Weiss, A.; Jerome, V.; Freitag, R. Comparison of strategies for the isolation of PCR-compatible, genomic DNA from a municipal biogas plants. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007, 15, 190–197. [Google Scholar] [CrossRef]
- Stach, J.E.; Bathe, S.; Clapp, J.P.; Burns, R.G. PCR-SSCP comparison of 16S rDNA sequence diversity in soil DNA obtained using different isolation and purification methods. FEMS Microbiol. Ecol. 2001, 36, 139–151. [Google Scholar]
- McIlroy, S.J.; Porter, K.; Seviour, R.J; Tillett, D. Extracting nucleic acids from activated sludge which reflect community population diversity. Antonie van Leeuwenhoek 2009, 96, 593–605. [Google Scholar] [CrossRef]
- McOrist, A.L.; Jackson, M.; Bird, A.R. A comparison of five methods for extraction of bacterial DNA from human faecal samples. J. Microbiol. Meth. 2002, 50, 131–139. [Google Scholar] [CrossRef]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of Complex Microbial populations by Denaturing Gradient Gel Electrophoresis Analysis of Polymerase Chain Reaction-Amplified Genes Coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar]
- Fischer, S.G.; Lerman, L.S. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc. Natl. Acad. Sci. USA 1983, 80, 1579–1583. [Google Scholar] [CrossRef]
- Durtschi, J.D.; Voelkerding, K.V.; Wittwer, C.T.; Seipp, M.T. Multiplex amplicon genotyping by high-resolution melting. J. Biomol.Tech. 2009, 20, 160–164. [Google Scholar]
- Relman, A. Universal bacterial 16S rDNA amplification and sequencing. In Diagnostic Molecular Microbiology, Principles and Applications; Persing, D.H., Smith, T.F., Tenover, F.C., White, T.J., Eds.; ASM: Washington, DC, USA, 1993; pp. 489–495. [Google Scholar]
- Petti, C.A.; Polage, C.R.; Schreckenberger, P. The role of 16S rRNA gene sequencing in identification of microorganisms misidentified by conventional methods. J. Clin. Microbiol. 2005, 43, 6123–6125. [Google Scholar] [CrossRef]
- Alvarez, A.J.; Buttner, M.P.; Stetzenbach, L.D. PCR for bioaerosol monitoring: sensitivity and environmental interference. Appl. Environ. Microbiol. 1995, 61, 3639–3644. [Google Scholar]
- Lantz, P.G.; Abu Al-Soud, W.A.; Knutsson, R.; Hahn-Hägerdal, B.; Radstrom, P. Biotechnical use of polymerase chain reaction for microbiological analysis of biological samples. Biotechnol. Annu. Rev. 2000, 5, 87–130. [Google Scholar] [CrossRef]
- Heuer, H.; Smalla, K. Application of denaturing gradient gel electrophoresis and temperature gradient gel electrophoresis for studying soil microbial communities. In Modern Soil Microbiology; van Elsas, J.D., Trevors, J.T., Wellington, E.M.H., Eds.; Marcel Dekker: New York, NY, USA, 1997; pp. 353–373. [Google Scholar]
- Chandler, D.P.; Frederickson, J.K.; Brockman, J. Effect of PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol. Ecol. 1997, 6, 475–482. [Google Scholar]
- Pomp, D.; Medrano, J.F. Organic solvents as facilitators of polymerase chain reaction. BioTechniques 1991, 10, 58–59. [Google Scholar]
- Sidhu, M.K.; Liao, M.J.; Rashidbaigi, A. Dimethyl sulfoxide improves RNA amplification. BioTechniques 1996, 21, 44–47. [Google Scholar]
- Nol, P.; Williamson, J.L.; Rocke, T.E.; Yuill, T.M. Detection of Clostridium botulinum type C cells in the gastrointestinal tracts of Mozambique Tilapia (Oreochromis mossambicus) by polymerase chain reaction. J. Wildl Dis. 2004, 40, 749–753. [Google Scholar] [CrossRef]
- Klammer, S.; Mondini, C.; Insam, H. Microbial community fingerprint of compost stored under different condition. Ann. Microbiol. 2005, 55, 299–305. [Google Scholar]
- Maarit-Niemi, R.; Heiskanen, I.; Wallenius, K.; Lindstrom, K. Extraction and purification of DNA in rhizosphere soil samples for PCR-DGGE analysis of bacterial consortia. J. Microbiol. Meth. 2001, 45, 155–165. [Google Scholar] [CrossRef]
- Bramucci, M.G.; Nagarajan, V. Industrial wastewater bioreactors: sources of novel microorganisms for biotechnology. Trends Biotechnol. 2000, 18, 501–505. [Google Scholar] [CrossRef]
- Sen, R; Chakrabarti, S. Biotechnology—applications to environmental remediation in resource exploitation. Curr. Sci. 2009, 97, 768–775. [Google Scholar]
- Layden, N.; Mavinic, D.; Kelly, H.; Moles, R.; Bartlett, J. Autothermal thermophilic aerobic digestion (ATAD)–Part II: Review of research and full-scale operating experiences. J. Environ. Eng. Sci. 2007, 6, 679–690. [Google Scholar] [CrossRef]
- Sonnleitner, B.; Fiechter, A. Bacterial diversity in thermophilic aerobic sewage sludge II. Types of organisms and their capacities. Eur. J. Appl. Microbiol. Biotechnol. 1983, 18, 174–180. [Google Scholar] [CrossRef]
- Sonnleitner, B.; Fiechter, A. Microbial flora studies in thermophilic aerobic sludge treatment. Conserv. Recycl. 1985, 8, 303–313. [Google Scholar] [CrossRef]
- Juteau, P.; Tremblay, D.; Villemur, R.; Bisaillon, J.G.; Beaudet, R. Analysis of the bacterial community inhabiting an aerobic thermophilic sequencing batch reactor (AT-SBR) treating swine waste. Appl. Microbiol. Biotechnol. 2004, 66, 115–122. [Google Scholar] [CrossRef]
- LaPara, T.M.; Nakatsu, C.H.; Pantea, L.; Alleman, J.E. Phylogenetic analysis of bacterial communities in mesophilic and thermophilic bioreactors treating pharmaceutical wastewater. Appl. Environ. Microbiol. 2000, 66, 3951–3959. [Google Scholar] [CrossRef]
- Piterina, A.V.; MacCausland, C.; Bartlett, J.; Pembroke, J.T. Microbial ecology of autothermal aerobic digestion (ATAD): diversity, dynamics and activity of bacterial communities involved in treatment of municipal wastewater. In Modern Multidisciplinary Applied Microbiology, Exploiting Microbes and Their Interactions; Mendez-Vilas, A., Ed.; Wiley-VCH: Weinheim, Germany, 2006; pp. 526–535. [Google Scholar]
- Piterina, A.V.; Barlett, J.; Pembroke, T.J. 13C-NMR assessment of the pattern of organic matter transformation during domestic wastewater treatment by Autothermal Aerobic Digestion (ATAD). Int. J. Environ. Res. Public Health 2009, 6, 2288–2306. [Google Scholar] [CrossRef]
- Miller, D.N.; Bryant, J.E.; Madsen, E.L.; Ghiorse, W.C. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 1999, 65, 4715–4724. [Google Scholar]
- Martin-Laurent, F.; Philippot, L.; Hallet, S.; Chaussod, R.; Germon, J.C.; Soulas, G.; Catroux, G. DNA extraction from soils: old bias for new microbial diversity analysis method. Appl. Environ. Microbiol. 2001, 67, 2354–2359. [Google Scholar] [CrossRef]
- Sambrook, J. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Muyzer, G.; Hottentrager, S.; Teske, A.; Wawer, C. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA. A new molecular approach to analyse the genetic diversity of mixed microbial communities. In Molecular Microbial Ecology Manual; Akkermans, A., Ed.; Kluwer Academic Publ.: Norwell, MA, USA, 1996; p. 23. [Google Scholar]
- Nubel, U.; Engelen, B.; Felske, A.; Snaidr, J.; Wieshuber, A.; Amann, R.I.; Ludwig, W.; Backhaus, H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 1996, 178, 5636–5643. [Google Scholar]
- Dahllof, I.; Baillie, H.; Kjelleberg, S. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl. Environ. Microbiol. 2000, 66, 3376–3380. [Google Scholar] [CrossRef]
- Böltner, D.; MacMahon, C.; Pembroke, J.T.; Strike, P.; Osborn, A.M. R391: a conjugative integrating mosaic comprised of phage, plasmid and transposon elements. J. Bacteriol. 2002, 184, 5158–5169. [Google Scholar] [CrossRef]
- McGrath, B.M; O’Halloran, J.A; Piterina, A.V.; Pembroke, J.T. Molecular tools to detect the IncJ elements: a family of integrating, antibiotic resistant mobile genetic elements. J. Microbiol. Methods 2006, 66, 32–42. [Google Scholar] [CrossRef]
- Wilson, I.G. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 1997, 63, 3741–3751. [Google Scholar]
- Demeke, T.; Adams, R.P. The effects of plant polysaccharides and buffer additives on PCR. Biotechniques 1992, 12, 332–334. [Google Scholar]
- Furrer, B.; Candrian, U.; Wieland, P.; Luthy, J. Improving PCR efficiency. Nature 1990, 26, 324. [Google Scholar]
- Kreader, C.A. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 1996, 62, 1102–1106. [Google Scholar]
- Sarkar, G.; Kapelner, S.; Sommer, S.S. Formamide can dramatically improve the specificity of PCR. Nucl. Acids Res. 1990, 25, 7465. [Google Scholar] [CrossRef]
- Rossen, L.; Norskov, P.; Holmstrom, K.; Rasmussen, O.F. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int. J. Food Microbiol. 1992, 17, 37–45. [Google Scholar] [CrossRef]
- Akane, A.; Matsubara, K.; Nakamura, H.; Takahashi, S.; Kimura, K. Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstains, a major inhibitor of polymerase chain reaction (PCR) amplification. J. Forensic Sci. 1994, 39, 362–372. [Google Scholar]
- Suarez, D. Molecular diagnostic techniques: can we identify influenza viruses, differentiate subtypes and determine pathogenicity potential of viruses by RT-PCR? Avian Dis. 1997, 47, 318–325. [Google Scholar]
- Sarkar, G.; Kapelner, S.; Sommer, S.S. Formamide can dramatically improve the specificity of PCR. Nucl. Acids Res. 1990, 18, 7465. [Google Scholar] [CrossRef]
- Abu Al-Soud, W.A.; Radstrom, P. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J. Clin. Microbiol. 2000, 38, 4463–4470. [Google Scholar]
- Chou, S.W. Optimizing polymerase chain reaction technology for clinical diagnosis. Clin. Chem. 1991, 37, 1893–1894. [Google Scholar]
- Kleiboeker, S.B. Applications of competitor RNA in diagnostic reverse transcription-PCR. J. Clin. Microbiol. 2003, 41, 2055–2061. [Google Scholar] [CrossRef]
- Sipos, R.; Székely, A.; Révész, S.; Márialigeti, K. Addressing PCR biases in environmental microbiology studies. Methods Mol. Bio 2010, 599, 37–58. [Google Scholar] [CrossRef]
- Don, R.H.; Cox, P.T.; Wainwright, B.J.; Baker, K.; Mattick, J.S. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucl. Acids Res. 1991, 19, 4008. [Google Scholar] [CrossRef]
- Hongoh, Y.; Yuzawa, H.; Ohkuma, M.; Kudo, T. Evaluation of primers and PCR conditions for the analysis of 16S rRNA genes from a natural environment. FEMS Microbiol. Lett. 2003, 25, 299–304. [Google Scholar]
- Massol-Deya, A.A.; Odelson, D.A.; Hickey, R.F.; Tiedje, J.M. Bacterial Community Fingerprinting of Amplified 16S and 16-23S Ribosomal DNA Gene Sequences and Restriction Endonuclease Analysis (ARDRA). In Molecular Microbial Ecology Manual; Akkermans, A.D.L., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1995; pp. 1–8. [Google Scholar]
- CLC bio’s Workbenches. ( http://www.clcbio.com).
- VecScreen. ( http://www.ncbi.nlm.nih.gov/VecScreen/VecScreen.html).
- Sequence manipulation suite. ( http://www.gobozzy.com/sequence-analysis-tools).
- Maidak, B.L; Cole, J.R.; Lilburn, T.G.; Parker, C.T., Jr; Saxman, P.R.; Farris, R.J.; Garrity, G.M.; Olsen, G.J.; Schmidt, T.M.; Tiedje, J.M. The RDP-II (Ribosomal Database Project). Nucl. Acids Res. 2001, 29, 173–174. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Wang, G.C.T.; Wang, Y. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology 1996, 142, 1107–1114. [Google Scholar] [CrossRef]
- Kuske, C.R.; Banton, K.L.; Adorada, D.L.; Stark, P.C.; Jackson, P.J. Small-scale DNA sample preparation method for field PCR detection of microbial cells and spores in soil. Appl. Environ. Microbiol. 1998, 64, 2463–2472. [Google Scholar]
- DellAnno, A.; Fabiano, M.; Duineveld, G.C.A.; Kok, A.; Danovaro, R. Nucleic acid (DNA, RNA) quantification and RNA/DNA ratio determination in marine sediments: comparison of spectrophotometric, fluorometric and High Performance Liquid Chromatography methods and estimation of detrital DNA. Appl. Environ. Microbiol. 1998, 64, 3238–3245. [Google Scholar]
- Jackson, C.R.; Harper, J.P.; Willoughby, D.; Roden, E.E.; Churchill, P.F. A simple, efficient method for the separation of humic substances and DNA from environmental samples. Appl. Environ. Microbiol. 1997, 63, 4993–4995. [Google Scholar]
- Radstrom, P.; Knutsson, R.; Wolffs, P.; Lövenklev, M.; Löfström, C. Pre-PCR processing: Strategies to generate PCR-compatible samples. Mol. Biotechnol. 2004, 26, 133–146. [Google Scholar] [CrossRef]
- Abu Al-Soud, W.; Râdström, P. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl. Environ. Microbiol. 1998, 64, 3748–3753. [Google Scholar]
- LaMontagne, M.G.; Michel, F.C., Jr.; Holden, P.A.; Reddy, C.A. Evaluation of extraction and purification methods for obtaining PCR-amplifiable DNA from compost for microbial community analysis. J. Microbiol. Meth. 2002, 49, 255–264. [Google Scholar] [CrossRef]
- Arbeli, Z.; Fuentes, C.L. Improved purification and PCR amplification of DNA from environmental samples. FEMS Microbiol. Lett. 2007, 272, 269–275. [Google Scholar] [CrossRef]
- Blanchard, M.M.; Tailon-Miller, P.; Nowotny, P.; Nowotny, V. PCR buffer optimization with a uniform temperature regimen to facicilate automation. PCR Meth. Applic. 1993, 2, 234–240. [Google Scholar]
- Innis, M.A.; Gelfand, D.H. Optimization of PCRs. In PCR protocols. A guide to methods and applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press Inc.: San Diego, CA, USA, 1990; p. 12. [Google Scholar]
- Loomis, W.D. Overcoming problems of phenolics and quinones in the isolation of plant enzymes and organelles. Meth. Enzymol. 1974, 31, 528–545. [Google Scholar] [CrossRef]
- Zhang, W.; Hu, G.Y.; Deisseroth, A. Improvement of PCR sequencing by formamide. Nucleic Acids Res. 1991, 19, 6649. [Google Scholar] [CrossRef]
- Loeffelholz, M.; Deng, H. PCR and Its Variations. In Advanced techniques in Diagnostic Microbiology; Tang, Y.W., Stratton, C.W., Eds.; Springer: New York, NY, USA, 2006; Volume 1, pp. 166–183. [Google Scholar]
- Wiedbrauk, D.L.; Werner, J.C.; Drevon, A.M. Inhibition of PCR by aqueous and vitreous fluids. J. Clin. Microbiol. 1995, 33, 2643–2646. [Google Scholar]
- Wilson, I.G.; Cooper, J.E.; Gilmour, A. Some factors inhibiting amplification of the Staphylococcus aureus enterotoxin C1 (sec1) by PCR. Int. J. Food Microbiol. 1994, 22, 55–62. [Google Scholar]
- Gibson, J.R.; Sutherland, K.; Owen, R.J. Inhibition of DNAse activity in PFGE analysis of DNA from Campylobacter jejuni. Lett. Appl. Microbiol. 1994, 19, 357–358. [Google Scholar] [CrossRef]
- Singer, G.A.; Hickey, D.A. Thermophilic prokaryotes have characteristic patterns of codon usage, amino acid composition and nucleotide content. Gene 2003, 23, 39–47. [Google Scholar] [CrossRef]
- Klappenbach, J.A.; Saxman, P.R.; Cole, J.R.; Schmidt, T.M. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 2001, 29, 181–184. [Google Scholar] [CrossRef]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).