Abstract
The mitochondrial genome (mitogenome) has been widely used for structural comparisons and phylogenetic analyses of Hemiptera groups at different taxonomic levels. However, little is known about the mitogenomic characteristics of species from Antheminia and Carpocoris, two morphologically similar genera in the Pentatomidae family, and their phylogenetic relationships need to be further confirmed. In this study, the mitogenomes of Antheminia varicornis (Jakovlev, 1874) and Carpocoris purpureipennis (De Geer, 1773) were sequenced and analyzed. Coupled with previously published mitogenomes of Pentatomidae, we performed a phylogenetic analysis. The mitogenomes of A. varicornis and C. purpureipennis are conserved in terms of genomic structure, base composition, codon usage, and tRNA secondary structure. Each mitogenome contains the typical 37 genes and a control region and all genes are arranged in the same order as in the ancestral insect mitogenome. Nucleotide composition is highly biased with the third codon in PCGs displaying the highest A + T content. Phylogenetic analysis strongly supports the sister relationship between A. varicornis and C. purpureipennis. The phylogenetic trees show a strong support for the monophyly of Asopinae and Phyllocephalinae, while the monophyly of Pentatominae and Podopinae was rejected. Our study enriches the mitochondrial genome database of the genera Antheminia and Carpocoris and provides a valuable resource for further phylogenetic and evolutionary analyses of the Pentatomidae.
1. Introduction
The typical insect mitochondrial genome (mitogenome) is a double-strand circular DNA molecule ranging from 14 kb to 20 kb in size, containing 37 genes (13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes), and a control region [1,2,3]. Due to its small size, maternal inheritance, low sequence recombination, and fast evolutionary rates [4,5], the mitogenome is considered a powerful marker for phylogenetic and evolutionary analyses [6,7,8,9,10,11]. In addition, comparing mitochondrial gene structure and genetic composition between species can provide crucial insights into the evolution of related species [3,12,13]. Benefiting from advances in high-throughput sequencing technology, an increasing number of mitogenomes have been sequenced among the Hemiptera [14,15,16,17] and have been widely used for mitochondrial structure comparison, phylogenetic analysis at different taxonomic levels, species delimitation, population genetic structure, and biogeographic studies [7,18,19,20,21,22,23,24,25,26,27,28,29].
Pentatomidae is one of the largest and most diverse families of Heteroptera, with a wide distribution around the world [30]. Most species are phytophagous, sucking sap from the stems, leaves, or fruits of their host plants, and pose a serious threat to a wide variety of valued crops, causing significant economic losses worldwide [31,32]. Since the publication of the first complete mitogenome of a Pentatomidae species, Nezara viridula (GenBank accession number NC_011755) [33], the number of mitogenomes in this family has continued to grow [20,22,34,35,36]. Detailed comparative analyses of the mitogenome and phylogenetic analyses have also been performed for the Pentatomidae family [37]. However, the coverage is still limited relative to the number of species, which is quite restrictive for clarifying the phylogenetic relationships of genera and species in the Pentatomidae family. Antheminia Mulsant and Rey, 1866, and Carpocoris Kolenati, 1846, are two morphologically similar genera within the Pentatomidae family; most species in both two genera are crop pests, with adults and nymphs sucking juice from inflorescences and young stems. The yellow spots will appear on the leaves after being sucked, and the inflorescence and tender ear will wither or even fade after being seriously damaged. Many species of Antheminia and Carpocoris mainly harm cash crops such as alfalfa, wheat, potato, radish, carrot, elm, and bayberry. A previous study based on small molecular fragments (1759 bp of 18S rRNA, 646 bp of 28S rRNA, 592 bp of 16S rRNA and 564 bp of COI) revealed a sister relationship between the two genera, but there is low node support [38]. Currently, little is known about the mitogenomic characteristics of the two genera. Their molecular data are yet to be supplemented. Their phylogenetic positions in the Pentatomidae family and the phylogenetic relationships between them still need to be further confirmed.
In this study, we sequenced and annotated the mitogenomes of Antheminia varicornis (Jakovlev, 1874) and Carpocoris purpureipennis (De Geer, 1773), representing the Antheminia and Carpocoris genera, respectively. We analyzed the genomic structure, base composition, codon usage, and tRNA secondary structure of the two mitogenomes. Coupled with published mitogenomes of Pentatomidae, we carried out a phylogenetic analysis to verify the phylogenetic positions of Antheminia and Carpocoris in the Pentatomidae family and their phylogenetic relationships. This study may increase our understanding of the relationships between the Antheminia and Carpocoris genera, and also verify the phylogeny and evolution of Pentatomidae.
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
The specimens of A. varicornis and C. purpureipennis were collected from Horqin Right Front Banner, Neimenggu Province, China (46.46 N, 120.31 E), on 11 July 2021 and Ziwu Mountain, Shaanxi Province, China (35.87 N, 108.55 E), on 27 July 2019, respectively. Since both species consist of unprotected invertebrates, no special permits were required to collect samples from these sites. All specimens were preserved in 100% ethanol and stored at −20 °C at the Institute of Entomology at Nankai University (Tianjin, China). All specimens were identified based on their morphology. Total genomic DNA was extracted from the thoracic muscle using a Universal Genomic DNA Kit (CWBIO, Beijing, China) and stored at −80 °C until downstream analyses.
2.2. Mitogenome Sequencing, Assembly and Annotation
The whole mitochondrial genomes were sequenced using the Illumina NovaSeq 6000 platform with a 150 bp paired-end read strategy at Novogene Co., Ltd. (Beijing, China). Low-quality reads were removed using fastp [39], and then approximately 2 Gb of clean data were obtained for each sample. The clean data from the sequencing reads were assembled using mitoZ 2.4 [40] with default settings and IDBA-UD 1.1.3 [41] with minimum and maximum k values of 40 and 120 bp, respectively. Transfer RNA (tRNA) genes and their secondary structures were identified on the MITOS2 webserver (http://mitos2.bioinf.uni-leipzig.de/index.py, accessed on 3 April 2023). The typical secondary structure for tRNAs were manually drawn according to MITOS2 predictions and using Adobe Illustrator 2021. Protein-coding genes (PCGs) and ribosomal RNA (rRNA) genes were annotated through alignment with homologous regions of previously published mitogenomes of Pentatominae in GenBank. Newly sequenced mitogenomes were submitted to GenBank (accession numbers: OR074478 and OR074479).
2.3. Bioinformatic Analyses
Mitogenome maps were drawn using the Proksee Server (https://proksee.ca, accessed on 29 May 2023) [42]. The base composition, codon usage, and relative synonymous codon usage (RSCU) values of A. varicornis and C. purpureipennis were calculated in MEGA X [43]. The bias of the nucleotide composition was measured by AT-skew [(A − T)/(A + T)] and GC-skew [(G − C)/(G + C)].
2.4. Phylogenetic Analyses
Phylogenetic analyses were performed using the newly sequenced mitogenomes of A. varicornis and C. purpureipennis, together with 28 Pentatomidae mitogenomes downloaded from GenBank (Table 1). Two species of Plataspidae were selected as outgroups (Table 1).

Table 1.
Taxonomic information and GenBank accession numbers of mitochondrial genomes downloaded from GenBank in this study.
The sequences of 13 PCGs and 2 rRNA genes were aligned using MAFFT 7.402 [44]. After removing the stop codon, the alignments of individual genes were then concatenated by PhyloSuite 1.2.2 [45] to generate two datasets: PCG123R (all three codon positions of the 13 PCGs and 2 rRNAs) and PCG12R (the first and second codon positions of the 13 PCGs and 2 rRNAs). The best-fit partitioning scheme and nucleotide substitution models were identified using PartitionFinder 2.0 [46] and the Bayesian Information Criterion (BIC). Phylogenetic analyses were conducted using the Bayesian inference (BI) and maximum likelihood (ML) methods based on the two datasets. BI analysis was performed using MrBayes 3.2.7a [47] with the best-fitting substitution model (Table 2). Two simultaneous Markov chain Monte Carlo (MCMC) runs of 10,000,000 generations were conducted, and trees were sampled every 1000 generations, with the first 25% discarded as burn-in. The convergence of runs was confirmed by checking whether the deviation of split frequencies was below 0.01. ML analysis was performed using IQ-TREE 2.2.0 [48] with 1000 bootstrap replicates under the best-fitting substitution model.

Table 2.
The best model for each partition of the two datasets.
3. Results and Discussion
3.1. Mitogenome Organization and Composition
The mitogenomes of A. varicornis and C. purpureipennis are 15,251 and 15,322 bp in size, respectively. Each mitogenome contains 37 typical genes (13 PCGs, 2 rRNAs, and 22 tRNAs) and a control region. Among these genes, four PCGs (ND1, ND4, ND4L, and ND5), eight tRNAs (trnC, trnF, trnH, trnL (UAG), trnP, trnQ, trnV and trnY), and two rRNAs (12S rRNA and 16S rRNA) are encoded on the minority strand (N strand), while the other 23 genes are encoded on the majority strand (J strand) (Figure 1, Table 3 and Table 4). The mitogenome of A. varicornis has a total of 26 bp space in seven gene overlaps, ranging in length from 1 to 8 bp; the longest overlap region fell between the tRNA-Trp and tRNA-Cys genes. In addition, there were sixteen 1–25 bp gene spacer regions, with a total length of 116 bp; the longest 25 bp intergenic spacer sequences were located between ND1 and tRNA-Ser. In the C. purpureipennis mitogenome, gene overlaps were found at eight gene junctions and involved a total of 35 bp; the longest 8 bp overlap was located between the tRNA-Trp and tRNA-Cys genes. Intergenic spacer sequences were found at 15 gene junctions and involved a total of 98 bp, ranging in length from 1 to 21 bp; the longest 21 bp intergenic spacer sequences were located between ND1 and tRNA-Ser. The number and arrangement of genes in both mitogenomes are conserved, consistent with those of ancestral insects [49].
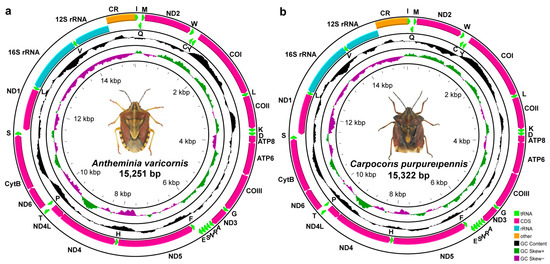
Figure 1.
Mitogenome maps of A. varicornis (a) and C. purpureipennis (b). The names of PCGs and rRNAs are indicated by standard abbreviations, while names of tRNAs are represented by a single letter abbreviation.

Table 3.
Organization of mitochondrial genome of A. varicornis.

Table 4.
Organization of mitochondrial genome of C. purpureipennis.
The nucleotide composition of the two mitogenomes is biased toward A + T, as in other Pentatomidae species [22,37]. The A + T content of the whole mitogenome is 76.7% for A. varicornis and 73.4% for C. purpureipennis. The A + T content of the protein-coding genes is 73.2% for A. varicornis and 72.9% for C. purpureipennis. The 12S rRNA and 16S rRNA exhibit a higher A + T content among the 37 typical genes in both mitogenomes. The A + T content of the third codon in PCGs is significantly higher than that of the first and second codons (Table 5). The whole mitogenome of A. varicornis exhibits negative AT-skew and GC-skew, while that of C. purpureipennis exhibits positive AT-skew and negative GC-skew. It is generally believed that asymmetric mutations at four bases and selection pressure are the two main reasons for the base composition preference of mitochondrial genome, which mainly come from the process of replication and gene transcription [50].

Table 5.
Nucleotide composition of mitochondrial genomes of A. varicornis and C. purpureipennis.
3.2. Protein-Coding Genes and Codon Usage
Most PCGs of the two mitogenomes begin with the standard start codon ATN (N represents one of four nucleotides, A, T, C, or G), while COI starts with TTG. In addition, the start codons of ATP8 are TTG and GTG in A. varicornis and C. purpureipennis, respectively. The start codon of ND1 in A. varicornis is TTG (Table 3 and Table 4). The termination codon in nine PCGs (ATP6, ATP8, COIII, CytB, ND1, ND2, ND4, ND4L and ND6) is TAA or TAG, while COI, COII, ND3, and ND5 have incomplete termination codons (T or TA) (Table 3 and Table 4) that are probably completed by post-transcriptional polyadenylation [51]. The total codon numbers (excluding the termination codons) in A. varicornis and C. purpureipennis are 3663 and 3669, respectively. The longest gene was the ND5 gene (1706 and 1705), and the shortest was the ATP8 gene (159 and 159) in A. varicornis and C. purpureipennis, respectively. The most frequently used codon families are Ile, Leu2, Met, and Phe, each numbering more than 300. The least frequently used codon family is Cys, with a total of 50 in both mitogenomes (Figure 2). The relative synonymous codon usage (RSCU) patterns for the two mitogenomes are similar, and the RSCU values are shown in Figure 3 and Table 6. For each amino acid, the most prevalently used codons are NNA and NNU (Figure 3, Table 6), which is consistent with the higher A + T content in the third codon in PCGs.
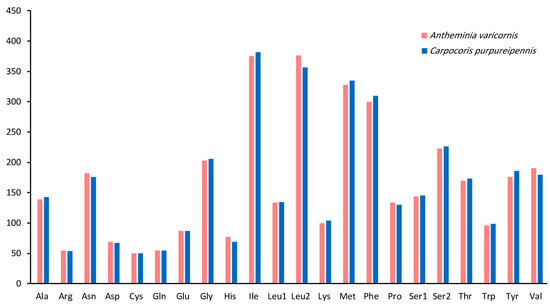
Figure 2.
Patterns of codon usage in the mitogenomes of A. varicornis and C. purpureipennis. The X-axis shows the codon families, and the Y-axis shows the total codons.
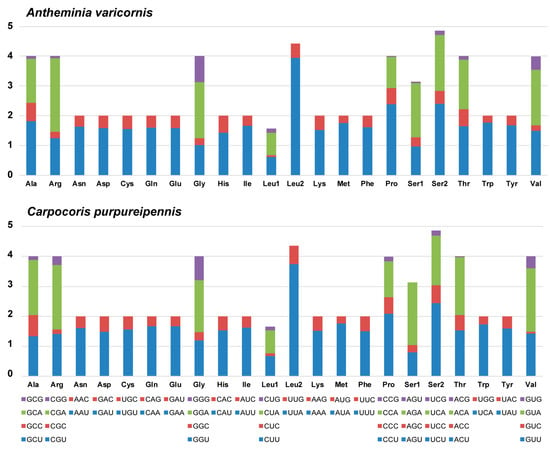
Figure 3.
The relative synonymous codon usage (RSCU) in the mitogenomes of A. varicornis and C. purpureipennis. The X-axis shows the codons, and the Y-axis shows RSCU values. The upper and lower color interpretation is shared.

Table 6.
Codon and the relative synonymous codon usage (RSCU) of the mitochondrial genomes (excluding the termination codons) of A. varicornis and C. purpureipennis.
3.3. tRNAs, rRNAs and Control Region
Typical sets of 22 tRNA genes ranging in length from 62 to 72 bp have been identified in the mitogenomes of A. varicornis and C. purpureipennis (Table 3 and Table 4), with variations in length. The A + T content of the concatenated tRNA genes is 75.7% and 73.9% for A. varicornis and C. purpureipennis, respectively. The nucleotide skews in the tRNA genes in the mitogenomes of the two species are consistent, with the concatenated tRNA genes exhibiting a positive AT-skew and a negative GC-skew (Table 5). Most tRNA genes can be folded into the typical cloverleaf secondary structure, while the dihydrouridine (DHU) arms of trnS (GCU) and trnV are very short, with only a single base pair. The non-Watson–Crick base pair G-U is common in tRNA genes from both species (Figure 4 and Figure 5). The size of A. varicornis ranged from 62 bp (tRNA-Cys) to 71 bp (tRNA-Asp) while the size of C. purpureipennis ranged from 63 bp (tRNA-Ala) to 72 bp (tRNA-Lys). The total length of the 22 tRNAs of A. varicornis were 1472 bp, and the total length of the 22 tRNAs of C. purpureipennis were 1463 bp, respectively. Both 12S and 16S rRNA genes exhibit similar positions and sizes in the mitogenomes of A. varicornis and C. purpureipennis (Table 3 and Table 4). The 12S rRNA exhibits a positive AT-skew and a negative GC-skew in both species. The 16S rRNA exhibits a negative AT-skew and a positive GC-skew in A. varicornis and a positive AT-skew and a negative GC-skew in C. purpureipennis (Table 5). The control regions of A. varicornis and C. purpureipennis are 595 and 682 bp in size (Table 3 and Table 4), with A + T contents of 70.4% and 70.5%, respectively (Table 5).
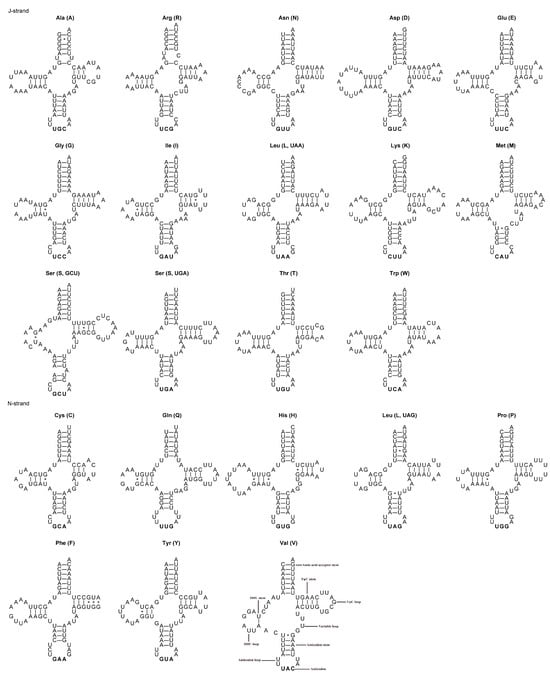
Figure 4.
Secondary structure of 22 tRNAs in A. varicornis. * represents a non-classical pairing of G=U.
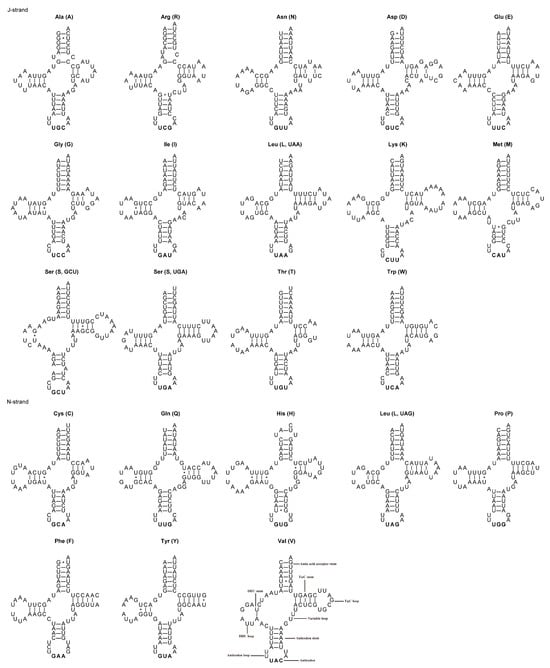
Figure 5.
Secondary structure of 22 tRNAs in C. purpureipennis. * represents a non-classical pairing of G=U.
3.4. Phylogenetic Relationships
A previous study, based on small molecular fragments (1759 bp of 18S rRNA, 646 bp of 28S rRNA, 592 bp of 16S rRNA and 564 bp of COI), revealed that the Antheminia genus forms a sister group with Carpocoris, but there is low node support [38]. In this study, we selected one species from each genus as a representative taxon, used mitogenome data to verify this sister relationship, and further explored their phylogenetic positions within the Pentatomidae family. Phylogenetic analyses were performed using the BI and ML methods based on the two datasets (PCG123R and PCG12R). All four phylogenetic trees show that A. varicornis forms a sister relationship with C. purpureipennis with high nodal support values (PP = 1 in BI trees; BS values = 100 in ML trees), which is consistent with the traditional taxonomy and the findings of a previous study [38].
In addition, all of the phylogenetic results support the fact that the two species form a sister group with Dolycoris baccarum, and then the three species together form a sister group with Rubiconia intermedia (Figure 6 and Figure 7). This result is consistent with previous results based on molecular and morphological evidence; accordingly, we support the previous proposal that the species in the Eysarcorini and Carpocorini are closely related [37]. Neojurtina typica is in the most basic position within Pentatomidae. The phylogenetic tree constructed through ML and BI analysis showed a strong support for the monophyly of Asopinae and Phyllocephalinae, while the monophyly of Pentatominae and Podopinae was rejected (Figure 6 and Figure 7).
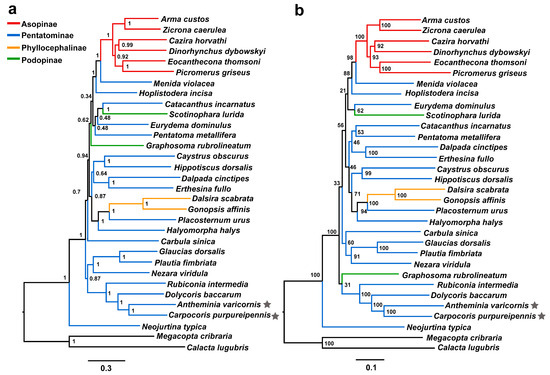
Figure 6.
Phylogenetic relationships of Pentatomidae based on dataset PCG123R. Pentagram, the mt genome sequences of A. varicornis and C. purpureipennis in this study. The black lines are the two outgroups used in this study. (a) BI tree, numbers at the nodes are posterior probabilities; (b) ML tree, numbers at the nodes are bootstrap values.
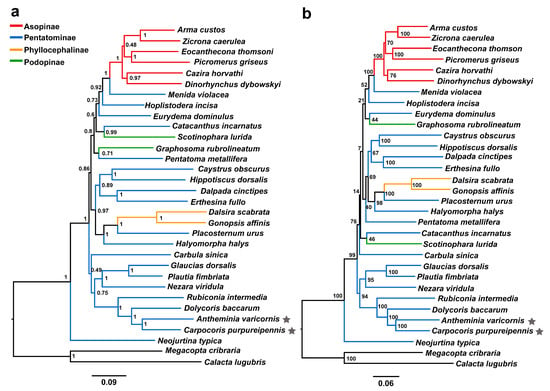
Figure 7.
Phylogenetic relationships of Pentatomidae based on dataset PCG12R. Pentagram, the mt genome sequences of A. varicornis and C. purpureipennis in this study. The black lines are the two outgroups used in this study. (a) BI tree, numbers at the nodes are posterior probabilities; (b) ML tree, numbers at the nodes are bootstrap values.
4. Conclusions
In previous studies, more attention has been paid to the phylogenetic relationships of the higher order members of Heteroptera, while less attention has been paid to the phylogenetic relationships within the subfamily. In this study, two mitochondrial genomes from Pentatominae were sequenced and added to the existing data; we sequenced and analyzed the mitogenomes of A. varicornis and C. purpureipennis. The two mitogenomes are conserved in genomic structure, base composition, codon usage, and tRNA secondary structure.
We performed a phylogenetic analysis based on the sequences of thirteen PCGs and two rRNA genes. Our results strongly support the sister relationship between A. varicornis and C. purpureipennis. Our results provide a valuable resource for further phylogenetic and evolutionary analyses of the Pentatomidae, which also reveal the relationships among four subfamilies within Pentatomidae. The phylogenetic trees show a strong support for the monophyly of Asopinae and Phyllocephalinae, while the monophyly of Pentatominae and Podopinae was rejected. More mitochondrial genomes and nuclear genes need to be sequenced to reveal the mitochondrial genome evolution and phylogenetic relationships of Pentatominae more comprehensively.
Author Contributions
Y.W., C.Z. and W.B. conceived and designed the research. Y.W. and R.Y. conducted experiments. Y.W., R.Y. and X.Z. analyzed data. Y.W., C.Z. and W.B. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (31820103013, 32130014).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank (accession numbers: OR074478 and OR074479) at https://www.ncbi.nlm.nih.gov/ (accessed on 1 June 2023).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [PubMed]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Brown, W.M.; George, M.; Wilson, A.C. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1979, 76, 1967–1971. [Google Scholar] [CrossRef]
- Curole, J.P.; Kocher, T.D. Mitogenomics: Digging deeper with complete mitochondrial genomes. Trends Ecol. Evol. 1999, 14, 394–398. [Google Scholar] [CrossRef]
- Song, F.; Li, H.; Liu, G.H.; Wang, W.; James, P.; Colwell, D.D.; Tran, A.; Gong, S.; Cai, W.; Shao, R. Mitochondrial Genome Fragmentation Unites the Parasitic Lice of Eutherian Mammals. Syst. Biol. 2019, 68, 430–440. [Google Scholar] [CrossRef]
- Du, Z.; Hasegawa, H.; Cooley, J.R.; Simon, C.; Yoshimura, J.; Cai, W.; Sota, T.; Li, H. Mitochondrial Genomics Reveals Shared Phylogeographic Patterns and Demographic History among Three Periodical Cicada Species Groups. Mol. Biol. Evol. 2019, 36, 1187–1200. [Google Scholar] [CrossRef]
- Camacho, M.A.; Cadar, D.; Horváth, B.; Merino-Viteri, A.; Murienne, J. Revised phylogeny from complete mitochondrial genomes of phyllostomid bats resolves subfamilial classification. Zool. J. Linn. Soc. 2022, 196, 1591–1607. [Google Scholar] [CrossRef]
- Ge, X.; Peng, L.; Vogler, A.P.; Morse, J.C.; Yang, L.; Sun, C.; Wang, B. Massive gene rearrangements of mitochondrial genomes and implications for the phylogeny of Trichoptera (Insecta). Syst. Entomol. 2022, 48, 278–295. [Google Scholar] [CrossRef]
- Nielsen, M.; Margaryan, A.; Nielsen, T.L.; Enghoff, H.; Allentoft, M.E. Complete mitochondrial genomes from museum specimens clarify millipede evolution in the Eastern Arc Mountains. Zool. J. Linn. Soc. 2022, 196, 924–939. [Google Scholar] [CrossRef]
- Kunde, M.N.; Barlow, A.; Klittich, A.M.; Yakupova, A.; Patel, R.P.; Fickel, J.; Forster, D.W. First mitogenome phylogeny of the sun bear Helarctos malayanus reveals a deep split between Indochinese and Sundaic lineages. Ecol. Evol. 2023, 13, e9969. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.P.; Liang, X.; Wang, X.; Hu, H.Y.; Huang, Y.X. Novel gene rearrangement pattern in mitochondrial genome of Ooencyrtusplautus Huang & Noyes, 1994: New gene order in Encyrtidae (Hymenoptera, Chalcidoidea). Zookeys 2022, 1124, 1–21. [Google Scholar] [PubMed]
- Pakrashi, A.; Patidar, A.; Singha, D.; Kumar, V.; Tyagi, K. Comparative analysis of the two suborders of Thysanoptera and characterization of the complete mitochondrial genome of Thrips parvispinus. Arch. Insect Biochem. Physiol. 2023, 114, e22010. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-Z.; Rédei, D.; Eger, J., Jr.; Wang, Y.-H.; Wu, H.-Y.; Carapezza, A.; Kment, P.; Cai, B.; Sun, X.-Y.; Guo, P.-L.; et al. Phylogeny and the colourful history of jewel bugs (Insecta: Hemiptera: Scutelleridae). Cladistics 2017, 34, 502–516. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.L.; Li, M.; Li, T.; Yuan, J.J.; Bu, W.J. A mitochondrial genome of Micronectidae and implications for its phylogenetic position. Int. J. Biol. Macromol. 2018, 119, 747–757. [Google Scholar] [CrossRef]
- Dong, X.; Wang, K.; Tang, Z.; Zhang, Y.; Yi, W.; Xue, H.; Zheng, C.; Bu, W. Phylogeny of Coreoidea based on mitochondrial genomes show the paraphyly of Coreidae and Alydidae. Arch. Insect Biochem. Physiol. 2022, 110, e21878. [Google Scholar] [CrossRef]
- Liu, X.; He, J.; Du, Z.; Zhang, R.; Cai, W.; Li, H. Weak genetic structure of flower thrips Frankliniella intonsa in China revealed by mitochondrial genomes. Int. J. Biol. Macromol. 2023, 231, 123301. [Google Scholar] [CrossRef]
- Li, H.; Leavengood, J.M.; Chapman, E.G., Jr.; Burkhardt, D.; Song, F.; Jiang, P.; Liu, J.; Zhou, X.; Cai, W. Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of true bugs. Proc. R. Soc. B Biol. Sci. 2017, 284, 20171223. [Google Scholar] [CrossRef]
- Yang, H.; Li, T.; Dang, K.; Bu, W. Compositional and mutational rate heterogeneity in mitochondrial genomes and its effect on the phylogenetic inferences of Cimicomorpha (Hemiptera: Heteroptera). BMC Genom. 2018, 19, 264. [Google Scholar] [CrossRef]
- Liu, Y.; Li, H.; Song, F.; Zhao, Y.; Wilson, J.J.; Cai, W. Higher-level phylogeny and evolutionary history of Pentatomomorpha (Hemiptera: Heteroptera) inferred from mitochondrial genome sequences. Syst. Entomol. 2019, 44, 810–819. [Google Scholar] [CrossRef]
- Zheng, C.; Ye, Z.; Zhu, X.; Zhang, H.; Dong, X.; Chen, P.; Bu, W. Integrative taxonomy uncovers hidden species diversity in the rheophilic genus Potamometra (Hemiptera: Gerridae). Zool. Scr. 2019, 49, 174–186. [Google Scholar] [CrossRef]
- Xu, S.; Wu, Y.; Liu, Y.; Zhao, P.; Chen, Z.; Song, F.; Li, H.; Cai, W. Comparative Mitogenomics and Phylogenetic Analyses of Pentatomoidea (Hemiptera: Heteroptera). Genes 2021, 12, 1306. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zheng, C.; Dong, X.; Zhang, H.; Ye, Z.; Xue, H.; Bu, W. Species boundary and phylogeographical pattern provide new insights into the management efforts of Megacopta cribraria (Hemiptera: Plataspidae), a bean bug invading North America. Pest Manag. Sci. 2022, 78, 4871–4881. [Google Scholar] [CrossRef] [PubMed]
- Francoso, E.; Zuntini, A.R.; Ricardo, P.C.; Araujo, N.D.; Silva, J.P.N.; Brown, M.J.F.; Arias, M.C. The complete mitochondrial genome of Trigonisca nataliae (Hymenoptera, Apidae) assemblage reveals heteroplasmy in the control region. Gene 2023, 881, 147621. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.X.; Wei, N.; Jost, M.; Wanke, S.; Rees, M.; Liu, Y.; Zhou, R.C. The Mitochondrial Genome of the Holoparasitic Plant Thonningia sanguinea Provides Insights into the Evolution of the Multichromosomal Structure. Genome Biol. Evol. 2023, 15, e155. [Google Scholar] [CrossRef] [PubMed]
- Ke, S.J.; Liu, D.K.; Tu, X.D.; He, X.; Zhang, M.M.; Zhu, M.J.; Zhang, D.Y.; Zhang, C.L.; Lan, S.R.; Liu, Z.J. Apostasia Mitochondrial Genome Analysis and Monocot Mitochondria Phylogenomics. Int. J. Mol. Sci. 2023, 24, 7837. [Google Scholar] [CrossRef]
- Kim, Y.K.; Jo, S.; Cheon, S.H.; Hong, J.R.; Kim, K.J. Ancient Horizontal Gene Transfers from Plastome to Mitogenome of a Nonphotosynthetic Orchid, Gastrodia pubilabiata (Epidendroideae, Orchidaceae). Int. J. Mol. Sci. 2023, 24, 11448. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.Y.; Li, J.L.; Zuo, Y.W.; He, P.; Zhang, H.; Zhang, X.X.; Wang, B.R.; Zhang, J.B.; Yu, J.; Deng, H.P. Complete mitochondrial genome of Thuja sutchuenensis and its implications on evolutionary analysis of complex mitogenome architecture in Cupressaceae. BMC Plant Biol. 2023, 23, 84. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, S.J.; Chen, J.J.; Chen, C.J.; Lin, X.J.; Peng, H.; Zhao, Q.; Wang, X.Y. Characterization and phylogenetic analysis of the complete mitochondrial genome sequence of Photinia serratifolia. Sci. Rep. 2023, 13, 770. [Google Scholar] [CrossRef]
- Rider, D.; Schwertner, C.; Vilimova, J.; Redei, D.; Kment, P.; Thomas, D. Higher Systematics of the Pentatomoidea; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Cesari, M.; Maistrello, L.; Ganzerli, F.; Dioli, P.; Rebecchi, L.; Guidetti, R. A pest alien invasion in progress: Potential pathways of origin of the brown marmorated stink bug Halyomorpha halys populations in Italy. J. Pest Sci. 2014, 88, 1–7. [Google Scholar] [CrossRef]
- Mi, Q.; Zhang, J.; Gould, E.; Chen, J.; Sun, Z.; Zhang, F. Biology, Ecology, and Management of Erthesina fullo (Hemiptera: Pentatomidae): A Review. Insects 2020, 11, 346. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.; Li, M.; Dong, P.; Cui, Y.; Xie, Q.; Bu, W. Comparative and phylogenomic studies on the mitochondrial genomes of Pentatomomorpha (Insecta: Hemiptera: Heteroptera). BMC Genom. 2008, 9, 610. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.L.; Yuan, M.L.; Shen, Y.Y. The complete mitochondrial genome of Dolycoris baccarum (Insecta: Hemiptera: Pentatomidae). Mitochondrial DNA 2013, 24, 469–471. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.L.; Zhang, Q.L.; Guo, Z.L.; Wang, J.; Shen, Y.Y. Comparative mitogenomic analysis of the superfamily Pentatomoidea (Insecta: Hemiptera: Heteroptera) and phylogenetic implications. BMC Genom. 2015, 16, 460. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wei, J.; Zhao, W.; Chen, C.; Gao, X.; Zhao, Q. The complete mitochondrial genome of Pentatoma rufipes (Hemiptera, Pentatomidae) and its phylogenetic implications. Zookeys 2021, 1042, 51–72. [Google Scholar] [CrossRef] [PubMed]
- Lian, D.; Wei, J.; Chen, C.; Niu, M.; Zhang, H.; Zhao, Q. Comparative analysis and phylogeny of mitochondrial genomes of Pentatomidae (Hemiptera: Pentatomoidea). Front. Genet. 2022, 13, 1045193. [Google Scholar] [CrossRef] [PubMed]
- Roca-Cusachs, M.; Schwertner, C.F.; Kim, J.; Eger, J.; Grazia, J.; Jung, S. Opening Pandora’s box: Molecular phylogeny of the stink bugs (Hemiptera: Heteroptera: Pentatomidae) reveals great incongruences in the current classification. Syst. Entomol. 2021, 47, 36–51. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.; Yiu, S.M.; Chin, F.Y. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2019, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Clary, D.O.; Wolstenholme, D.R. The mitochondrial DNA molecule of Drosophila yakuba: Nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 1985, 22, 252–271. [Google Scholar] [CrossRef]
- Hassanin, A.; Leger, N.; Deutsch, J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetic inferences. Syst. Biol. 2005, 54, 277–298. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).