Abstract
Pluteus is a species-rich genus of saprotrophic agaric in the family Pluteaceae and is widely distributed in tropical, subtropical, and temperate areas throughout the world. Some species in this genus are threatened species according to the International Union for Conservation of Nature (IUCN) red list. During investigations of agarics in northern Thailand, four Pluteus taxa were collected. Morphological characteristics and phylogenic analyses were investigated. Two new species, namely P. chandrasikuliae and P. saisamorniae, were introduced. Pluteus chandrasikuliae is characterized by its relatively large basidiomata, an applanate, dark brown scaly pileus with a cutis pileipellis, two types of hymenial cystidia viz. irregular, as well as diverticulate cells and lageniform cells. In accordance with the phylogenetic results, this new species belongs to the Pluteus sect. Celluloderma. Moreover, P. saisamorniae is distinguished by a plano-convex with a broad umbo, dark brown minute squamules pileus, light brown lamellar edges, greyish orange stipe covered with brown granules, subglobose to broadly ellipsoid basidiospores, an abundance of thin- to thick-walled cheilocystidia and pleurocystidia, a trichohymeniderm pileipellis, and brown caulocystidia in clusters. Pluteus saisamorniae is a member of Pluteus sect. Hispidoderma. Additionally, P. losulus and P. septocystidiatus were discovered in Thailand for the first time and they belong to Pluteus sect. Pluteus. Comprehensive descriptions along with illustrations, photographs, phylogenetic trees showing their positions, and a comparison with phenetically similar taxa are provided.
1. Introduction
Pluteus Fr. is the largest genus within the family Pluteaceae Kotl. & Pouzar. This genus currently comprises about 500 accepted species [1]. Morphologically, Singer [2] recognized the Pluteus species with combinations of free lamellae, inverse hymenophoral trama, pinkish brown spore prints, and the absence of volva. This species is inamyloid with cyanophilic basidiospores and usually without annulus. Modern molecular studies involving nuclear ribosomal genes have been used to circumscribe the species within the genus Pluteus. Pileipellis structure and basidiospore size are key characteristics that are used to separate Pluteus from other related genera, namely Volvariella and Volvopluteus. Pluteus is monophyletic and currently includes species with an annulus traditionally classified in the genus Chamaeota [3]. The type species of the genus has been designated as P. cervinus; however, this type specimen does not exist in the established records. Thus, the confusion associated with identifying this species has resulted in the creation of a complex of several species [2,4,5]. Recently, many specimens of P. cervinus collected from Germany, where the species has originally been described, have been re-examined. Consequently, the lectotype and epitype of P. cervinus were designated by Justo and colleagues [6] with the use of accepted morphological descriptions provided by Vellinga [5] and molecular data [7].
Historically, various infrageneric classifications within the genus were proposed by Singer in 1986 and Vellinga and Schreurs in 1985 [2,8]. Based on materials collected from different continents, various taxonomic concepts have been applied by local mycologists resulting in a variety of species-complexes within this genus. Molecular phylogeny has been applied together with morphology to clarify the species boundary of taxa obtained from different geographical locations [6,7]. Moreover, DNA sequences have also been used to revise certain taxonomic schemes. Accordingly, three sections have been recognized: Pluteus with metuloid pleucystidia forming a cutis pileipellis; Celluloderma Fayod with non-metuloid pleurocystidia, pileipellis usually composed of short clavate or spheropedunculate cells and mixed or not with elongated cystidioid elements; and Hispidoderma Fayod with non-metuloid pleurocystidia forming hymeniderm or trichoderm pileipellis with long, elongated elements [3]. Furthermore, it is noteworthy to mention that multigene phylogenetic analyses were used to elucidate the Holarctic species and infrageneric delimitation, while the use of ITS sequences proved to be sufficient for species recognition, even among specific species-complexes within the genus [3,6,7].
Pluteus is a saprotrophic genus that can usually be found on the rotting logs of both angiosperms and gymnosperms [9]. It is distributed worldwide and has been widely documented in many continents such as Africa [10,11,12,13], Europe [5,9,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35], and North and South America [36,37,38,39,40,41,42,43,44]. However, two Pluteus species, P. bressollensis Eyssart., Ducousso & Buyck and P. fenzlii (Schulzer) Corriol & P. A. Moreau, are listed in the global IUCN Red List due to their limited population and the fact that their habitats have been adversely affected by climate change [45,46].
Specimens of the Pluteus species have been recorded in some countries of Asia. In China, forty-one species of Pluteus have been documented [47,48]; over thirty species have been documented in Japan [6,7,49,50,51,52,53]; at least thirteen species in the Republic of Korea [54,55,56,57,58]; over twenty species in Sri Lanka [59,60,61]; and seventeen species in both India [62,63] and Vietnam [64]. In Thailand, only eight species of Pluteus, viz. P. admirabilis (Peck) Peck, P. aglaeotheles (Berk. & Broome) Sacc., P. aurantiorugosus (Trog) Sacc., P. cervinus (Schaeff.) P. Kumm, P. cinereofuscus J.E. Lange, P. leoninus (Schaeff.) P. Kumm., P. spinulosus Murrill, and P. subcervinus (Berk. & Broome) Sacc., have been listed on the checklist of Basidiomycota for the country but without voucher specimens [65]. Notably, Thailand is located in a tropical region with a high diversity of vegetation. Since it is classified as a biodiversity hot spot, the species diversity of Pluteus should be high. In order to fulfill the data of macrofungi within the country, surveys of agaric macrofungi in Si Satchanalai National Park, Sukhothai Province, and on the campus of Chiang Mai University were conducted during the period between May and August in the years 2018 and 2020. As a result of our fieldwork, various specimens of agarics were collected and two new species and two new records of Pluteus are presented herein.
2. Materials and Methods
2.1. Sample Collection and Morphological Study
Specimens were collected in Chiang Mai University, Chiang Mai Province, and Si Satchanalai National Park, Sukhothai Province, under permission of the Department of National Parks, Wildlife and Plant Conservation, Bangkok, Thailand (document No. 0907.4/13696). All macro-morphological descriptions are based on fresh basibiomes. Color notations follow Kornerup and Wanscher [66]. Photographs of fresh material were taken in situ. Microscopic features were obtained from dried material rehydrated in 95% ethanol followed by distilled water, 3% aqueous potassium hydroxide (KOH), and/or 3% aqueous Congo Red or Melzer’s reagent under a microscope (Olympus BX43, Japan) attached with drawing tube. Measurement of the basidiospores exclude an apiculus. Spore statistics were determined for the quotient of spore length by spore width (Q), x denotes the arithmetic mean and the range calculated from 25 spores taken from the number of specimens (s) involved. Means are reported in the form of mean ± one standard deviation. All holotype collections were deposited in Bangkok Forest Herbarium (BKF), Bangkok, Thailand. Isotype collections were kept in the herbarium of Pibulsongkram Rajabhat University (PSRU), Phitsanulok Province, and the Herbarium of Sustainable Development of the Biological Resources Laboratory, Faculty of Science, Chiang Mai University (SDBR-CMU), Chiang Mai Province, Thailand.
2.2. Molecular Procedures and Phylogenetic Analyses
Genomic DNA was extracted from each dried specimen using a DNA Extraction Mini Kit (FAVORGEN, Ping-Tung, Taiwan) following the manufacturer’s protocol. The ITS and nrLSU regions were amplified by polymerase chain reactions (PCRs) using ITS1/ITS4 primers [67] and LROR/LRO5 primers [68], respectively. The amplification of both ITS and nrLSU regions consisted of an initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 52 °C for 45 s, and an extension at 72 °C for 1 min and 72 °C for 10 min on a peqSTAR thermal cycler (PEQLAB Ltd., Farehamm, UK). PCR products were checked and purified by a PCR clean-up Gel Extraction NucleoSpin® Gel and PCR Clean-up Kit (Macherey-Nagel, Duren, Germany). Final PCR products were sent to 1st Base Company Co., Ltd. (Kembangan, Malaysia) for sequencing. The obtained sequences were used to query GenBank via BLAST (http://blast.ddbj.nig.ac.jp/top-e.html, 21 December 2021).
Phylogenetic analysis was carried out based on only the ITS sequences because the amount of available sequence data in the nrLSU gene is practically limited. Sequences from this study along with those obtained from previous studies and the GenBank database were selected and are provided in supplementary Table S1. Multiple sequence alignment was performed using MUSCLE [69]. The alignments of ITS sequences of Pluteus sect. Celluloderma, sect. Hispidoderma, and sect. Pluteus were deposited in TreeBASE under the study ID numbers 29242, 29243, and 29244, respectively. A phylogenetic tree was constructed under the maximum likelihood (ML) and Bayesian inference (BI) methods. The ML analysis was carried out using RAxML-HPC2 on XSEDE (8.2.10) in CIPRES Science Gateway V. 3.3 [70] using the GTRCAT model with 25 categories and 1000 bootstrap (BS) replications. The optimum nucleotide substitution model was obtained using jModeltest v.2.3 [71] under the Akaike information criterion (AIC) method. The BI analysis was performed using MrBayes 3.2.6 software for Microsoft Windows [72] with the selected optimal model being the GTR + I+G model. Bayesian posterior probabilities (PPs) were obtained from the 50% majority rule consensus of the trees kept. The tree topologies were visualized in FigTree v1.4.0 [73].
3. Results
3.1. Phylogenetic Relationships
Three phylogenetic trees (Pluteus sect. Celluloderma, sect. Hispidoderma, and sect. Pluteus) were constructed. The topologies of each phylogram obtained from the ML and BI analyses were found to be similar. Therefore, we have presented only the phylogram obtained from ML analysis. Firstly, the ITS sequence dataset of Pluteus sect. Celluloderma consisted of 72 taxa and the aligned dataset comprised 814 characters. ML analysis yielded a best scoring tree with a final ML optimization likelihood value of −10,484.6331. The matrix contained 557 distinct alignment patterns with 20.54% undetermined characters or gaps. Estimated base frequencies were recorded as follows: A = 0.2210, C = 0.2338, G = 0.2378, T = 0.3072; substitution rates AC = 1.1947, AG = 4.1403, AT = 1.3521, CG = 0.6129, CT = 5.8670, GT = 1.0000. The gamma distribution shape parameter alpha was equal to 0.3169 and the tree-length was equal to 4.9548. In addition, the final average standard deviation of the split frequencies at the end of the total MCMC generations was calculated as 0.00791 by BI analysis. A phylogram of Pluteus sect. Celluloderma is shown in Figure 1. The results indicate that the sequences of two specimens, BKF10214 and NW1565 (introduced as P. chandrasrikuliae), formed a monophyletic clade with high BS (100%) and PP (1.0) support and were clearly distinguished from the previously known species of Pluteus sect. Celluloderma.
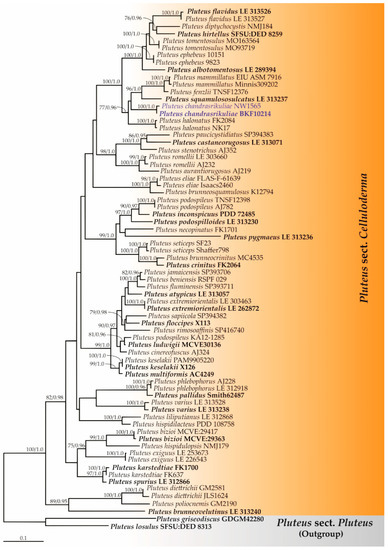
Figure 1.
Phylogram derived from maximum likelihood analysis of Pluteus sect. Celluloderma consists of 72 taxa of ITS sequences. Pluteus griseodiscus and P. losulus were used as the outgroup. The numbers above branches represent bootstrap percentages (left) and Bayesian posterior probabilities (right). Bootstrap values ≥75% and Bayesian posterior probabilities ≥0.90 are shown. The scale bar represents the expected number of nucleotide substitutions per site. Sequences obtained from this study are in blue. Type specimens are in bold.
Secondly, the ITS sequence dataset of Pluteus sect. Hispidoderma consisted of 46 taxa with the aligned dataset comprising 770 characters. ML analysis yielded a best scoring tree with a final ML optimization likelihood value of −5852.5497. The matrix contained 460 distinct alignment patterns with 18.64% undetermined characters or gaps. Estimated base frequencies were recorded as follows: A = 0.2255, C = 0.2596, G = 0.2361, T = 0.2786; substitution rates AC = 1.5265, AG = 3.1440, AT = 1.4790, CG = 0.5951, CT = 5.0949, GT = 1.0000, and gamma distribution shape parameter alpha = 0.3183. The tree-length value was equal to 3.1449, and the final average standard deviation of the split frequencies at the end of the total MCMC generations was 0.00857. A phylogram of Pluteus sect. Hispidoderma is shown in Figure 2. A phylogram has assigned the specimens BKF10213 and NW1567 as a new species that has been described herein as P. saisamorniae. Pluteus saisamorniae formed a monophyletic clade with high BS (100%) and PP (1.0) supports and formed a sister taxon to P. pantherinus with high statistical support (100% BS and 1.0 PP).
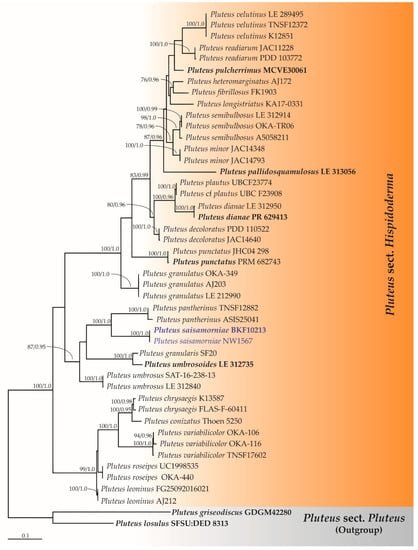
Figure 2.
Phylogram derived from maximum likelihood analysis of Pluteus sect. Hispidoderma consists of 46 taxa of ITS sequences. Pluteus griseodiscus and P. losulus were used as the outgroup. The numbers above branches represent bootstrap percentages (left) and Bayesian posterior probabilities (right). Bootstrap values ≥75% and Bayesian posterior probabilities ≥0.90 are shown. The scale bar represents the expected number of nucleotide substitutions per site. Sequences obtained from this study are in blue. Type specimens are in bold.
Lastly, the ITS sequence dataset of Pluteus sect. Pluteus consisted of 59 taxa with the aligned dataset comprising 694 characters. The final ML optimization likelihood value was −4801.7434. The matrix contained 366 distinct alignment patterns with 11.75% undetermined characters or gaps. Estimated base frequencies were recorded as follows: A = 0.2330, C = 0.2120, G = 0.2147, T = 0.3393; substitution rates AC = 1.4448, AG = 5.5472, AT = 2.7851, CG = 0.7953, CT = 9.3026, GT = 1.0000, and gamma distribution shape parameter alpha = 0.3015. The tree-length value was equal to 2.5212 and the average standard deviation of the split frequencies of the BI analysis was 0.00625. A phylogram of Pluteus sect. Pluteus is shown in Figure 3. The results indicate that the sequences of two specimens, SDBR-CMU0842 and BKF10212, were placed in the monophyletic clade of P. losulus and P. septocystidiatus with high BS (100%) and BP (1.0) supporting values.
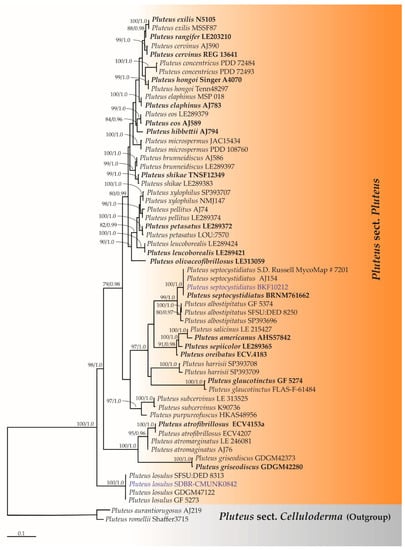
Figure 3.
Phylogram derived from maximum likelihood analysis of Pluteus sect. Pluteus consists of 59 taxa of ITS sequences. Pluteus aurantiorugosus and P. romellii were used as the outgroup. The numbers above branches represent bootstrap percentages (left) and Bayesian posterior probabilities (right). Bootstrap values ≥75% and Bayesian posterior probabilities ≥0.90 are shown. The scale bar represents the expected number of nucleotide substitutions per site. Sequences obtained from this study are in blue. Type specimens are in bold.
3.2. Taxonomic Description of New Species
Pluteus chandrasrikuliae Wannathes, J. Kumla & N. Suwannarach sp. nov. (Figure 4).
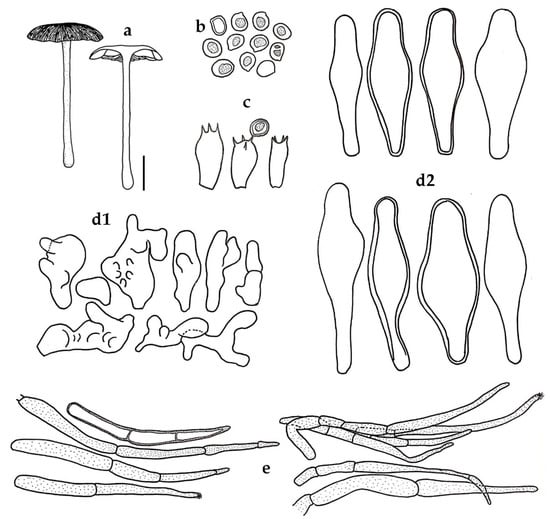
Figure 4.
Pluteus chandrasrikuliae BKF 10214 (holotype): (a) basidiomes; (b) basidiospores; (c) basidia; hymenial cystidia: (d1) irregular shaped cystidia; (d2) lageniform cystidia; (e) pileipellis element. Scale bars: (a) = 20 mm; (b,c) = 10 μm; (e) = 25 μm.
MycoBank: MB838336.
Diagnosis: Characterized by relatively large basidiomata with brown furfuraceous pileus, white glabrous stipe, two types of hymenial cystidia: (a) irregular diverticulate cells; (b) lageniform cells, covering the sides and edges of lamellae, cutis pileipellis with acute narrowly fusoid terminal cell.
Etymology: ‘chandrasrikuliae’ referring to the name of Anong Chandrasrikul (1935–2020) in honor of her pioneering contribution to Thai mycology.
Holotype: THAILAND, Sukhothai province, Si Satchanalai National Park, 99°48′44″ E 17°55′12″ N elevation 277 m, on decaying wood, August, 2018, N. Wannathes, J. Kumla & N. Suwannarach, BKF 10214 (isotype: NW1020).
Description: Pileus 35–40 mm diam., plano-convex to applanate, surface dull, dry, disc hirsute, with tiny fibrillose scales, furfuraceous to subglabrous elsewhere, striate at margin; disc dark brown (6F5-6), margin light brown (5D5-6) or paler. Context thin, soft, greyish white. Lamellae free, close, with 2 series of lamellulae, broad (2–4 mm), greyish orange (5B4-5). Stipe 65–80 × 2.5–3 mm cylindrical, central, fistulose, surface dull, dry, glabrous with a few silky fibrils, white overall.
Basidiospores 6–7(–8) × 4–6 μm (x = 6.54 ± 0.58 × 5.29 ± 0.51, Q = 1.17–1.33, q = 1.24 ± 0.06, s = 2) subglobose to broadly ellipsoid, smooth, hyaline, inamyloid, thick-walled. Basidia 12–17 × 6–8 μm, clavate to cylindrical, thin-walled, unclamped. Lamellar edge sterile. Cheilocystidia and Pleurocystidia similar, common on the sides and edges of lamellae composed of two types of cells: (a) irregular shaped cells, often branched, diverticulate with multiple bifurcations 11–32 × 7–18 μm, hyaline to pale yellow, inamyloid, thin- to thick-walled; (b) lageniform to broadly lageniform, 46–58 × 12–20 μm, hyaline, inamyloid, thin-walled to thick-walled (up to 1.5 μm). Pileipellis a cutis composed of parallel cylindrical hyphae 10–14 μm diam., repent, hyaline or with pale brown cytoplasmic pigments, non-incrusted, non-gelatinous; terminal cells over disc clustered, erect, elsewhere repent to suberect, 15–110 × 4–8 μm, narrowly fusoid, acute, with pale brown cytoplasmic pigment, sometimes with some encrusting pigment at apex, thick-walled (up to 1.5 μm). Pileus trama regular, hyphae 2–7 μm diam., cylindrical, hyaline, inamyloid, non-gelatinous, thin-walled. Lamellar trama convergent, hyphae 3–6 μm diam., cylindrical, hyaline, inamyloid, non-gelatinous, thin-walled. Stipitipellis a cutis of repent hyphae 4–15 μm diam., cylindrical, hyaline, inamylid, non-incrusted, non-gelatinous, thin-walled. Caulocystidia absent. Clamp connections absent in the examined tissues.
Habit, habitat, and known distribution: Solitary on decaying wood in bamboo thickets, deciduous dipterocarp-oak forests in Thailand.
Additional specimen examined: THAILAND, Phitsanulok Province, Pibulsongkram Ratjabhat University, herbal garden beside Faculty of Science and Technology, 100°12′54″ E 16°49′43″ N elevation 28 m, on decaying wood, September, 2020, NW 1565.
GenBank accession numbers: holotype (BKF 10214) ITS: MN492649, nrLSU: OM250115; additional specimen (NW 1565) ITS: OM257409, nrLSU: OM250229.
Notes: Pluteus chandrasrikuliae is characterized by a relatively large appearance with applanate pileus, a dark brown scaly disc, white stipe, subglobose to broadly ellipsoid basidiospores with mean dimensions of 6.5 × 5.3 μm and two types of hymenial cystidia as follows: (1) irregular, diverticulate cells and (2) lageniform cells, a cutis-type pileipellis with fusoid-acute, brown terminal cells, lacking caulocystidia and a clamp connection.
According to our phylogenetic analyses inferred from ITS sequences (Figure 1), P. chandrasrikuliae formed a distinct clade as a sister group of several taxa in the ephebeus clade [40]. The remarkable characteristics of P. chandrasrikuliae are as follows: irregular, diverticulate hymenial cystidia indicating phylogenetic distance among Pluteus in sect. Celluloderma. Notably, the irregular, diverticulate cystidia are unusual and rare characteristics that are found in the genus.
Morphologically, P. chandrasrikul is most similar to P. hirtellus and P. riberaltensis due to its macroscopic features, a cutis to trichoderm pileipellis with long, acute, brown terminal cells and the absence of caulocystidia. However, P. hirtellus differs by appearing globose to subglubose (avQ = 1.7), having slightly smaller basidiospores with mean dimensions of 5.3 × 4.96 μm, and the absence of irregular, diverticulate hymenial cystidia [10]. Pluteus riberaltensis differs by forming globose (avQ = 1.08) basidiospores and is absent of irregular, diverticulate hymenial cystidia [39]. Based on certain macroscopic features, such as a dark brown to brown pileus and white stipe, P. chandrasrikul is close to P. ephebeus, P. hispidulus var. cephalocystis, P. escharites, and P. albostipitatus. Pluteus ephebeus forms larger basidiomata with a pileus of 35–70 mm in diam., and a stipe that is 45–95 × 4–8 mm and covered with brown fibrils. It is utriform to fusiform or lageniform hymenial cytidia but lacks diverticulate cystidia [5]. Pluteus hispidulus var. cephalocystis forms smaller basidiomata with a pileus of 7–23 mm in diam., a stipe of 17–40 × 1–3 mm, a conical to applanate-umbonate pileus, and the absence of pleurocystidia [5,21]. Pluteus escharites differs from the new species by forming smaller basidiospores (5–6 × 4.5–5 μm). It is versiculose with ellipsoid to pyriform cheilocystidia and lacks pleurocystidia [59]. Pluteus albostipitatus forms larger basidiospores (7.5–10 × 6.2–7.5 μm). It is ventricose with a rounded to subcapitate apex, thin- to relatively thick-walled pleurocystidia, and clavate with thin-walled cheilocystidia [74]. The new species is somewhat close to P. atrofusens in terms of its cutis pileipellis, its spore size, and the fact that it has thin- to thick-wall hymenial cysdidia without hooks. However, it differs from the latter in terms of the shape of the cystidia, a smaller basidiomata with a pileus of 17–23 mm in diam, a stipe of 45–51 mm long, and the presence of cualocystidia [75].
Pluteus saisamorniae Wannathes, J. Kumla & N. Suwannarach sp. nov. (Figure 5).
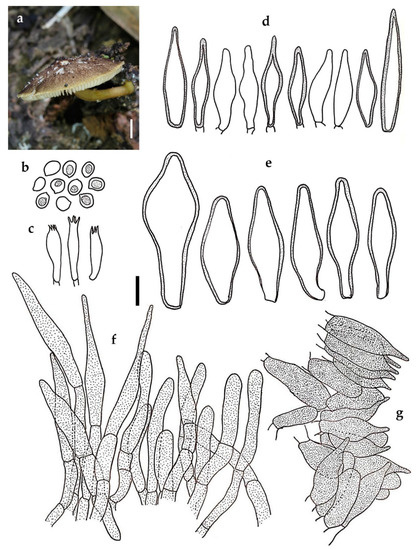
Figure 5.
Pluteus saisamorniae BKF 10213 (holotype): (a) habit; (b) basidiospores; (c) basidia; (d) cheilocystidia; (e) pleurocystidia; (f) pileipellis element; (g) caulocystidia. Scale bars: (a) = 5 mm; (b–g) = 10 μm.
MycoBank: MB838337.
Diagnosis: Characterized by relatively small basidiomata with squamules, greyish brown to light brown pileus, greyish orange stipe covered by brown minutely dotted, pinkish white lamellar with light brown edges, subglobose to broadly ellipsoid basidiospores, abundant of thin- to thick-walled cheilo- and pleurocystidia, trichohymeniderm pileipellis with fusiform to cylindrical terminal cells, and brown clustered caulocystidia.
Etymology: ‘saisamorniae’ referring to the name of Emeritus Professor Saisamorm Lumyong in honor of her 70th birthday and her contribution of over 30 years to Thai mycology.
Holotype: THAILAND, Sukhothai Province, Si Satchanalai National Park, 99°48′44″ E 17°55′12″ N elevation 277 m, on decaying wood, August, 2018, N. Wannathes, J. Kumla & N. Suwannarach, BKF 10213 (isotype: NW695).
Description: Pileus 25–30 mm diam., plano-convex with subumbonate, surface dull, dry, fibrillose, fully covered by dark brown minute squamules, margin not striate; disc greyish brown (6F3-4), margin light brown (6D7-8) or paler. Context thin, soft, pinkish white. Lamellae free, close, with 2 series of lamellulae, ventricose, broad (5 mm), pinkish white (9A2), with light brown lamellar edge. Stipe 28–30 × 2.5–3 mm cylindrical, central, fistulose, surface dull, dry, minutely dotted brownish toward the base, greyish orange (5B5) overall.
Basidiospores (5–)6–7 × 4.5–6 μm (x = 6.20 ± 0.47 × 5.05 ± 0.38, Q = 1.08–1.30, q = 1.23 ± 0.05, s = 2) subglobose to broadly ellipsoid, smooth, hyaline, inamyloid, thick-walled. Basidia 16–19 × 4–5 μm, clavate to cylindrical, thin-walled, 4-spored, unclamped. Lamellar edge sterile. Cheilocystidia common, 28–44 × 6–8 μm, fusiform to narrowly fusiform, hyaline to pale yellow, inamyloid, thin- to thick-walled (up to 1.5 μm). Pleurocystidia common, 17–54 × 10–20 μm, fusiform, lageniform to broadly lageniform, hyaline to pale yellow, inamyloid, thick-walled. Pileipellis a trichohymeniderm, with terminal cell 30–46 × 4–7 μm, narrowly fusoid, cylindrical, acute or round toward the apex, with pale yellow cytoplasmic pigment, thin-walled. Pileus trama interwoven, hyphae 5–16 μm diam., cylindrical, inflated, hyaline, inamyloid, non-gelatinous, thin-walled. Lamellar trama convergent, hyphae 3–10 μm diam., cylindrical, hyaline, inamyloid, non-gelatinous, thin-walled, with numerous refractive oleiferous hyphae interspersed. Stipitipellis a cutis with caulocystidia, hyphae 5–18 μm diam., cylindrical, pale yellow to hyaline, inamylid, non-incrustrated, non-gelatinous, thin-walled. Caulocystidia numerous, group in clusters, fusiform with acute apex, clavate to cylindrical, brown, thin-walled. Clamp connections absent in the examined tissues.
Habit, habitat, and known distribution: Solitary on decaying wood in bamboo thickets, deciduous dipterocarp-oak forests in Thailand.
Additional specimen examined: THAILAND, Phitsanulok Province, Pibulsongkram Ratjabhat University, herbal garden beside Faculty of Science and Technology, 100°12′54″ E 16°49′43″ N elevation 28 m, on decaying wood, September, 2020, NW 1567.
GenBank accession numbers: holotype (BKF 10213) ITS: MN492646, nrLSU: OM250111; additional specimen (NW 1567) ITS: OM257410, nrLSU: OM250370.
Notes: Pluteus saisamorniae is characterized by relatively small basidiomata that appear plano-convex with a broad umbo pileus, fully covered with dark brown minute squamules, light brown lamellar edges, greyish orange stipe covered with brown granules, subglobose to broadly ellipsoid basidiospores with mean dimensions of 6.2 × 5.1 μm, abundant with cheilo- and pleurocystidia displaying fusiform to lageniform shape, a trichohymeniderm pileipellis with fusiform to cylindrical terminal cells, clustered brown caulocystidia, and the absence of a clamp connection.
Pluteus granularis Peck, the species described as being from the USA, is macro-morphologically similar to P. saisamorniae since they both form a relatively small, brown granular surface of basidiomata. However, they are noticeably different according to their microscopic morphology. As such, P. granularis forms elongate basidiospores with mean dimensions of 6.5 × 4.8 μm, possesses apical mucilage or with forked appendages as pleurocystidia, and displays thin-walled broadly rounded apex cheilocystidia [76]. A phylogenetic study indicated that the P. granularis SF 20 clade was associated with P. saisamorniae with notable distances (Figure 2). The new species is also morphologically similar to P. umbrosus and P. umbrosoides in terms of the color and surface of the pileus. It has lamellar edges with a dark color and the basidiospores are similar in size and shape. However, both species differ by having larger basidiomata as well as having distinctly longer terminal cells of both the pileipellis (65–180 μm long) and caulocystidia (74–230 μm long) [9]. Pluteus podospilloides shares some morphological features such as color, shape, and surface of the pileus and stipe. It also has dark brown gill edges but differs by having smaller basidiomata with a pileus of 9–12 mm, a stipe of 10–13 × 1–1.5 mm, smaller basidoispores with mean dimensions of 4.8 × 4.4 μm and with a hymeniderm to epithelium pileipellis [64].
According to the phylogenetic analyses of ITS (Figure 2), P. saisamorniae is closely related to P. pantherinus Courtec. & M. Uchida, which has been described from Japan. However, the latter species is distinct by its larger basidiomata (pileus 50–60 mm diam. and stipe 60–80 × 5 mm), as well as by appearing fibrous and irregularly dotted with large and small white spots on the ocher background of the pileus, with white stipe and globose to subglobose basidiospores of 5.5–6.5 × 5–6 μm [77].
3.3. Taxonomic Description of New Record
Pluteus losulus Justo, Myco. Progr. 10(4): 473. 2011. (Figure 6).

Figure 6.
Pluteus losulus SDBR-CMUNK0842: (a) habit; (b) basidiospores; (c) basidia; (d) cheilocystidia; (e) pleurocystidia; (f) pileipellis element; (g) caulocystidia. Scale bars: (a) = 2 mm; (b–e, g) = 10 μm; (f) = 25 μm.
≡ Pluteus cervinus var. ealaensis Beeli, Bull. Soc. R. Bot. Belg. 61:81. 1928.
(nom.nov., non Pluteus ealaensis Beeli, Bull. Soc. R. Bot. Belg. 61(1): 80. 1928)
Description: Pileus 52–95 mm diam., broadly convex-umbonate to plano-convex with a shallow broad umbo; margin decurved, split in age; surface dull, dry, radially streaked, appressed-fibrillose, greyish brown (7E3) with a dark greyish brown (7F4) disc and streaks. Context 2–4 mm thick, soft, white. Lamellae free, close to crowded with 3 series of lamellulae, broad (up to 7 mm), initially white and becoming greyish red (7B3). Stipe 70–80 × 6–9 mm, central, cylindrical above an enlarged base, solid, pliant; surface dull, dry, glabrous above, base appressed-fibrillose, white overall when young, in age base becoming greyish brown (7E3).
Basidiospores 6–7(–8) × (5.5)6–6.5 μm (x = 6.74 ± 0.51 × 6.12 ± 0.25, Q = 1.00–1.23, q = 1.10 ± 0.06, s = 1) globose to subglobose, smooth, hyaline, inamyloid, thick-walled. Basidia 25–30 × 9–10 μm, clavate, thin-walled, 4-spored. Lamellar edge sterile. Cheilocystidia common, 19–55 × 9–16 μm, clavate to subclavate, hyaline, inamyloid. Pleurocystidia common, 54–65 × 11–22 μm, ventricose to utriform, with 3–4 apical hooks, straight to recurved hook, hyaline, inamyloid, thick-walled (up to 2 μm). Pileipellis a cutis of repent hyphae, with terminal element 37–90 × 6–11 μm, cylindrical, acute or round toward the apex, with brown cytoplasmic pigment, thin-walled. Pileus trama regular, hyphae (3–)5–17 μm diam., cylindrical, hyaline, inamyloid, non-gelatinous, thin-walled. Lamellar trama convergent, hyphae (2–)4–11 μm diam., cylindrical, hyaline, inamyloid, non-gelatinous, thin-walled. Stipitipellis a cutis with caulocystidia, hyphae 5–13 μm diam., cylindrical, hyaline or with brown cytoplasmic pigment, inamylid, non-incrustrated, non-gelatinous, thin-walled. Caulocystidia scattered, 42–73 × 10–11 μm, cylindrical, fusiform, acute or round toward the apex, with brown cytoplasmic pigment, thin-walled. Clamp connections present in all tissues.
Habit, habitat, and known distribution: Solitary or gregarious on rotting wood in bamboo thickets, deciduous dipterocarp-oak forests, secondary forest and coffee plantations in DR Congo, Principe Island, and Thailand.
Specimen examined: THAILAND, Chiang Mai Province, Chiang Mai University campus, 18°48′8″ N 98°57′25″ E, elevation 331 m, on rotting wood, August 2020, N. Suwannarach & J. Kumla, SDBR-CMUNK0842.
GenBank accession numbers: ITS: OM250105, nrLSU: OM250110.
Notes: Pluteus losulus is recognized by relatively large basidiomata that are plano-convex with a broad umbo pileus, fully covered with dark greyish brown appressed-fibrillose, white to greyish brown stipe, globose to subglobose basidiospores with mean dimensions of 6.7 × 6.1 μm, simple clavate, thin-walled cheilocystidia, apical hooked pleurocystidia, a cutis pileipellis with cylindrical to fusiform terminal cells, and the presence of a clamp connection. Accordingly, these features would indicate placement in sect. Pluteus. Our Thai material matches nicely with the African material that had designated this species as P. losulus by Justo [3], while also sharing a resemblance with the recently reported São Tomé material [10].
The ITS sequences of the Thai material were identical to those of African (GF 5273), São Tomé (SFSU:DED 8313), and Chinese (GDGM47122) materials with 100%, 100%, and 99.85%, respectively. Moreover, the nrLSU sequence of Thai material is similar to African material (GF 5273) with 100%. According to the ITS phylogram of the Pluteus sect. Pluteus (Figure 3), a sequence of the Thai material was determined to be clad with the sequences of P. losulus with strong statistical support (100% bootstrap support and 1.0 Bayesian posterior probabilities).
Pluteus septocystidiatus Ševčíková, Antonín & Borovička. Sydowia 66(2): 230. 2014. (Figure 7).
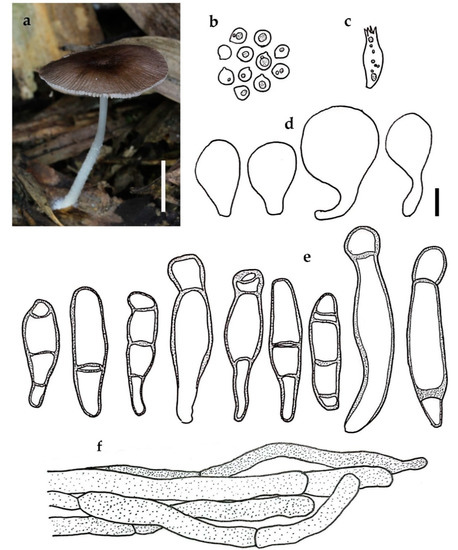
Figure 7.
Pluteus setocystidiatus BKF 10212: (a) habit; (b) basidiospores; (c) basidium; (d) cheilocystidia; (e) pleurocystidia; (f) pileipellis element. Scale bars: (a) = 10 mm; (b–f) = 10 μm.
Description: Pileus 10–25 mm diam. plano-convex with slightly depressed at center, striate at margin, otherwise smooth, glabrous, chocolate brown (6F4) at disc, brown (6E4–5) slightly paler towards margin. Context thin, soft, white. Lamellae free subdistant with 2–3 series of lamelluae, dark blond (5D4). Stipe 20–40 × 3–4 mm. cylindrical, central, hollow, finely floccose-tomentose half way from base, white.
Basidiospores 6–7 × 5–6 μm (x = 6.70 ± 0.46 × 5.60 ± 0.50, Q = 1.09–1.40, q = 1.20 ± 0.08, s = 1) subglobose to broadly ellipsoid, smooth, hyaline, inamyloid, thick-walled. Basidia 20–23 × 7–8 μm, clavate, thin-walled, 4-spored, unclamped. Lamellar edge sterile. Cheilocystidia common, 26–47 × 16–28 μm, broadly clavate to vesiculose, hyaline, inamyloid, thin-walled. Pleurocystidia common, 35–75 × 9–14 μm, fusiform, narrow fusiform, clavate, multiseptate, hyaline to pale yellow, inamyloid, relatively thick-walled. Pileipellis a cutis, hyphae 5–10 μm diam., cylindrical, with pale brown cytoplasmic pigment, inamyloid, thin-walled. Pileus trama interwoven. Lamellar trama convergent, cylindrical, hyaline, inamyloid, non-gelatinous, thin-walled. Stipitipellis a cutis, hyphae 3–12 μm diam., cylindrical, hyaline, inamylid, non-incrustrated, non-gelatinous, thin-walled. Caulocystidia absent. Clamp connections absent in the examined tissues.
Habit, habitat, and known distribution: Solitary on decaying wood in bamboo thickets, deciduous dipterocarp-oak forests in South Korea, Thailand, and USA.
Specimen examined: THAILAND, Sukhothai Province, Si Satchanalai National Park, 99°48′44″ E 17°55′12″ N elevation 277 m, on decaying wood, August, 2018, N. Wannathes, J. Kumla & N. Suwannarach, BKF 10212.
GenBank accession numbers: ITS: MN483327, nrLSU: OM250113.
Notes: Pluteus septocystidiatus is characterized by appearing slightly depressed, brown, halfway striate pileus, white cylindrical stipe, subglobose to broadly ellipsoid basidiospores with mean dimensions of 6.7 × 5.6 μm, thick-walled multiseptate pleucystidia without hooks, and a cutis pileipellis.
From the protologue [58], P. septocystidiatus has been described as being from the Republic of Korea and has been determined to be distinct from P. albostipitatus, a closely related species sharing similar morphology and ITS sequences, by the formation of obvious multiseptate and thick-walled pleurocystidia. The material from Thailand matches quite nicely with the description of the holotype. However, the Thailand collection expands knowledge about a variability in the basidiospore size of this species.
The ITS sequences of the Thai material exhibit 99.24% similarity with the sequences of the holotype. According to the ITS phylogram of Pluteus sect. Pluteus (Figure 3), a sequence of Thai collection formed a clade with P. septocystidiatus with high statistical support (100% BS and 1.0 PP). Accordingly, it is recognized as a sister group of P. albostipitatus and P. densifibrillosus as has been reported in the protologue [48].
4. Discussion
Infrageneric classification of Pluteus is mainly based on pleurocystidia and pileipellis structures. In this work, two new species, P. chandrasikuliae and P. saisamorniae, have been described from Thailand, while P. losulus and P. septocystidiatus have been recognized as a new record of Thailand. Both molecular phylogeny and morphological data prove their classification and identification. Based on its non-mentuloid cystidia and cutis pileipellis, P. chandrasikuliae belongs to sect. Celluloderma. This classification is concordant with the prevailing molecular evidence. The analyses of ITS sequences (Figure 1) have confirmed that P. chandrasikuliae is distinct from related taxa in the sect. Cellulodema. Its sequences fell in the ephebeus clade, which has accommodated taxa with a cutis or cutis-like pileipellis [30]. Likewise, P. saisamorniae belongs to the sect. Hispidoderma based on its non-mentuloid cystidia and trichoderm pileipellis, which conforms with the ITS phylogram presented in Figure 2.
The genus Pluteus is geographically widely distributed and has been well-documented throughout many continents. However, information on the species diversity of Pluteus in Thailand has been limited. In 2011, only eight species had previously been reported without voucher specimens and molecular data [65]. The results from this work are the first report of Pluteus species in Thailand, supported by molecular evidence. Accordingly, the numbers of species recorded in the country have increased. Thus, to our knowledge, the Pluteus species recorded in Thailand has been raised to 12 species. Importantly, four species from our study were confirmed by both morphological and molecular evidence, while eight Pluteus species listed by Chandrasrikul et al. [65] require further confirmation by molecular data. However, the number of Pluteus species that have been found in Thailand is very low compared to those reported in other tropical countries throughout Asia where species diversity of the genus has been well-documented. These countries include India, Sri Lanka, and Vietnam [59,60,61,62,63,64]. Future work on the species diversity of Pluteus in Thailand is needed to account for the prevailing climate change crisis, which has caused changes to fungal habitats and may have affected the diversity of the genus.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/d14030156/s1, Table S1: Details of sequences used in phylogenetic analysis.
Author Contributions
Conceptualization, N.W., N.S. and J.K.; methodology N.W., N.S. and J.K.; software, J.K.; validation, N.W., N.S. and J.K.; formal analysis, N.W. and J.K.; investigation, N.W., N.S., J.K. and S.K.; resources, N.W., N.S. and J.K.; data curation, N.W., N.S. and J.K.; writing—original draft preparation, N.W. and J.K.; writing—review and editing, N.S. and J.K.; project administration, N.W.; funding acquisition, N.W., N.S. and J.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Plant Genetic Conservation Project under the Royal initiative of Her Royal Highness Princess Maha Chakri Sirindhorn, Pibulsongkram Rajabhat University, and was partially supported by Chiang Mai University, Thailand.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The DNA sequence data obtained from this study have been deposited in GenBank under accession numbers ITS (MN483327, MN492649, MN492646, OM250105, OM257409 and OM257410) and nrLSU (OM250110, OM250111, OM250113, OM250115, OM250229 and OM250370).
Acknowledgments
The authors are grateful to the staff of Si Satchanalai National Park for their excellent field assistance and to Russell Kirk Hollis for kind help with the English correction. The authors are also grateful to Netethip Khamkiti for phylogram preparation. The first author thanks the Science Center and Faculty of Science and Technology, Pibulsongkram Rajabhat University, for providing instruments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- He, M.Q.; Zhao, R.L.; Hyde, K.D.; Begerow, D.; Kemler, M.; Yurkov, A.; McKenzie, E.H.C.; Raspé, O.; Kakishima, M.; SánchezRamírez, S.; et al. Notes, outline and divergence times of Basidiomycota. Fungal Divers. 2019, 99, 105–367. [Google Scholar]
- Singer, R. The Agaricales in Modern Taxonomy, 4th ed.; Koeltz Scientific Books: Koenigstein, Germany, 1986; pp. 454–459. [Google Scholar]
- Justo, A.; Vizzini, A.; Minnis, A.M.; Menolli, N., Jr.; Capelari, M.; Rodríguez, O.; Malysheva, E.; Contu, M.; Ghinone, S.; Hibbett, D.S. Phylogeny of Pluteaceae (Agaricales, Basidiomycota): Taxonomy and character evolution. Fungal Biol.-UK 2011, 115, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.H. Speciation in higher fungi in relation to modern generic concepts. Mycologia 1968, 60, 742–755. [Google Scholar] [CrossRef]
- Vellinga, E.C. Pluteus . In Flora Agaricina Neerlandica; Bas, C., Kuyper, T.W., Noordeloos, M.E., Vellinga, E.C., Eds.; A.A. Balkema: Rotterdam, The Netherlands, 1990; Volume 2, pp. 31–55. [Google Scholar]
- Justo, A.; Minnis, A.M.; Ghignone, S.; Menolli, N.; Capelari, M.; Rodríguez, O.; Malysheva, E.; Contu, M.; Vizzini, A. Species recognition in Pluteus and Volvopluteus (Pluteaceae, Agaricales): Morphology, geography and phylogeny. Mycol. Prog. 2011, 10, 453–479. [Google Scholar] [CrossRef]
- Justo, A.; Malysheva, E.; Bulyonkova, T.; Vellinga, E.C.; Cobian, G.; Nguyen, N.; Minnis, A.M.; Hibbett, D.S. Molecular phylogeny and phylogeography of Holarctic species of Pluteus section Pluteus (Agaricales: Pluteaceae), with description of twelve new species. Phytotaxa 2014, 180, 1–85. [Google Scholar] [CrossRef]
- Vellinga, E.C.; Schreurs, J. Pluteus Fr. in Western Europe. Persoonia 1985, 12, 337–373. [Google Scholar]
- Kaygusuz, O.; Türkekul, I.; Knudsen, H.; Çolak, Ö.F. New records of Pluteus section Hispidoderma in Turkey based on morphological characteristics and molecular data. Phytotaxa 2019, 413, 175–206. [Google Scholar] [CrossRef]
- Desjardin, D.E.; Perry, B.A. The genus Pluteus (Basidiomycota, Agaricales) from Republic of São Tomé and Príncipe, West Africa. Mycosphere 2018, 8, 1317–1391. [Google Scholar] [CrossRef]
- Menolli, N., Jr.; Justo, A.; Arrillaga, P.; Pradeep, C.K.; Minnis, A.M.; Capelari, M. Taxonomy and phylogeny of Pluteus glaucotinctus sensu lato (Agaricales, Basidiomycota), a multicontinental species complex. Phytotaxa 2014, 188, 78–90. [Google Scholar] [CrossRef]
- Malysheva, E.F.; Malysheva, V.F.; Justo, A. Observation on Pluteus (Pluteaceae) diversity in South Siberia, Russia: Morphological and molecular data. Mycol. Prog. 2016, 15, 861–882. [Google Scholar] [CrossRef]
- Pegler, D.N. A preliminary Agaric Flora of East Africa; Kew Bulletin Additional Series 6; Her Majesty’s Stationery Office: Richmond, UK, 1977; pp. 1–615. [Google Scholar]
- Ferisin1, G.; Dovana, F.; Justo, A. Pluteus bizioi (Agaricales, Pluteaceae), a new species from Italy. Phytotaxa 2019, 408, 099–108. [Google Scholar] [CrossRef]
- Kaygusuz, O.; Gezer, K.; Şeker, M. Four new records of Pluteus Fr. from interesting habitats in the Aegean region of Turkey. Bot. Lett. 2016, 163, 251–259. [Google Scholar] [CrossRef]
- Kaygusuz, O.; Knudsen, H.; Menolli, N., Jr.; Türkekul, I. Pluteus anatolicus (Pluteaceae, Agaricales): A new species of Pluteus sect. Celluloderma from Turkey based on both morphological and molecular evidence. Phytotaxa 2021, 482, 240–250. [Google Scholar] [CrossRef]
- Malysheva, E.F.; Morozova, O.V.; Zvyagina, E.A. New records of the annulate Pluteus in European and Asian Russia. Acta Mycol. 2007, 42, 153–160. [Google Scholar] [CrossRef]
- Ševčíková, H.; Malysheva, E.F.; Justo, A.; Heilmamm-Clausen, J.; Tomšovský, M. Pluteus dianae and P. punctatus resurrected, with first records from eastern and northern Europe. Mycotaxon 2020, 135, 245–274. [Google Scholar] [CrossRef]
- Vizzini, A.; Ercole, E. A new annulate Pluteus variety from Italy. Mycologia 2011, 103, 904–911. [Google Scholar] [CrossRef][Green Version]
- Radu, M.-I.; Şesan, T.-E. Pluteus aurantiorugosus (Trog) Sacc. and Coprinus patouillardii Quél.—New records of macrofungi for Romania. Acta Mycol. 2013, 48, 189–196. [Google Scholar] [CrossRef]
- Orton, P.D. Bristish Fungus Flora Agarics and Boleti 4—Pluteaceae: Pluteus & Volvariella; Royal Botanic Garden: Edinburgh, UK, 1986; pp. 1–99. [Google Scholar]
- Orton, P.D. New check list of British Agarics and Boleti, part III (Note on genera and species in the list). Trans. Brit. Mycol. Soc. 1960, 43, 159–439. [Google Scholar] [CrossRef]
- Citérin, M.; Eyssartier, G. Clé analytique du genre Pluteus Fr. Doc. Mycol. 1998, 28, 47–67. [Google Scholar]
- Kaygusuz, O.; Justo, A.; Knudsen, H.; Ševčíková, H.; Türkekul, I. Pluteus lauracearum (Agaricales, Basidiomycota), a new species of Pluteus sect. Hispidoderma from thermophilic Laurus forests. Phytotaxa 2021, 523, 126–140. [Google Scholar] [CrossRef]
- Kaygusuz, O.; Turkekul, I.; Knudsen, H.; Menolli, N., Jr. Volvopluteus and Pluteus section Pluteus (Agaricales: Pluteaceae) in Turkey based on morphological and molecular data. Turk. J. Bot. 2021, 45, 224–242. [Google Scholar] [CrossRef]
- Ševčíková, H.; Bororvička, J. Pluteus rugosidiscus (Basidiomycota, Pluteaceae), first record of this North American species in Europe. Nova Hedwigia 2019, 108, 227–241. [Google Scholar] [CrossRef]
- Bonnard, J. Pluteus lidipocystis sp. nov. Mycol. Helv. 1986, 2, 35–42. [Google Scholar]
- Bonnard, J. Pluteus brunneoradiatus sp. nov. Mycol. Helv. 1987, 2, 141–154. [Google Scholar]
- Bonnard, J. Les cystides de la section Pluteus (Agaricales). Mycol. Helv. 1988, 3, 53–72. [Google Scholar]
- Bonnard, J. Pluteus primus sp. nov. (Agaricales, Basidiomycetes). Mycol. Helv. 1991, 4, 169–178. [Google Scholar]
- Bonnard, J. Clé provisoire des Plutées européens à boucles. Mycol. Helv. 1993, 6, 203–205. [Google Scholar]
- Bonnard, J. Pluteus pellitus désignation d’un néotype (Section Pluteus, Agaricales, Basidiomycetes). Mycol. Helv. 1995, 7, 97–103. [Google Scholar]
- Bonnard, J. Pluteus albineus sp. nov. (Agaricales, Basidiomycetes). Mycol. Helv. 2001, 11, 131–136. [Google Scholar]
- Justo, A.; Castro, M.L. Observations in Pluteus section Pluteus in Spain: Two new records for Europe. Mycotaxon 2007, 102, 209–220. [Google Scholar]
- Justo, A.; Castro, M.L. An annotated checklist of Pluteus in the Iberian Peninsula and Balearic Islands. Mycotaxon 2007, 102, 231–234. [Google Scholar]
- Campi, M.G.; Maubet, Y.; Cristaldo, E.; Grassi, E.; Menolli, N., Jr. Pluteus Fr. (Pluteaceae, Agaricales) in Paraguay: Morphological studies and new records. Check List 2019, 15, 313–322. [Google Scholar] [CrossRef]
- Justo, A.; Battistin, E.; Angelini, C. Two new species of Pluteus section Celluloderma from the Dominican Republic. Mycotaxon 2012, 120, 11–21. [Google Scholar] [CrossRef]
- Homola, R.L. Section Celluloderma of the Genus Pluteus in North America. Mycologia 1972, 64, 1211–1257. [Google Scholar] [CrossRef]
- Menolli, N., Jr.; Asai, T.; Capelari, M. Records and new species of Pluteus from Brazil based on morphological and moleculardata. Mycology 2010, 1, 130–153. [Google Scholar] [CrossRef]
- Menolli, N., Jr.; Justo, A.; Capelari, M. Phylogeny of Pluteus section Celluloderma including eight new species from Brazil. Mycologia 2015, 107, 1205–1220. [Google Scholar] [CrossRef]
- Menolli, N., Jr.; Justo, A.; Capelari, M. Pluteus section Hispidoderma in Brazil with new records based on morphological and molecular Data. Cryptogam. Mycol. 2015, 36, 331–354. [Google Scholar] [CrossRef]
- Minnis, A.M.; Sundberg, W.J. Pluteus section Celluloderma in the U.S.A. N. Am. Fungi 2010, 5, 1–107. [Google Scholar] [CrossRef][Green Version]
- Murrill, W.A. The Agaricaceae of Tropical North America—IV. Mycologia 1911, 3, 271–282. [Google Scholar]
- Minnis, A.M.; Sundberg, W.J.; Methven, A.S.; Sipes, S.D.; Nickrent, D.L. Annulate Pluteus species: A study of the genus Chamaeota in the United States. Mycotaxon 2006, 96, 31–39. [Google Scholar]
- Brandrud, T.E.; Krisai-Greilhuber, I.; Kunca, V. Pluteus fenzlii . In The IUCN Red List of Threatened Species 2019; GBIF: Copenhagen, Denmark, 2019; pp. 1–8. [Google Scholar]
- Leonard, P.L. Pluteus bressollensis . In The IUCN Red List of Threatened Species 2019; GBIF: Copenhagen, Denmark, 2019; pp. 1–6. [Google Scholar]
- Hosen, M.I.; Liang, X.; Xu, J.; Li, T.H. Pluteus squarrosus sp. nov. (Pluteus sect. Celluloderma, Pluteaceae) from northeast China. Nord. J. Bot. 2019, 37, e02427. [Google Scholar] [CrossRef]
- Xu, J.; Li, T.H.; Justo, A.; Ge, Z.W. Two new species of Pluteus (Agaricales, Pluteaceae) from China. Phtyotaxa 2015, 233, 61–68. [Google Scholar] [CrossRef][Green Version]
- Hosaka1, K.; Kobayashi, T.; Castellano, M.A.; Orihara, T. The status of voucher specimens of mushroom species thought to be extinct from Japan. Bull. Natl. Mus. Nat. Sci. Ser. B 2018, 44, 53–66. [Google Scholar]
- Lezzi, T.; Vizzin, A.; Ercole, E.; Magliozzi, V.; Justo, A. Phylogenetic and morphological comparison of Pluteus variabilicolor and P. castri (Basidiomycota, Agaricales). IMA Fungus 2014, 5, 415–423. [Google Scholar] [CrossRef]
- Takahashi, H. Notes on new Agaricales of Japan 2. Mycoscience 2001, 42, 347–353. [Google Scholar] [CrossRef]
- Takahashi, H. Pluteus romellii (Agaricales, Basidiomycetes), new to Japan, found in Odawara (in Japanese with English summary). Nat Hist. Rep. Kanagawa 2001, 22, 21–23. [Google Scholar]
- Takehashi, S.; Kasuya, T. First record of Pluteus chrysophaeus and reexamination of Pluteus leoninus from Japan. Mycoscience 2007, 48, 321–325. [Google Scholar] [CrossRef]
- Lee, J.N.; Lee, H.K.; Min, K.H.; Park, W.H.; Kim, Y.S. Studies on genus Pluteus of Korea. Korean Mycol. 1992, 20, 296–301. [Google Scholar]
- Cho, S.; Eun, K.Y.; Jo, J.; Won, H.S.; Oh, S.H.; Kim, C.S. Macrofungal diversity of urbanized areas in southern part of Korea. J. Asia Pac. Biodivers. 2020, 13, 189–197. [Google Scholar] [CrossRef]
- Park, M.S.; Cho, H.J.; Kim, N.K.; Park, J.Y.; Lee, H.; Park, K.H.; Kim, M.J.; Kim, J.J.; Kim, C.; Lim, Y.W. Ten new recorded species of macrofungi on Ulleung Island, Korea. Mycobiology 2017, 45, 286–296. [Google Scholar] [CrossRef]
- Lee, T.S. The full list of recorded mushrooms in Korea. Kor. J. Mycol. 1990, 18, 233–259. [Google Scholar]
- Ševčíková, H.; Antonín, V.; Borovička, J. Pluteus septocystidiatus, a new species with unique pleurocystidia. Sydowia 2014, 66, 229–239. [Google Scholar]
- Pegler, D.N. Agaric Flora of Sri Lanka; Kew Bulletin Additional Series 12; Her Majesty’s Stationery Office: Richmond, UK, 1986; pp. 1–519. [Google Scholar]
- Berkeley, M.J.; Broome, C.E. The fungi of Ceylon. J. Linn. Soc., Bot. 1871, 11, 494–567. [Google Scholar] [CrossRef]
- Petch, T. Ceylon pink-spored agarics. Roy. Bot. Gard. (Peradeniya) 1924, 9, 201–216. [Google Scholar]
- Pradeep, C.K.; Vrinda, K.B. New and noteworthy species of Pluteus (Pluteaceae, Agaricales) from Kerala State, India. Persoonia 2006, 19, 95–99. [Google Scholar]
- Pradeep, C.K.; Justo, A.; Vrinda, K.B.; Shibu, V.P. Two new species of Pluteus (Pluteaceae, Agaricales) from India and additional observations on Pluteus chrysaegis. Mycol. Prog. 2012, 11, 869–878. [Google Scholar] [CrossRef]
- Malysheva, E.; Malysheva, V.; Alexandrova, A.; Morozova, O. Observations on Pluteaceae in Vietnam. 2. One new record and ten new species of Pluteus. Phytotaxa 2020, 461, 79–107. [Google Scholar] [CrossRef]
- Chandrasrikul, A.; Suwanarit, P.; Sangwanit, U.; Lunyong, S.; Payapanon, A.; Sanoamuang, N.; Pukahuta, C.; Petcharat, V.; Sardsud, U.; Duengkae, K.; et al. Checklist of Mushrooms (Basidiomycetes) in Thailand; Office of Natural Resources and Environmental Policy and Planning: Bangkok, Thailand, 2011; pp. 112–113. [Google Scholar]
- Kornerup, A.; Wanscher, J.H. Methuen Handbook of Colour, 3rd ed.; Eyre Methuen: London, UK, 1978; pp. 1–252. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Stamatakis, A. Raxml-vi-hpc: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Hoehna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree Tree Figure Drawing Tool Version 131, Institute of Evolutionary 623 Biology, University of Edinburgh. Available online: http://treebioedacuk/software/figtree/ (accessed on 25 December 2021).
- Menolli, N., Jr.; Meijer, A.A.R.; Capelari, M. The genus Pluteus (Pluteaceae, Agaricales) from the state of Paraná, Brazil. Nova Hedwigia 2015, 100, 101–157. [Google Scholar] [CrossRef]
- Takahashi, H.; Kasuya, T. Type study of Pluteus atrofuscens (Agaricales, Pluteaeceae). Mycoscience 2010, 51, 81–83. [Google Scholar] [CrossRef]
- Singer, R. Contributions towards a monograph of the genus Pluteus. Trans. Brit. Mycol. Soc. 1956, 39, 145–232. [Google Scholar] [CrossRef]
- Imazeki, R.; Otani, Y.; Hongo, T. Fungi of Japan, 2nd ed.; Yama-Kei Publishers: Tokyo, Japan, 1989; pp. 179–180. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).