Phylotranscriptomic and Evolutionary Analyses of Oedogoniales (Chlorophyceae, Chlorophyta)
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultures
2.2. Library Preparation and Sequencing
2.3. Quality Control, De Novo Assembly, and Sequence Annotation
2.4. Orthologous Group Identification and Phylotranscriptomic Analysis
2.5. Evolutionary Analyses Based on Phylogenetic Analysis by Maximum Likelihood
2.6. Gene Ontology Enrichment and Kyoto Encyclopedia of Genes and Genomes Pathway Analyses of Rapidly Evolving Genes
3. Results
3.1. De Novo Transcriptome Assembly and Ortholog Detection
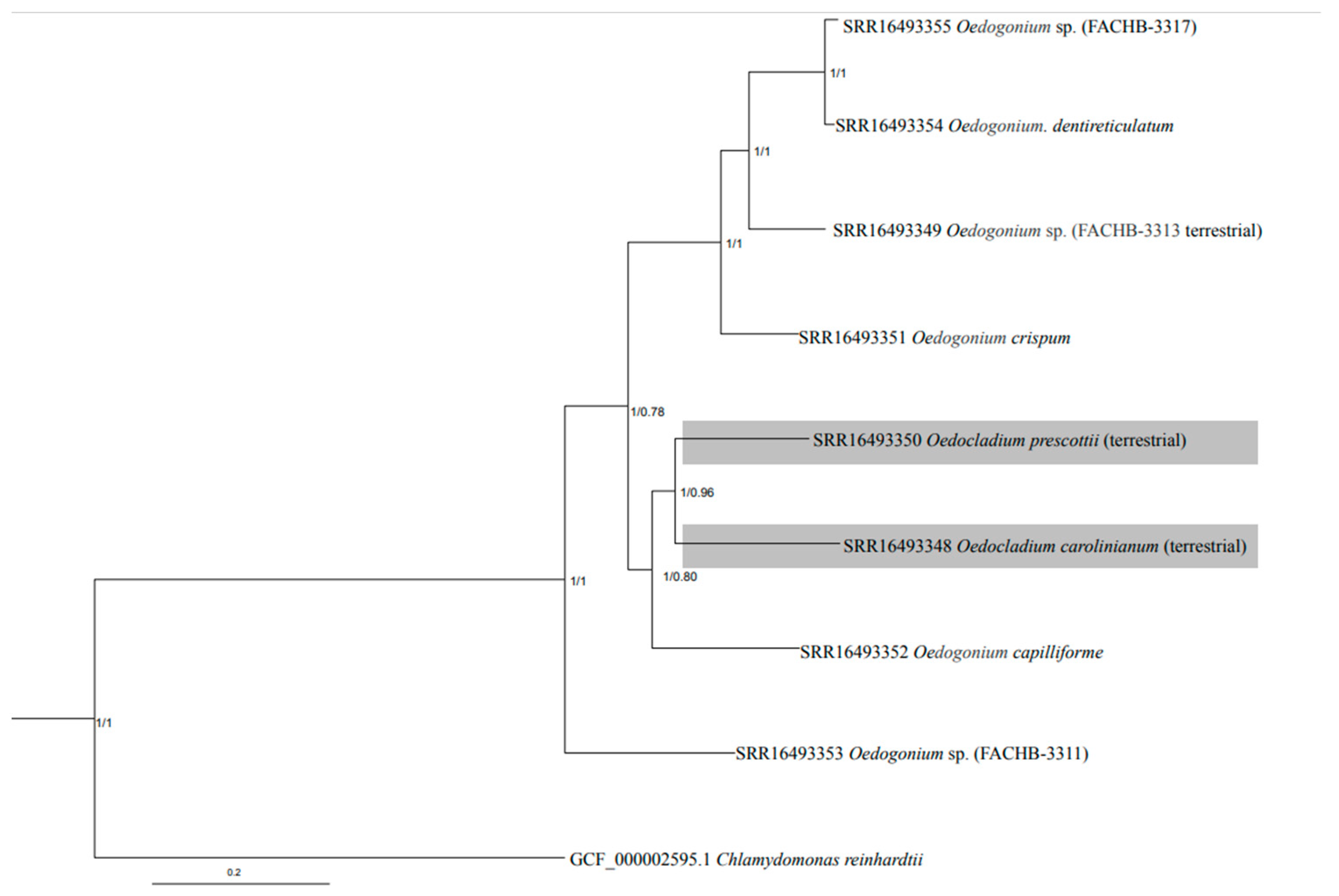
3.2. Phylotranscriptomic Analyses
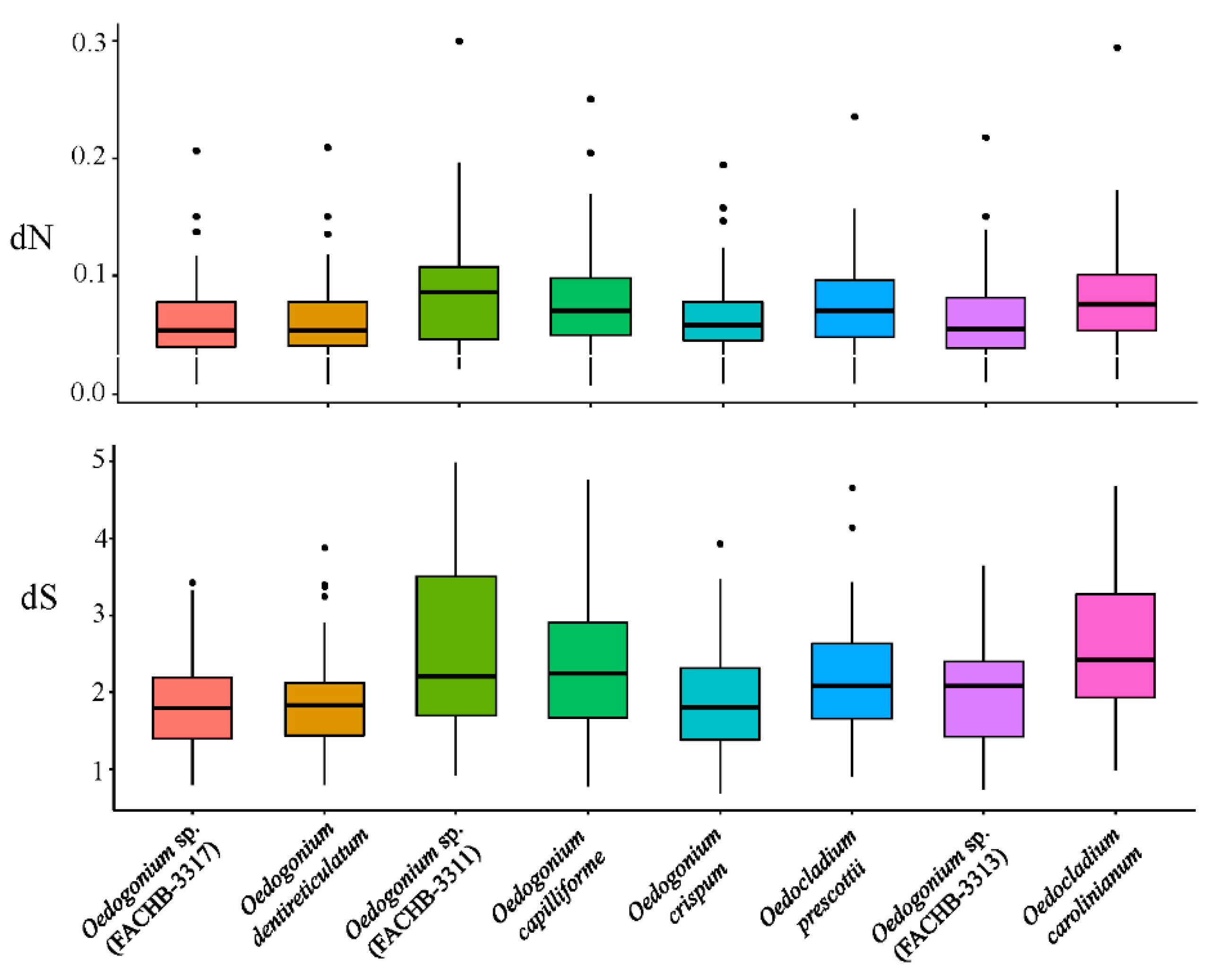
3.3. Evolutionary Analyses Based on PAML, GO Enrichment, and KEGG Pathway Analyses of Rapidly Evolving Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Agardh, C.A. Synopsis Algarum Scandinaviae: Adjecta Dispositione Universali Algarum; Kessinger Publishing, LLC: Whitefish, MT, USA, 1817. [Google Scholar]
- Bary, A. Ueber die Algengattungen Oedogonium und Bulbochaete. Naturf. Ges. Frankf. 1854, 1, 29–105. [Google Scholar]
- Stahl, E. Oedocladium protonema, eine neue Oedogoniaceen-Gattung. Wiss Bot. 1891, 23, 339–348. [Google Scholar]
- Hirn, K.E. Monographie und Iconographie der Oedogoniaceen. Acta Soc. Scienti. Fennicae. 1900, 27, 1–395. [Google Scholar]
- Tiffany, L.H. North American Flora: Oedogoniales; New York Botanical Garden: New York, NY, USA, 1937. [Google Scholar]
- Gemeinhardt, K. Oedogoniales. In Dr. L. Rabenhorst’s Kryptogamen-Flora von Deutschland, Oesterreich und der Schweiz; Rabenhorst, L., Ed.; Akademische Verlagsgesellschaft: Leipzig, Germany, 1939. [Google Scholar]
- Islam, A.K.M.N.; Sarma, P. Two new species of terrestrial Oedogonium from East Pakistan. Trans. Am. Micros. Soc. 1963, 82, 74–77. [Google Scholar] [CrossRef]
- Gauthier-Lièvre, L. Oedogoniacées Africaines; Cramer: Stuttgart, Gertmany, 1964; pp. 1–104. [Google Scholar]
- Jao, C.C. Monographia Oedogoniales Sinicae; Beijing Science Press: Beijing, China, 1979. [Google Scholar]
- Mrozińska, T. Oedogoniophyceae: Oedogoniales. In Süßwasserflora von Mitteleuropa 14; Chlorophyta VI; Gustav Fischer Verlag: Stuttgart, Germany, 1985. [Google Scholar]
- van Den Hoek, C.; Mann, D.G.; Jahns, U.M. Algae: An Introduction to Phycology; Cambridge University Press: Cambridge, UK, 1995. [Google Scholar]
- Graham, L.E.; Wilcox, L.W. Algae; Prentice Hall: Upper Saddle River, NJ, USA, 2000. [Google Scholar]
- Liu, G.X.; Hu, Z.Y. Predominant occurrence of apical cell divisions in Oedogoniam pakistanense and its phylogenetic significance. Phycologia 2004, 43, 669–671. [Google Scholar] [CrossRef]
- Wittrock, V.B. Prodromus Monographiae Oedogoniearum. Nova Acta Regiae Soc. Sci. Ups. 1874, 3, 1–64. [Google Scholar]
- Booton, G.C.; Floyd, G.L.; Fuerst, P.A. Origins and affinities of the filamentous green algal orders Chaetophorales and Oedogoniales based on 18S rRNA gene sequences. J. Phycol. 1998, 34, 312–318. [Google Scholar] [CrossRef]
- Buchheim, M.A.; Michalopulos, E.A.; Buchheim, J.A. Phylogeny of the Chlorophyceae with special reference to the Sphaeropleales: A study of 18S and 26S rDNA data. J. Phycol. 2001, 37, 819–835. [Google Scholar] [CrossRef]
- Krienitz, L.; Hegewald, E.; Hepperle, D.; Wolf, M. The systematics of coccoid green algae. 18S rRNA gene sequence data versus morphology. Biologia 2003, 58, 437–446. [Google Scholar]
- Shoup, S.; Lewis, L.A. Polyphyletic origin of parallel basal bodies in swimming cells of Chlorophycean green algae (Chlorophyta). J. Phycol. 2003, 39, 789–796. [Google Scholar] [CrossRef]
- Alberghina, J.S.; Vigna, M.S.; Confalonieri, V.A. Phylogenetic position of the Oedogoniales within the green algae (Chlorophyta) and the evolution of the absolute orientation of the flagellar apparatus. Plant. Syst. Evol. 2006, 261, 151–163. [Google Scholar] [CrossRef]
- Mei, H.; Luo, W.; Liu, G.X.; Hu, Z.Y. Phylogeny of Oedogoniales (Chlorophyceae, Chlorophyta) inferred from 18S rDNA sequences with emphasis on the relationships in the genus Oedogonium based on ITS-2 sequences. Plant. Syst. Evol. 2007, 265, 179–191. [Google Scholar] [CrossRef]
- Xiong, Q.; Hu, Y.; Liu, B.; Zhu, H.; Liu, G.; Hu, Z. Chloroplast genomes and phylogenetic analysis of two species of Oedocladium (Oedogoniales, Chlorophyta). Eur. J. Phycol. 2021, 56, 403–415. [Google Scholar] [CrossRef]
- Xiong, Q.; Hu, Y.; Lv, W.; Wang, Q.; Liu, G.; Hu, Z. Chloroplast genomes of five Oedogonium species: Genome structure, phylogenetic analysis and adaptive evolution. BMC Genom. 2021, 22, 707. [Google Scholar] [CrossRef]
- Maddison, W.P. Gene trees in species trees. Syst. Biol. 1997, 46, 523–536. [Google Scholar] [CrossRef]
- Huynen, M.A.; Bork, P. Measuring genome evolution. Proc. Natl. Acad. Sci. USA 1998, 95, 5849–5856. [Google Scholar] [CrossRef]
- Doolittle, W.F. Phylogenetic classification and the universal tree. Science 1999, 284, 2124–2128. [Google Scholar] [CrossRef]
- Degnan, J.H.; Rosenberg, N.A. Discordance of species trees with their most likely gene trees. PLoS Genet. 2006, 2, e68. [Google Scholar] [CrossRef]
- Zou, J.; Yue, W.; Li, L.; Wang, X.; Lu, J.; Duan, B.; Liu, J. DNA barcoding of recently diversified tree species: A case study on spruces based on 20 DNA fragments from three different genomes. Trees 2016, 30, 959–969. [Google Scholar] [CrossRef]
- Gernandt, D.S.; Dugua, X.A.; Vázquez-Lobo, A.; Willyard, A.; Letelier, A.M.; Pérez de la Rosa, J.A.; Pinero, D.; Liston, A. Multi-locus phylogenetics, lineage sorting, and reticulation in Pinus subsection Australes. Am. J. Bot. 2018, 105, 711–725. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Rogozin, I.B.; Grishin, N.V.; Koonin, E.V. Genome trees and the tree of life. Trends Genet. 2002, 18, 472–479. [Google Scholar] [CrossRef]
- Mirarab, S.; Reaz, R.; Bayzid, M.S.; Zimmermann, T.; Swenson, M.S.; Warnow, T. ASTRAL: Genome-scale coalescent-based species tree estimation. Bioinformatics 2014, 30, i541–i548. [Google Scholar] [CrossRef] [PubMed]
- Davidson, R.; Vachaspati, P.; Mirarab, S.; Warnow, T. Phylogenomic species tree estimation in the presence of incomplete lineage sorting and horizontal gene transfer. BMC Genom. 2015, 16, S1. [Google Scholar] [CrossRef] [PubMed]
- Mirarab, S. Species tree estimation using ASTRAL: Practical considerations. arXiv 2019, arXiv:1904.03826. [Google Scholar]
- Deng, H.; Zhang, G.Q.; Lin, M.; Wang, Y.; Liu, Z.J. Mining from transcriptomes: 315 single-copy orthologous genes concatenated for the phylogenetic analyses of Orchidaceae. Ecol. Evol. 2015, 5, 3800–3807. [Google Scholar] [CrossRef]
- Wen, J.; Xiong, Z.; Nie, Z.L.; Mao, L.; Zhu, Y.; Kan, X.Z.; Fang, X.D. Transcriptome sequences resolve deep relationships of the grape family. PLoS ONE 2013, 8, e74394. [Google Scholar]
- Cavalier-Smith, T.; Chao, E.E.; Lewis, R. 187-gene phylogeny of protozoan phylum Amoebozoa reveals a new class (Cutosea) of deep-branching, ultrastructurally unique, enveloped marine Lobosa and clarifies amoeba evolution. Mol. Phylogenet. Evol. 2016, 99, 275–296. [Google Scholar] [CrossRef]
- Cavalier-Smith, T.; Chao, E.E.; Snell, E.A.; Berney, C.; Fiore-Donno, A.M.; Lewis, R. Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts (animals, fungi, choanozoans) and Amoebozoa. Mol. Phylogenet. Evol. 2014, 81, 71–85. [Google Scholar] [CrossRef]
- Saunders, G.W.; Jackson, C.; Salomaki, E.D. Phylogenetic analyses of transcriptome data resolve familial assignments for genera of the red-algal Acrochaetiales-Palmariales complex (Nemaliophycidae). Mol. Phylogenet. Evol. 2018, 119, 151–159. [Google Scholar] [CrossRef]
- Yin, Y.; Johns, M.A.; Cao, H.; Rupani, M. A survey of plant and algal genomes and transcriptomes reveals new insights into the evolution and function of the cellulose synthase superfamily. BMC Genom. 2014, 15, 260. [Google Scholar] [CrossRef]
- Hu, Y.; Xing, W.; Song, H.; Hu, Z.; Liu, G. Comparison of colonial volvocine algae based on phylotranscriptomic analysis of gene family evolution and natural selection. Eur. J. Phycol. 2019, 54, 1–13. [Google Scholar] [CrossRef]
- Dunn, C.W.; Giribet, G.; Edgecombe, G.D.; Hejnol, A. Animal phylogeny and its evolutionary implications. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 371–395. [Google Scholar] [CrossRef]
- Giribet, G. New animal phylogeny: Future challenges for animal phylogeny in the age of phylogenomics. Org. Divers. Evol. 2015, 16, 419–426. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Logacheva, M.D.; Schelkunov, M.I.; Penin, A.A. Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biol. Evol. 2011, 3, 1296–1303. [Google Scholar] [CrossRef]
- McNeal, J.R.; Kuehl, J.V.; Boore, J.L.; De Pamphilis, C.W. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 2007, 7, 57. [Google Scholar] [CrossRef]
- Yang, Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 1998, 15, 568–573. [Google Scholar] [CrossRef]
- Davies, T.J.; Savolainen, V.; Chase, M.W.; Moat, J.; Barraclough, T.G. Environmental energy and evolutionary rates in flowering plants. Proc. R. Soc. B 2004, 271, 2195–2200. [Google Scholar] [CrossRef]
- Clarke, A.; Gaston, K.J. Climate, energy and diversity. Proc. Biol. Sci. 2006, 273, 2257–2266. [Google Scholar] [CrossRef]
- Marcelino, V.R.; Ma, C.; Jackson, C.J.; Larkum, A.; Verbruggen, H. Evolutionary dynamics of chloroplast genomes in low light: A case study of the endolithic green alga Ostreobium quekettii. Genome Biol. Evol. 2016, 8, 2939–2951. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 972–1973. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Lam-Tung, N.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, 232–235. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1196. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Zhang, C.; Rabiee, M.; Sayyari, E.; Mirarab, S. ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 2018, 19, 15–30. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger sets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Philip, J.; Binns, D.; Chang, H.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2016, 34, 2115–2122. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Pollock, D.D.; Zwickl, D.J.; McGuire, J.A.; Hillis, D.M. Increased tax on sampling is advantageous for phylogenetic inference. Syst. Biol. 2002, 51, 664–671. [Google Scholar] [CrossRef]
- Buchheim, M.A.; Sutherland, D.M.; Schleicher, T.; Förster, F.; Wolf, M. Phylogeny of Oedogoniales, Chaetophorales and Chaetopeltidales (Chlorophyceae): Inferences from sequence-structure analysis of ITS2. Ann. Bot. 2012, 109, 109–116. [Google Scholar] [CrossRef]
- Caisová, L.; Marin, B.; Melkonian, M. A consensus secondary structure of ITS2 in the Chlorophyta identified by phylogenetic reconstruction. Protist 2013, 164, 482–496. [Google Scholar] [CrossRef]
- Liu, B.; Hu, Y.; Hu, Z.; Liu, G.; Zhu, H. Taxonomic scheme of the order Chaetophorales (Chlorophyceae, Chlorophyta) based on chloroplast genomes. BMC Genom. 2020, 21, 442. [Google Scholar] [CrossRef]
- Brouard, J.S.; Turmel, M.; Otis, C.; Lemieux, C. Proliferation of group II introns in the chloroplast genome of the green alga Oedocladium carolinianum (Chlorophyceae). PeerJ 2016, 4, e2627. [Google Scholar] [CrossRef][Green Version]
- Liu, B.; Zhu, H.; Dong, X.; Yan, Q.; Liu, G.; Hu, Z. Reassessment of Suitable Markers for Taxonomy of Chaetophorales (Chlorophyceae, Chlorophyta) based on Chloroplast Genomes. J. Eukaryot. Microbiol. 2021, 68, e12858. [Google Scholar] [CrossRef]
- Smith, D.R. Mutation rates in plastid genomes. They are lower than you might think. Genome Biol. Evol. 2015, 7, 1227–1234. [Google Scholar] [CrossRef]
- Guisinger, M.M.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc. Natl. Acad. Sci. USA 2008, 105, 18424–18429. [Google Scholar] [CrossRef] [PubMed]
- Kato-Minoura, T.; Karino, K.; Akimoto, N.; Yoshiga, N.; Ehara, M.; Aoki, S. Phylogenetic analysis of NAP, an unconventional actin of the Volvocales. Plant Syst. Evol. 2015, 301, 1725–1733. [Google Scholar] [CrossRef]
- Vigeland, M.D.; Spannagl, M.; Asp, T.; Paina, C.; Rudi, H.; Rognli, O.-A.; Fjellheim, S.; Sandve, S.R. Evidence for adaptive evolution of low-temperature stress response genes in a Pooideae grass ancestor. New Phytol. 2013, 199, 1060–1068. [Google Scholar] [CrossRef] [PubMed]
- McGowen, M.R.; Grossman, L.I.; Wildman, D.E. Dolphin genome provides evidence for adaptive evolution of nervous system genes and a molecular rate slowdown. Proc. R. Soc. B 2012, 279, 3643–3651. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, C.; Gong, X. Degeneration of photosynthetic capacity in mixotrophic plants, Chimaphila japonica and Pyrola decorata (Ericaceae). Plant Divers. 2017, 39, 80–88. [Google Scholar] [CrossRef]
- Laurin, M. How Vertebrates Left Water; University of California Press: Berkeley, CA, USA, 2010. [Google Scholar]

| Species | Number of Raw Reads | Number of Clean Reads | Sequence Assembled by Tri-ity | Complete BUSCOs | Number of Coding Sequence Predicted by TransDecoder | |||
|---|---|---|---|---|---|---|---|---|
| Number of Contigs | Average Contig Length | N50 Length | Orthogroups | |||||
| Oedogonium sp. (FACHB-3313, terrestrial) | 66,116,070 | 66,020,778 | 82,567 | 1084.35 | 2165 | 28,539 | 92.7% | 50,244 |
| Oedocladium carolinianum (terrestrial) | 49,070,110 | 49,038,062 | 112,614 | 959.71 | 1897 | 36,030 | 92.3% | 63,044 |
| Oedogonium capilliforme | 67,815,700 | 67,680,428 | 118,267 | 842.10 | 1439 | 37,039 | 93.0% | 59,861 |
| Oedocladium prescottii (terrestrial) | 64,034,464 | 64,005,336 | 139,202 | 1204.96 | 2693 | 66,000 | 91.9% | 118,126 |
| Oedogonium sp. (FACHB-3311) | 60,835,586 | 60,730,652 | 94,883 | 849.91 | 1305 | 34,131 | 89.3% | 64,417 |
| Oedogonium dentireticulatum | 52,801,814 | 52,695,338 | 100,195 | 838.23 | 1371 | 30,859 | 93.7% | 48,137 |
| Oedogonium crispum | 66,019,080 | 65,947,940 | 143,592 | 753.29 | 1085 | 33,704 | 92.1% | 51,654 |
| Oedogonium sp. (FACHB-3317) | 69,832,706 | 69,639,876 | 69,335 | 785.33 | 1129 | 21,432 | 90.2% | 29,750 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, Q.; Hu, Y.; Dong, X.; Chen, Y.; Liu, G.; Hu, Z. Phylotranscriptomic and Evolutionary Analyses of Oedogoniales (Chlorophyceae, Chlorophyta). Diversity 2022, 14, 157. https://doi.org/10.3390/d14030157
Xiong Q, Hu Y, Dong X, Chen Y, Liu G, Hu Z. Phylotranscriptomic and Evolutionary Analyses of Oedogoniales (Chlorophyceae, Chlorophyta). Diversity. 2022; 14(3):157. https://doi.org/10.3390/d14030157
Chicago/Turabian StyleXiong, Qian, Yuxin Hu, Xiaoqi Dong, Yangliang Chen, Guoxiang Liu, and Zhengyu Hu. 2022. "Phylotranscriptomic and Evolutionary Analyses of Oedogoniales (Chlorophyceae, Chlorophyta)" Diversity 14, no. 3: 157. https://doi.org/10.3390/d14030157
APA StyleXiong, Q., Hu, Y., Dong, X., Chen, Y., Liu, G., & Hu, Z. (2022). Phylotranscriptomic and Evolutionary Analyses of Oedogoniales (Chlorophyceae, Chlorophyta). Diversity, 14(3), 157. https://doi.org/10.3390/d14030157






