Abstract
Chironomids can inhabit a large variety of water bodies. They contribute to the process of biological purification of water bodies, and they are a high-quality food for commercial fish. Any comprehensive study of biodiversity in water bodies begins with the investigation of chironomids, which are typically variable and difficult to identify through morphology. Similar species are called sibling-species. For precise identification, we used a comprehensive approach, including morphology, cytogenetics, and molecular genetics. In one sample from Kurchatskoe Lake (Tyumen reg.), with mineralized water of 7‰, we found three Chironomus species. Karyological analysis revealed seven banding sequences in Chironomus agilis2, eight in Ch. balatonicus, and seven in Camptochironomus tentans. The combination of balD1.2 was found in all Ch. balatonicus larvae. All the found banding sequences are typical for the studied region, and have previously been recorded in European and Altai populations. All the estimated genetic distances of COI gene sequences in the studied larvae of each species are much lower than the commonly accepted threshold of 3% in species of the genus Chironomus.
1. Introduction
The study of lakes with increased mineralization arouses increasing interest, because there we can find hydrobionts that are not characteristic of that region, or their unusual communities. The research area is located in the south of Western Siberia, in the Tobolo-Ishim forest-steppe [1]. It is noted that most of the lakes are non-flowing, shallow, and growing over with vegetation; the shores are often swampy. Intensive evaporation of water and low rainfall contributes to increased mineralization [2]. Lakes with a mineralization of 1–10‰ make up about 28% of all Ishim Plain lakes [3]; at the moment of sampling, the water mineralization of Kurchatskoe Lake was 7‰.
Previously, Chironomus had not been studied in Kurchatskoe or nearby lakes. In different lakes of the Tobolo-Ishim forest-steppe were noted Chironomus plumosus (Linnaeus, 1758), Ch. cingulatus (Meigen, 1830), Ch. dorsalis (Meigen, 1818), Ch. anthracinus (Zetterstedt, 1860), and Ch. heterodentatus (Konstantinov, 1956) [2,4]. In most previous studies, the species were specified as Ch. gr. plumosus or Chironomus sp. [5,6]. Only Camptochironomus tentans (Fabricius, 1805) was previously noted in the Tyumen region [4,5]. In all listed investigations, the species identification of Chironomus was conducted by morphology. As is known, the Chironomus genus is characterized by sibling-species which are difficult to identify by morphology alone [7,8]. Cytogenetics helps to resolve this problem, as many new species of Chironomus have been recognized among cryptic species using chromosomal analysis [7,9]. Most studies recommend using multiple approaches including morphology, cytogenetic, and molecular-genetic analysis [10,11,12]. Following these recommendations, we found, in Kurchatskoe Lake (in one sample), Ch. agilis2 (Kiknadze, Siirin & Filippova, 1991), Ch. balatonicus (Dévai, Wülker & Scholl, 1983), and C. tentans, which are common in the Palearctic [8,13,14].
Ch. agilis2 and Ch. balatonicus belong to the sibling-species of the Ch. plumosus group: Ch. agilis (Shobanov & Djomin, 1988), Ch. agilis2, Ch. balatonicus, Ch. bonus (Shilova & Dzhvarsheishvili, 1974), Ch. borokensis (Kerkis, Filippova, Shobanov, Gunderina et Kiknadze, 1988), Ch. entis (Shobanov, 1989), Ch. sp. J (Kiknadze, Shilova & Kerkis, 1991), Ch. muratensis, Ch. nudiventris (Ryser, Scholl & Wülker, 1983), Ch. plumosus, and Ch. usenicus (Loginova & Belyanina, 1994) [13]. The first description of Ch. agilis2 was performed by karyotype and characterized by a low level of chromosomal polymorphism (0.04 inv. per individual) [15]. By morphological and karyotype characteristics, the species of Ch. agilis and Ch. agilis2 are similar, the main difference being the presence in Ch. agilis2 of a heterochromatinized centromeric disc, and a new banding sequence in arm C [15]. The species with heterochromatinized centromere discs are typical of the Altai and Yakutian reservoirs [8,13,14]. Previously, Ch. agilis2 was found in the Novosibirsk, Altai, and Kurgan regions, as well as Kazakhstan [9,15,16].
The original description of Ch. balatonicus was from the Balaton Lake in Hungary [17], but the karyotype was known before as a new race of Ch. plumosus [18,19]. The species is characterized by a wide range of habitats in the Palearctic. It was found in Hungary, Bulgaria, Poland [17,20,21], Russia (Kaliningrad, Leningrad, Moscow, Yaroslavl, Volgograd, Omsk, Penza, Novosibirsk, and Altai regions), Kazakhstan, etc. [7,9,18,19,22,23,24]. A feature of the Ch. balatonicus karyotype is the presence of fixated pericentric inversions in chromosomes CD and EF [9,25,26]. In the banding pool sequence of Ch. balatonicus, more than 59 sequences were noted [23], and then three endemic sequences from the Vistula Lagoon, with the average mineralization of water at 3.8 ‰, were added [22]. Arm D is the most polymorphic and an increase in the frequency of occurrence of the balD2 sequence is associated with the pollution of water [21].
Camptochironomus tentans is a common species in the Palearctic. Previously, it was found in England, Germany, Switzerland, Austria, Bulgaria, Armenia, Kazakhstan, and Russia, in the Kaliningrad [27], Ural, Irkutsk, Tyva, Yakutia, and Altai regions [7,27,28,29,30,31,32,33,34,35,36,37,38,39,40]. C. tentans has three sibling-species, C. pallidivittatus in the Holarctic [41], C. dilutus in North America [42], and C. biwaprimus, which is endemic to Japan [43]. In the C. tentans banding pool sequences, about 62 sequences in the Holarctic, and 43 in the Palearctic, were noted [13,41]. Later, four new sequences were found in the Saratov reg. [44].
It is known that the recorded species possess a high level of heterozygosity and heterozygotes per individual; 65–100% and 0.8–2.3, respectively, for Ch. balatonicus [23,45], and 95–99% and 2.0–2.6 for C. tentans [13,27]. The distribution of banding sequences and compositions of zygotic combinations over the areal has a regularity; it is more dependent on conditions in water bodies than on geographical location [13,24,46].
The current study aims to precisely identify the cryptic species of Chironomus larvae from Kurchatskoe Lake with mineralized water using a variety of approaches, including morphology, cytogenetics, and molecular genetics, as well as a study of the level of chromosomal polymorphism in the identified species.
2. Materials and Methods
Fourth instar larvae of Ch. agilis2, Ch. balatonicus, and C. tentans were collected from Kurchatskoe Lake (55°58′43.4″ N 67°40′50.4″ E) in the Armizon district, Tyumen region, Russia, in October 2021. The mineralization of water was 7‰. The depth of the sample location was 0.3 m. The bottom was sand. One larva of Ch. agilis2, eight larvae of Ch. balatonicus, and one larva of C. tentans were found in one sample.
All larvae were studied karyologically by ethanol-orcein technique [47]. The age was determined by the standard method [48]. A Micromed-6C (LOMO, St. Petersburg, Russia) light microscope equipped with a standard (kit) oil objective x100 and a camera ToupCam5.1 (China) was used for microscopy analysis.
To identify chromosome banding sequences, existing cytomaps [7,9,15,23,25,42,49,50,51,52] were used.
Larvae of Ch. agilis2 and C. tentans, and one larva of Ch. balatonicus, studied karyologically, were taken for the total DNA extraction using the «M-sorb-OOM» (Sintol, Moscow, Moscow region, Russia) kit with magnet particles, according to the manufacturer’s protocol. For amplification of COI (cytochrome oxidase subunit I), we used primers LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) (Eurogen, Moscow, Moscow region, Russia) [53]. An amplification reaction was carried out in 25 μL reaction mixture (1x buffer, 1.5 μM MgCl2, 0.5 mM of each primer, 0.2 μM dNTP of each nucleotide, 17.55 μL deionized water, 1 μL template DNA, and 1 unit Taq-polymerase (Evrogen, Moscow, Moscow region, Russia). PCR was performed at 94 °C (3 min), followed by 30 cycles at 94 °C (15 s), 50 °C (45 s), 72 °C (60 s), and a final extension at 72 °C (8 min). PCR products were visualized on 1% agarose gels and later purified by ethanol and ammonium acetate (3 M). Both strands were sequenced on an Applied Biosystems 3500 DNA sequencer (Thermo Scientific, Waltham, MA, USA) following the manufacturer’s instructions.
For the alignment of COI nucleotide sequences, we used MUSCLE in the MEGA6 software [54]. MEGA6 was used to calculate pairwise genetic distances (p-distance) with codon position preferences: 1st, 2nd, 3rd, and noncoding sites. The Bayesian analysis was performed using the program MrBayes v.3.2.6 [55,56], with the previously suggested settings [57,58,59], for 1,000,000 iterations and 1000 iterations of burn-in, nst = 6 (GTR + I + G). The phylogenetic trees resulting from the Bayesian inference analyses were visualized and edited using FigTree v.1.4.3 [60].
In addition, forty-three COI gene sequences of the genus Chironomus from “GenBank” and “Barcode of Life Data Systems” (BOLD) were analyzed. Accession numbers of used sequences in GenBank and BOLD: Ch. agilis (Shobanov & Djomin, 1988) (MZ656297), Chironomus agilis2 (Kiknadze, Siirin & Filippova, 1991) (AF192190), Ch. balatonicus (Dévai, Wülker & Scholl, 1983) (JN016827, JN016826, AF192193), C. biwaprimus (Sasa & Kawai 1987) (AF110166), Ch. acutiventris (Wülker, Ryser et Scholl, 1983) (AF192200), Ch. annularius (Meigen, 1818) (AF192189), Ch. anthracinus (Zetterstedt, 1860) (KF278222), Ch. aprilinus (Meigen, 1818) (KC250746), Ch. bernensis (Wülker & Klötzli, 1973) (AF192188), Ch. borokensis (Kerkis, Filippova, Schobanov, Gunderina et Kiknadze, 1988) (AB740261), Ch. cingulatus (Meigen, 1830) (AF192191, MZ660558), Ch. commutatus (Keyl, 1960) (AF192187.1), Ch. curabilis (Belyanina, Sigareva et Loginova, 1990) (JN016810.1), C. dilutus (Shobanov, Kiknadze et Butler, 1999) (KF278335.1), Ch. entis (Shobanov, 1989) (KM571024.1), Ch. heterodentatus (Konstantinov, 1956) (AF192199.1), Ch. heteropilicornis (Wülker, 1996) (MK795770.1), Ch. luridus (Strenzke, 1959) (AF192203), Ch. maturus (Johannsen, 1908) (DQ648204), Ch. melanotus (Keyl, 1961) (OL546775), Ch. nipponensis (Tokunaga, 1940) (DQ648206), Ch. novosibiricus (Kiknadze, Siirin et Kerkis, 1993) (AF192197), Ch. nuditarsis (Keyl, 1961) (KY225345), C. pallidivittatus (Malloch, 1915) (AF110164), Ch. piger (Strenzke, 1959) (AF192202), Ch. pilicornis (Fabricius, 1787) (HM860166), Ch. plumosus (Linnaeus, 1758) (KF278217.1), Ch. pseudothummi (Strenzke, 1959) (KC250754), Ch. riparius (Meigen, 1804) (KR756187.1), Ch. salinarius (Kieffer, 1915) (KR641621), Ch. sokolovae (Istomina, Kiknadze & Siirin, 1999) (MW471100), Ch. sororius (Wülker, 1973) (MZ324811), C. tentans (Fabricius, 1805) (AF110157, AF110156, AF110158, MZ658107, MN942989), Ch. tuvanicus (Kiknadze, Siirin et Wülker, 1993) (AF192196.1), Ch. usenicus (Loginova & Belyanina, 1994) (JN016820.1), and Ch. whitseli (Sublette et Sublette, 1974) (KR683438.1). The COI sequence of Drosophila melanogaster (Meigen, 1830) (HQ551913) was used as an outgroup in the phylogenetic analysis.
3. Results
3.1. Morphological Characters of Ch. agilis2 from Kurchatskoe Lake
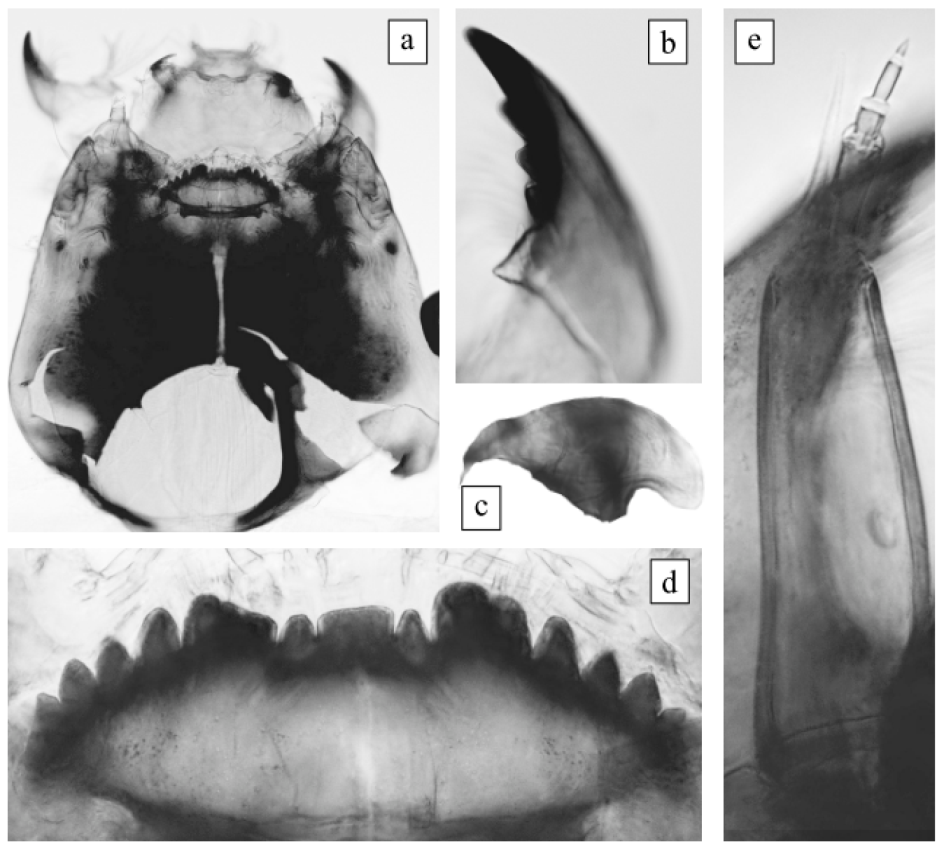
The morphological characteristics of the larva satisfied the original description of Ch. agilis2 [15]. Photos of the morphology of the larva are presented for the first time in this study (Figure 1). Body length was 14 mm. Head capsule was yellow (light brownish). Occipital sclerites were dark; on the lateral side there was an intensely colored area, and borders were blurred. Basal segment of the antenna was wedge-shaped; blade was extended to the middle of the fourth or base of the fifth segment. The ring organ was located at a length of 2/3 (almost 1/2) of the basal segment length. Ventromental plates with small teeth were on the front edge. The larva in our preparation had a damaged mentum, but it had a light brownish coloration and its characteristics corresponded to those of the Ch. plumosus group. All four teeth of the mandible were dark brown in color.
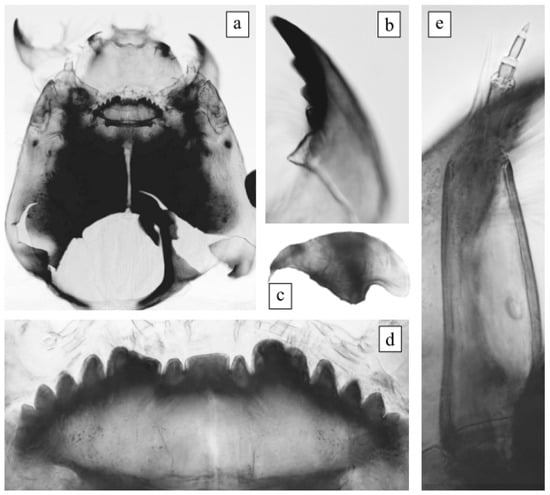
Figure 1.
Larva morphology of Ch. agilis2 from Kurchatskoe Lake. (a) Head capsule, (b) mandible (fragment), (c) ventromental plate, (d) mentum, and (e) antenna.
3.2. Karyotypes of Chironomus from Kurchatskoe Lake
3.2.1. Karyotype of Chironomus agilis2
The species was described as cytospecies [15]. By karyotype characteristics, Ch. agilis2 was very similar to Ch. agilis, but differed from them by the size of its heterochromatinized centromeric region and banding sequences in arm C [9,15].
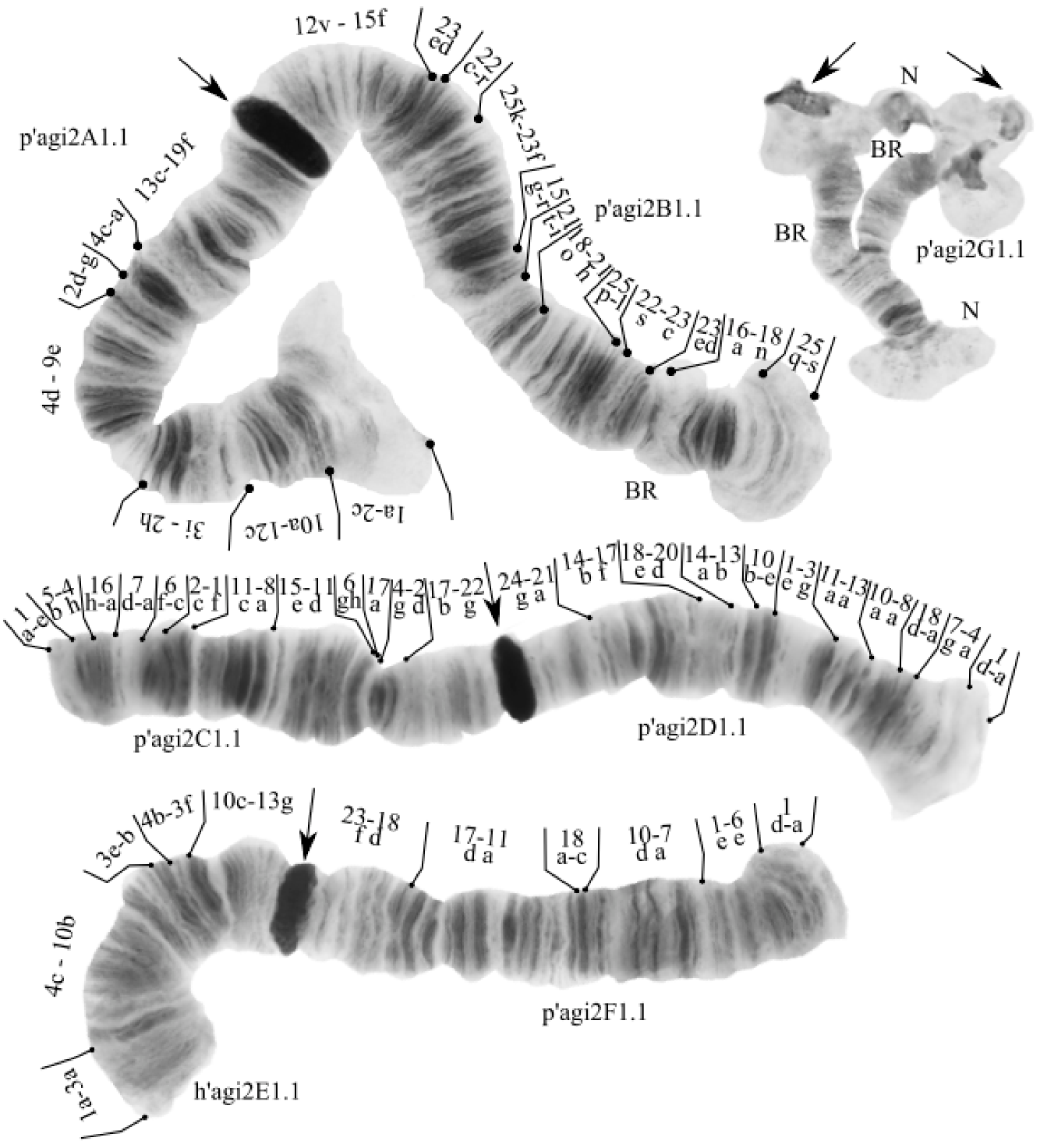
The species has a 2n = 8 set of chromosomes. By the chromosome arm combination AB CD EF G, the species belongs to the Chironomus “thummi” cytocomplex. Two nucleoli were found in arm G, and Balbiani rings were found in arms B and G (Figure 2).
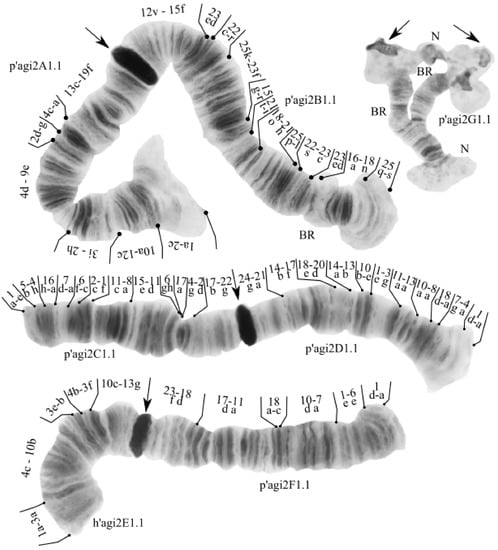
Figure 2.
Karyotype of Chironomus agilis2 from Kurchatskoe Lake, Tyumen, Russia. Arrows indicate centromeric bands; p’agi2A1, p’agi2B1, etc., genotypic combinations of banding sequences in chromosome arms; BR, Balbiani rings; N, nucleolus; and prefix p’ for Palearctic sequences, h’ for Holarctic sequences.
One zygotic combination was found: agi2A1.1. B1.1. C1.1. D1.1. E1.1. F1.1. G1.1. All banding sequences were previously found in different populations [9,15,61].
Arm A—one banding sequence: agi2A1 1a-2c 10a-12c 3i-2h 4d-9e 2d-g 4c-a 13a-19f C [15,61].
Arm B—one banding sequence: agi2B1 25s-q 18n-16a 22ab 23c-22s 25l-p 21h-18o 21i-t 15r-g 23f-25k 22r-c 23de 15f-12v C [61,62,63].
Arm C—one banding sequence: agi2C1 1a-e 5b-4h 16h-a 7d-a 6f-c 2c-1f 5c-6b 11c-8a 15e-11d 6gh 17a 4g-2d 17b-22g C [15,25].
Arm D—one banding sequence: agi2D1 1a-d 4a-7g 18a-d 8a-10a 13a-11a 3g-1e 10e-b 13b-14a 20d-18e 17f-14b 21a-24g C [25].
Arm E—one banding sequence: agi2E1 1a-3a 4c-10b 3e-b 4b-3f 10c-13g [26].
Arm F—one banding sequence: agi2F1 1a-d 6e-1e 7a-10d 18c-a 11a-17d 18d-23f C [9,15].
Arm G—one banding sequence: agi2G1. Not mapped.
3.2.2. Karyotype of Chironomus balatonicus
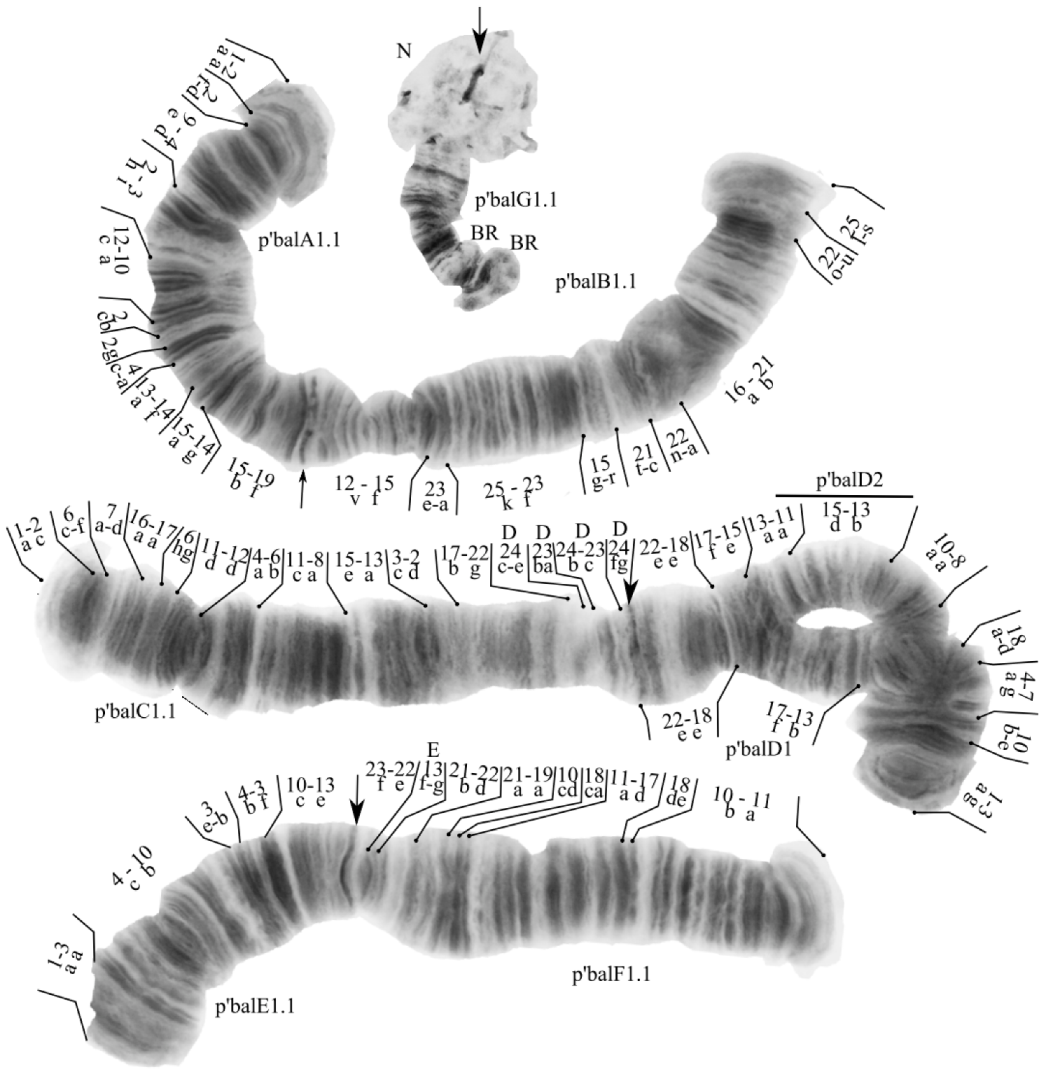
The species has a 2n = 8 set of chromosomes. By the chromosome arm combination AB CD EF G, the species belongs to the Chironomus “thummi” cytocomplex. The chromosomes AB and CD are metacentric, EF is submetacentric, and G is telocentric. One nucleolus was found in arm G. Balbiani rings were found in arms B and G (Figure 3). In all studied larvae there was found one banding sequence: balA1.1. B1.1.C1.1.D1.2.E1.1.F1.1.G1.1. All eight banding sequences were previously published [7,9,17,23].
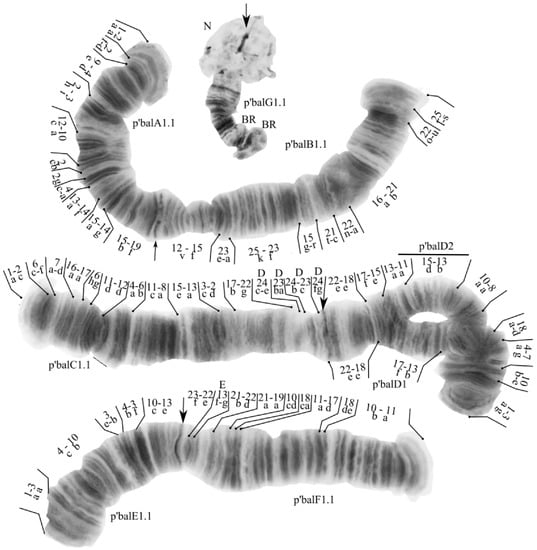
Figure 3.
Karyotype of Chironomus balatonicus from Kurchatskoe lake, Tyumen, Russia. Arrows indicate centromeric bands; p’balA1, p’balB1, etc., genotypic combinations of banding sequences in chromosome arms; BR, Balbiani rings; N, nucleolus; and prefix p’ for Palearctic sequences.
Arm A—one banding sequence: p’balA1 1a-2a 2f-d 9e-4d 2h-3i 12c-10a 2cb 2g 4c-a 13a-14f 15a-14g 15b-19f C.
Arm B—one banding sequence: p’balB1 25s-l 22u-o 21b-16a 22a-n 21c-t 15r-g 23f-25k 23a-e 15f-12v C.
Arm C—one banding sequence: 1a-2c 6c-f 7a-d 16a-17a 6hg 11d-12d 4a-6b 11c-8a 15e-13a 3c-2d 17b-22g [D24c-e D23ba D24b-D23c D24fg C] [9,25]. A similar inversion was found in Chironomus oculatus [64].
Arm D—two banding sequences: p’balD1 1a-3g 10b-e 4a-7g 18a-d 8a-10a 13a-11a 13b-17f 18e-22e C was found in heterozygous state with p’balD2 1a-3g 10b-e 4a-7g 18a-d 8a-10a 15d-13b 11a-13a 15e-17f 18e-22e C.
Arm E—one banding sequence: p’balE1 1a-3a 4c-10b 3e-b 4b-3f 10c-13e C.
Arm F—one banding sequence: p’balF1 1a-10b 18ed 17d-11a 18a-c 10dc 19a-21a [22d-21b E13gf 22e-23f C].
Arm G—one banding sequence: p’balG1. Not mapped.
3.2.3. Karyotype of Camptochironomus tentans
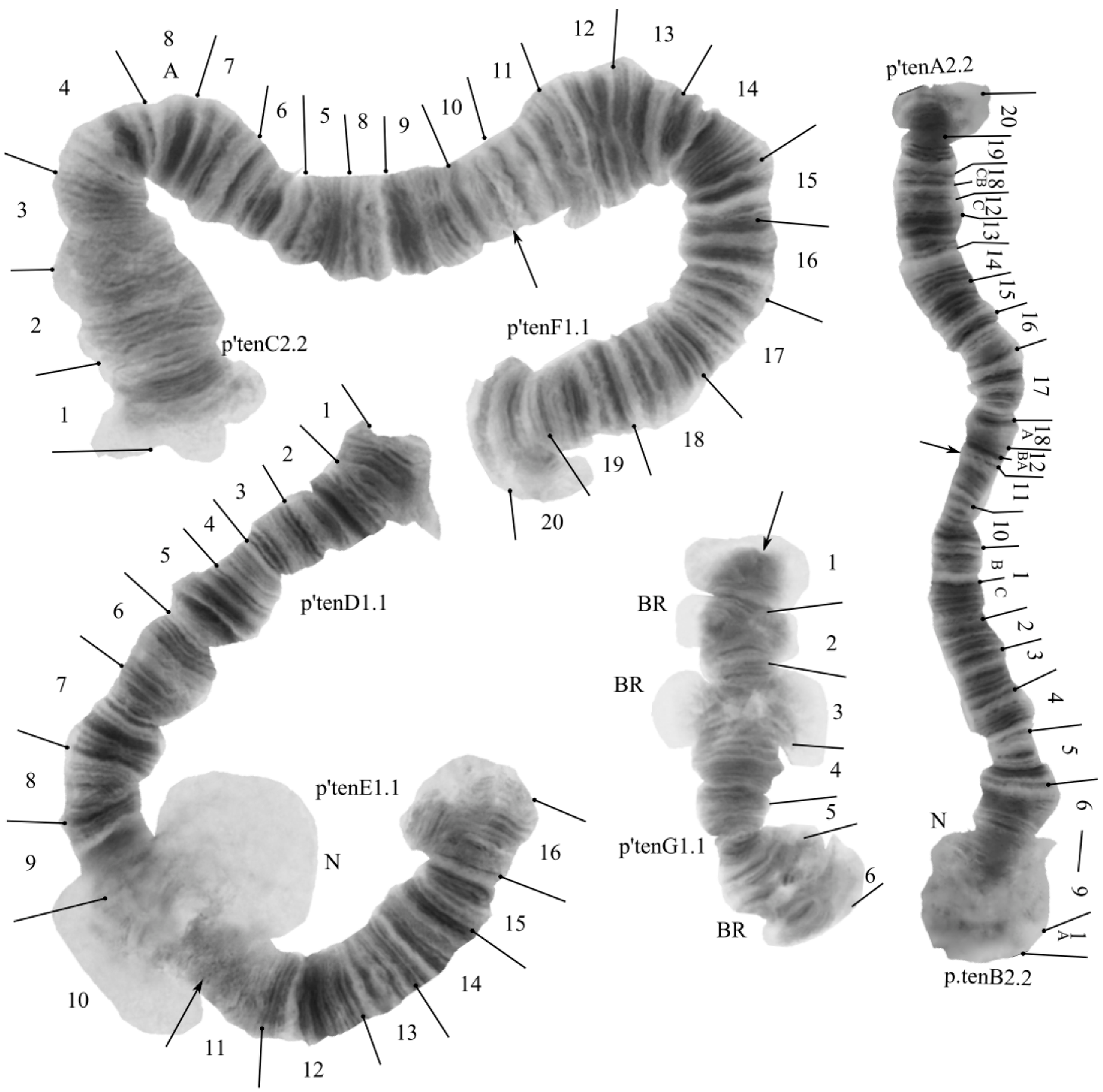
The species has a 2n = 8 set of chromosomes. By the chromosome arm combination AB CF DE G, the species belongs to the «camptochironomus» cytocomplex. Two nucleoli were found in arms B and D. Three Balbiani rings were found in arm G (Figure 4). In the studied larva there was found one banding sequence: tenA2.2. B1.1.C1.1.F1.1.D1.1.E1.1.G1.1. All banding sequences were previously published [7,36,44].
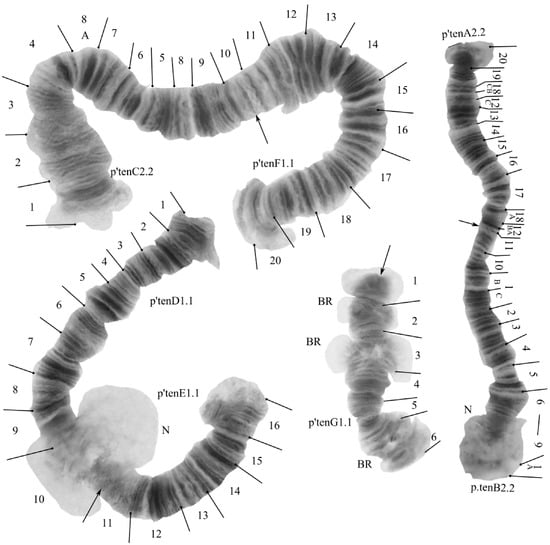
Figure 4.
Karyotype of Camptochironomus tentans from Kurchatskoe Lake, Tyumen reg., Russia. Arrows indicate centromeric bands; p’tenA2, p’tenB1, etc., genotypic combinations of banding sequences in chromosome arms; BR, Balbiani rings; N, nucleolus; and prefix p’ for Palearctic sequences.
Arm A—one banding sequence: p’tenA2 20CBA 19CBA 18CBA 12C 13ABC 14ABC 15ABC 16ABC 17ABC 18A 12BA.
Arm B—one banding sequence: p’tenB2 1A 9BA 8CBA 7CBA 6CBA 5CBA 4CBA 3CBA 2CBA 1CB 9C 10ABC 11ABC.
Arm C—one banding sequence: p’tenC2 1ABC 2ABC 3ABC 4ABC 8A 7CBA 6CBA 5CBA 8BC 9ABC 10ABC.
Arm F—one banding sequence: p’tenF1 20CBA 19CBA 18CBA 17CBA 16CBA 15CBA 14CBA 13CBA 12CBA 11CBA.
Arm D—one banding sequence: p’tenD1 1ABC 2ABC 3ABC 4ABC 5ABC 6ABC 7ABC 8ABC 9ABC 10ABC.
Arm E—one banding sequence: p’tenE1 16CBA 15CBA 14BCA 13CBA 12CBA 11CBA 10CBA.
Arm G—one banding sequence: p’tenG1 1ABC 2ABC 3ABC 4ABC 5ABC 6ABC.
3.3. COI Gene Sequences and Phylogenetic Analysis of Chironomus from Kurchatskoe Lake
Characteristics of the obtained COI gene sequences for studied species:
- Ch. agilis2: GenBank accession number OP435179; length 675 bp; percentage of nucleotides A:24, C:21, G:18, and T:37.
- Ch. balatonicus: GenBank accession number OP435180; length 677 bp; percentage of nucleotides A:25, C:20, G:17, T:38.
- C. tentans: GenBank accession number OP434795; length 671 bp; percentage of nucleotides A:27, C:19, G:16, T:38.
3.3.1. Genetic Distances between Chironomus Obtained with K2P
In GenBank, we found one COI gene sequence (AF192190) in Ch. agilis2 from the Novosibirsk region, identified by karyology [65,66]. The pairwise genetic distance between the sequences of Ch. agilis2 from the Novosibirsk reg. and Tyumen reg. was 1.0% (Table 1), which is much lower than the accepted 3% interspecific threshold [11,12,67,68,69]. The genetic distance between sequences of Ch. agilis2 (OP435179) and Ch. agilis (MZ656297) from Finland, identified by morphology [70], was 17.2%. At the same time, the distance between Ch. agilis (MZ656297) and Ch. cingulatus (MZ660558) from Finland was 0%, and Ch. cingulatus (AF192191) from Novosibirsk reg. was 13.9%. Ch. agilis (MZ656297) was closest to Ch. heterodentatus (AF192199), with a distance of 1.9%.

Table 1.
The pairwise genetic distances (K2P) between COI gene sequences of Chironomus species.
For Ch. balatonicus. we used sequences from the Saratov reg. (JN016826. JN016827) and Novosibirsk reg. (AF192193). Specimens were studied cytogenetically [65,71]. The distance between sequences of Ch. balatonicus from the Tyumen reg. and Saratov reg. was 0%; between the Tyumen reg. and Novosibirsk reg. the distance was 0.8% (Table 1).
The smallest genetic distance was between sequences of C. tentans (OP434795) from the Tyumen reg. and C. tentans (MZ658107) from Finland, at 0.0% (Table 1). The distance between sequences of C. tentans (OP434795) from the Tyumen reg. and C. tentans from the Altai reg. (Russia) and Yakutia reg. (Russia) was 0.2%. C. tentans (AF110158) from Sweden, identified cytogenetically [43], was 0.4%. The distance between C. tentans (OP434795) and C. pallidivittatus (AF110164) from the Yakutia reg. was 1.1%.
The distance between C. tentans (OP434795) from the Tyumen reg. (Russia) and C. tentans (MN942989) from China [72] was 4.5%. The distance between C. tentans (MN942989) from China and C. biwaprimus (AF110166) from Japan [43] was 1%.
3.3.2. Analysis of the Phylogenetic Tree
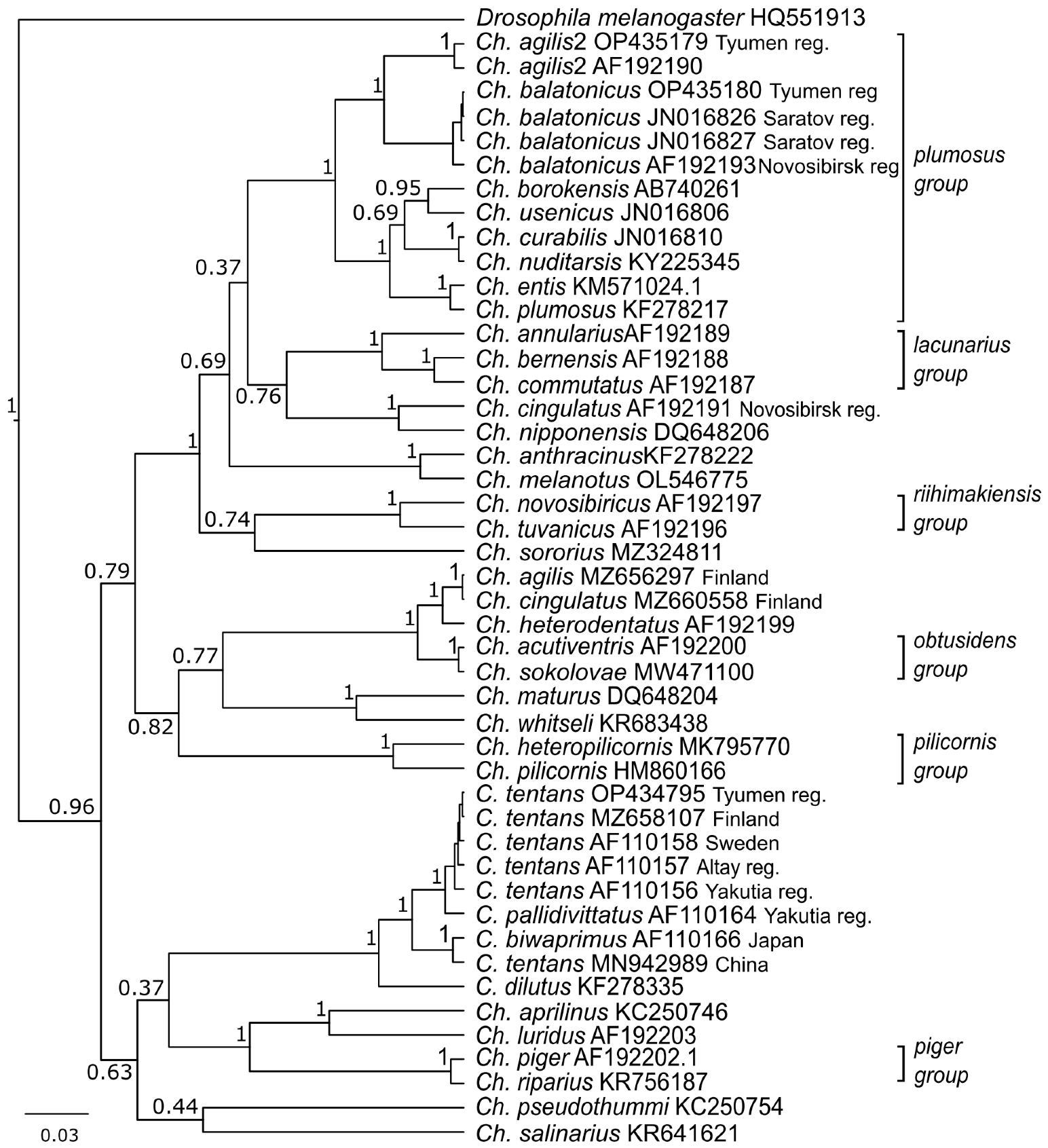
On the phylogenetic tree, gene sequences of Ch. agilis2 from the Tyumen reg. (OP435179) and Novosibirsk reg. (AF192190) formed a separate cluster from Ch. agilis (MZ656297) and Ch. cingulatus (MZ660558) from Finland (Figure 5). Here we see a repeat of the situation with genetic (K2P) distances (Table 1). The sequences of Ch. balatonicus from the Tyumen reg. (OP435180), Saratov reg. (JN016826. JN016826), and Novosibirsk reg. (AF192193) formed a separate cluster, and, with Ch. agilis2 (OP435179. AF192190), they were combined in the Chironomus plumosus group cluster.
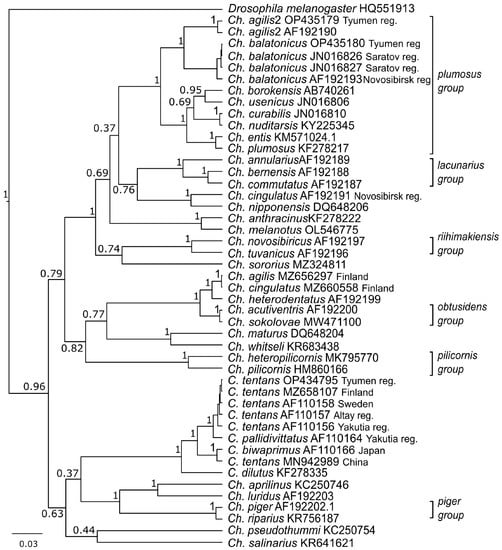
Figure 5.
Bayesian tree of the analyzed samples of Chironomus (Camptochironomus) spp., inferred from COI gene sequences. Species name. GenBank accession numbers and group name are shown to the right of the branches. Support p-values are given if they exceed 0.3.
In C. tentans, we distinguished two clusters. The first cluster included C. tentans sequences from the Tyumen reg. (OP434795), Finland (MZ658107), Sweden (AF110158), the Altai reg. (AF110157), and the Yakutia reg. (AF110164). The second cluster included sequences of C. tentans from China (MN942989) and C. biwaprimus from Japan (AF110166).
4. Discussion
Most investigations into the biodiversity of water bodies (rivers, lakes, ponds, and temporary reservoirs) begin with the study of chironomids. They contribute significantly to biomass and provide food for industrial fish. They build tube-shaped houses; as a result, the area of interaction between bottom sediments and water increases and microbiological processes intensify. They also surpass Dreissena polymorpha (Pallas, 1771) in relative sedimentation activity, all of which contributes to the biological purification of water [73].
The ecological plasticity determined by morphological and cytogenetic variability allows them to inhabit various reservoirs [7,24,46]; at the same time, this makes them difficult to identify.
In one sample location at Kurchatskoe Lake, with the mineralization of water at 7‰, near Tyumen city, we found a few Chironomus larvae. By using only morphological characters, we cannot identify all larvae with high precision. All morphological characteristics, in general, correspond to the species Ch. agilis2 [15], Ch. balatonicus [7], and C. tentans [7,42]. Papers studying chironomids recommend a multiple approach that includes cytogenetic and molecular-genetic analysis [10,11,12].
Using karyotype characteristics, especially the presence of a heterochromatinized centromeric disc, we identified one of the species as Ch. agilis2 [15]. The level of polymorphism in Ch. agilis2 is known to be very low [15], and we did not find any alternative banding sequences. Molecular-genetic analysis showed the genetic distance between Ch. agilis2 from the Tyumen reg. (OP435179) and Novosibirsk reg. (AF192190) to be 1%, which is much lower than the 3% accepted interspecific threshold. The first description indicated a high affinity between Ch. agilis2 and Ch. agilis at the cytogenetic and biochemical levels [15]; however, in the Bayesian three (Figure 5), they are in different branches. A detailed study showed that Ch. agilis (MZ656297) and Ch. cingulatus (MZ660558) from Finland are in the same branch as Ch. heterodentatus (AF192199), studied cytogenetically [65,66], with a distance of 1.9%. The distance between sequences of Ch. cingulatus from Finland (MZ660558), identified by morphology [70], and from the Novosibirsk reg., identified by karyology [65,66], was 13.9%. We assume that this was a case of misidentification of sibling-species by morphology, as previously shown by Ch. melanotus [59].
A high level of chromosomal polymorphism is typical for Ch. balatonicus [23,45]. In all the Ch. balatonicus larvae founded in Kurchatskoe Lake, we found one zygotic combination. The main feature of this finding is the presence of balD1 and balD2 banding sequences in only heterozygous states. The frequency of balD1.2 in the Altai reg. population is 8–18%; in Novosibirsk reg. it is 0–29%, in Kazakhstan it is 0–6%, and in Lithuania it is 50% [23]. We know that the quantity of individuals is too small for statistical analysis, but we can assume that there is an excess of heterozygotes for balD1.2 in Kurchatskoe Lake. It was shown that, in the distribution of banding sequences and the compositions of zygotic combinations in range of habitat depend more on conditions in water bodies than on geographical localization [13,24,46]. In addition, in experiments with infection by the entomopathogenic bacterium Bacillus thuringiensis israelensis, it was found that the presence of balD1.1 and balD2.2 reduces the possibility of infection [74]. Based on these data, we suggest the presence balD1.2 combination has adaptive significance for larvae in the mineralized water in Kurchatskoe Lake. On the phylogenetic tree, all sequences of Ch. balatonicus form one cluster. The genetic distances between Ch. balatonicus from Kurchatskoe Lake (OP435180) and other sequences do not exceed the 3% accepted interspecific threshold. This low level of chromosomal polymorphism could be a result of the proximity of the range boundary of the species [23].
In one larva of C. tentans we found one zygotic combination with sequences in a homozygous state, which is atypical for this species, as it was noted that the quantity of heterozygous larvae varies from 78 to 100% [36,44]. Alternate sequences tenA2 and tenC2 typically predominated in Ural and West Siberian populations, reaching 85% in the Altai reg. and Kazakhstan populations [36]. Sequence tenB2 is common in European populations [36]. The sequences tenD1, tenE1, and tenF1 are typical in all studied regions [36,44]. The genetic distances and position on the phylogenetic tree suggest that C. tentans from Kurchatskoe Lake has no differences from the Finnish population, and has minimal distance between the Altai reg. and Yakutian populations.
The distance between sequences of C. tentans from Kurchatskoe Lake (OP434795) and China (MN942989) was more than 3%, which suggests misidentification of the species. On the phylogenetic tree, we can see that the distance between C. tentans from China (MN942989) and C. biwaprimus (AF110166) was about 1% (Table 1), and, on the phylogenetic tree, they form a distinct branch (Figure 5). Similar results were obtained previously in Camptochironomus with a Holarctic distribution [43]. It is noted that C. biwaprimus is endemic to Japan, and has been described on morphological grounds as being close to C. tentans [43,75].
5. Conclusions
Tobolo-Ishim forest-steppe lakes, small and closed, are studied on a regular basis. They are interesting in their conditions, the first being mineralization. About 63% of them are slightly mineralized > 1‰, 28% are mineralized 1–10‰, and 8% are highly mineralized > 10‰ [3]. Larvae were sampled from the previously unstudied Kurchatskoe Lake, with mineralization of 7‰. Using a comprehensive approach, including morphology, cytogenetics, and molecular genetics, we found three species of Chironomus. In all larvae of Ch. agilis2, Ch. balatonicus, and C. tentans, common in Europe and the Altai reg, banding sequences were found. Usually high-heterozygous, Ch. balatonicus and C. tentans possess 2 heterozygotes per individual; in Kurchatskoe Lake they were low-heterozygous. Low chromosomal polymorphism in Ch. balatonicus is characteristic in the studied Kurchatskoe Lake and for the areal boundary Kazakhstan population [23]. All the estimated genetic distances of COI gene sequences of the studied species were much lower than the commonly accepted threshold of 3% for species of the genus Chironomus. Only in C. tentans from China the distance was 4.5%. We suggest that additional research with the use of cytogenetic analysis is required.
Author Contributions
V.B. and E.M. developed the concept of this study; V.S. collected the samples; E.M. designed and carried out the molecular analyses; V.B. performed project administration and carried out the karyological and statistical analysis; V.B wrote the paper, with input from E.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was performed in the framework of the state assignment: theme No. 121050500046-8 and 121051100099-5. Research of V.A. Stolbov was funded by the Tyumen Oblast Government. as part of the West-Siberian Interregional Science and Education Center’s project No. 89-DON (2).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data supporting reported results can be obtained upon request from the corresponding author (V.V.B.).
Acknowledgments
The authors are grateful to B.A. Levin and M.Kh. Karmokov (IEMT RAS) for their help and consultations during all stages of the investigation and manuscript preparation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Azarov, V.I.; Lezin, V.A.; Krivenko, V.G. Lakes of the Tobolo-Ishim forest-steppe. In Wetlands of Russia.V. 1. Wetlands of International Importance; Krivenko, V.G., Ed.; Wetlands International Publications: Moscow, Russia, 1998; pp. 158–172. [Google Scholar]
- Aleshina, O.A.; Larin, S.I.; Katanaeva, V.G. To assess the state of macrozoobenthos in the lakes of the middle forest-steppe of the Tyumen Ishim region. In Geoecological Problems of the Tyumen Region; Kalinin, V.M., Ed.; Vektor Buk: Tyumen, Russia, 2006; pp. 246–253. [Google Scholar]
- Kozlov, O.V. Crustaceans of Ecosystems of Small Lakes under Conditions of Anthropogenic Stress (on the Example of Reservoirs of the Ishim Plain); Moscow State University: Moscow, Russia, 2005; p. 54. [Google Scholar]
- Stepanova, V.B.; Sharapova, T.A. The fauna of chironomids of Western Siberia. Bull. Ecol. For. Landsc. Stud. 2001, 2, 117–124. [Google Scholar]
- Aleshina, O.A.; Katanaeva, V.G. Distribution and organization of macrozoobenthos in brackish lakes of the Ishim forest-steppe. Bull. Tyumen State Univ. Ecol. Nat. Manag. 2010, 7, 66–75. [Google Scholar]
- Krasnenko, A.S. The Structure of the Macrozoobenthos Population of Reservoirs in the South of the Tyumen Region (on the Example of the Ishim District). Ph.D. Thesis, Omsk State Pedagogical Institute, Omsk, Russia, 2010. [Google Scholar]
- Kiknadze, I.I.; Shilova, A.I.; Shobanov, N.A.; Zelentsov, N.I.; Grebenyuk, L.P.; Istomina, A.G.; Pratasov, V.A. Karyoypes and Larval Morphology in Tribe Chironomini. Atlas; Nauka Publish: Novosibirsk, Russia, 1991; pp. 1–115. [Google Scholar]
- Kiknadze, I.I.; Istomina, A.G.; Gunderina, L.I.; Aimanova, K.G.; Savvinov, D.D. Banding Sequence Pools of Chironomid of Yakutian Permafrost. Tribe Chironomini; Nauka: Novosibirsk, Russia, 1996; pp. 1–166. [Google Scholar]
- Kiknadze, I.I.; Istomina, A.G.; Golygina, V.V.; Gunderina, L.I. Karyotypes of Palearctic and Holarctic Species of the Genus Chironomus [Electronic Resource]; Kiknadze, I.I., Ed.; Russian Academy of Sciences: Moscow, Russia; Siberian Branch: Novosibirsk, Russia; Federal Research Center Institute of Cytology and Genetics: Novosibirsk, Russia; Academic Publishing House “GEO”: Novosibirsk, Russia, 2016; ISBN 9785990885325. [Google Scholar]
- DeSalle, R.; Egan, M.G.; Siddall, M. The unholy trinity: Taxonomy, species delimitation and DNA barcoding. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1905–1916. [Google Scholar] [CrossRef] [PubMed]
- Proulx, I.; Martin, J.; Carew, M.; Hare, L. Using various lines of evidence to identify Chironomus species (Diptera: Chironomidae) in eastern Canadian lakes. Zootaxa 2013, 3741, 401–458. [Google Scholar] [CrossRef] [PubMed]
- Ekrem, T.; Willassen, E.; Stur, E. A comprehensive DNA sequence library is essential for identification with DNA barcodes. Mol. Phylogenet. Evol. 2007, 43, 530–542. [Google Scholar] [CrossRef]
- Kiknadze, I.I.; Istomina, A.G. Karyotypes and Chromosomal Polymorphisms in Siberian Chironomids (Diptera, Chironomidae). Sib. Ecol. J. 2000, 4, 445–460. [Google Scholar]
- Kiknadze, I.I. The role of chromosomal polymorphism in divergence of populations and species of the genus Chironomus (Diptera). Entomol. Rev. 2008, 88, 509–524. [Google Scholar] [CrossRef]
- Kiknadze, I.I.; Siirin, M.T.; Filippova, M.; Gunderina, L.I.; Kalachikov, S.M. The change of the pericentromeric heterochromatin mass in one of important ways of the chironomid evolution. Tsitologiia 1991, 33, 90–98. [Google Scholar]
- Golygina, V.V.; Kiknadze, I.I.; Broshkov, A.D.; Karamysheva, T.V.; Rubtsov, N.B. Structural features of centromeres in poletene chromosomes of chironomidae (Diptera) species. VOGiS Her. 2010, 14, 622–631. [Google Scholar]
- Dévai, G.; Wülker, W.F.; Scholl, A. Revision der Gattung Chironomus Meigen (Diptera). IX. C. balatonicus sp. n. aus Flachsee Balaton. Acta Zool. Acad. Sci. Hung. 1983, 29, 357–374. [Google Scholar]
- Maksimova, F.L.; Petrova, N.A. Geographical variability of the karyotype of Chironomus plumosus (Diptera, Chironomidae). Zool. Zhurnal 1978, 57, 1816–1826. [Google Scholar]
- Maksimova, F.L. The inversion polymorphism of Chironomus plumosus nature populations. New Data Karyosyst. Diptera Proc. Zool. Inst. 1980, 95, 31–39. [Google Scholar]
- Dévai, G.; Miskolczi, M.; Wülker, W.F. Standardization of chromosome arms B, C and D in Chironomus (Diptera, Chironomidae). Acta Biol. Debricina. Suppl. Oecologica Hung. 1989, 2, 79–92. [Google Scholar]
- Michailova, P.; Petrova, N.A. Cytogenetic characteristics of Chironomus balatonicus Devai, Wulker, Scholl (Diptera, Chironomidae) from Chernobyl region. Cytobios 1994, 79, 15–29. [Google Scholar] [PubMed]
- Markiyanova, M.F. The main parameters of the cytogenetic structure of the population of Chironomus balatonicus Dévai et al. from the Vislinsky Bay of the Baltic Sea. Bull. I. Kant Russ. State Univ. 2009, 7, 42–49. [Google Scholar]
- Golygina, V.V.; Istomina, A.G.G.; Rakisheva, A.Z.; Kiknadze, I.I. New banding sequences in the Chironomus balatonicus karyofund. Tsitologiya 1996, 38, 869–883. [Google Scholar]
- Gunderina, L.I.; Kiknadze, I.I.; Golygina, V.V. Differentiation of the Cytogenetic structure of natural populations in the plumosus group of sibling species Chironomus balatonicus, Chironomus entis, Chironomus muratensis, Chironomus nudiventris (Chironomidae: Diptera). Genetica 1999, 35, 606–614. [Google Scholar]
- Golygina, V.V.; Kiknadze, I.I. A revision of chromosome II (CD) mapping in Chironomus plumosus (Linnaeus, 1758) group (Diptera, Chironomidae). Comp. Cytogenet. 2012, 6, 249–266. [Google Scholar] [CrossRef]
- Golygina, V.V.; Kiknadze, I.I. The revision of chromosome III (EF) mapping in Chironomus plumosus (Linnaeus, 1758) group (Diptera, Chironomidae). Comp. Cytogenet. 2018, 12, 201–222. [Google Scholar] [CrossRef]
- Petrova, N.A.; Vinokurova, N.V.; Danilova, M.V.; Sharton, A.Y. Inversion polymorphism in the population of Camptochironomus tentans from Kaliningrad city. Tsitologiya 2011, 53, 580–585. [Google Scholar]
- Beermann, W. Chromomeren Konstanz und spezifische Modifikationen der Chromosomen Struktur inder Entwicklung und Organdiffernzierung von Chronomus tentans. Chromosoma 1952, 5, 139–198. [Google Scholar] [CrossRef]
- Beermann, W. Zytologische Analyse eines Camptochironomus–Artbastards: 1. Kreuzungsergebnisse und die Evolution des Karyotypus. Chromosoma 1955, 7, 198–259. [Google Scholar] [CrossRef]
- Acton, A.B. Chromosome inversions in natural populations of chirnomus tentans. J. Genet. 1957, 55, 61–94. [Google Scholar] [CrossRef]
- Acton, A.B. A cytological comparison of Nearctic and Palearctic representatives of Chironomus tentans. Proc. Linn. Soc. 1958, 169, 129–131. [Google Scholar]
- Acton, A.B. A study of the differences between widely separated populations of Chironomus (=Tendipes) tentans (Diptera). Proc. R. Soc. London. Ser. B. Biol. Sci. 1959, 151, 277–296. [Google Scholar] [CrossRef]
- Michailova, P.V. The polytene chromosomes and their significance to the family Chironomidae, Diptera. Acta Zool. Fenn. 1989, 186, 1–107. [Google Scholar]
- Kiknadze, I.I.; Aimanova, K.T.; Gunderina, L.I.; Philippova, M.A.; Istomina, A.G. Chromosomal polymorphism in the Ural and Siberian populations of Camptochironomus tentans. Zool. Zhurnal 1993, 72, 59–75. [Google Scholar]
- Gunderina, L.I.; Kiknadze, I.I.; Aimanova, K.G.; Istomina, A.G.; Proviz, V.I.; Salova, T.A.; Rakisheva, A.Z.; Butler, M.G. Cytogenetic differentiation of natural and laboratory populations of Camptochironomus tentans (Fabricius) (Chironomidae: Diptera). Russ. J. Genet. 1996, 32, 45–57. [Google Scholar]
- Kiknadze, I.I.; Aimanova, K.G.; Gunderina, L.I.; Butler, M.G.; Cooper, J.K. Geographic variation in the polytene chromosome banding pattern of the Holarctic midge Chironomus (Camptochironomus) tentans (Fabricius). Can. J. Zool. 1996, 74, 171–191. [Google Scholar] [CrossRef]
- Kiknadze, I.I.; Butler, M.G.; Aimanova, K.G.; Andreeva, E.N.; Martin, J.; Gunderina, L.I. Divergent cytogenetic evolution in Nearctic and Palearctic populations of sibling species in the subgenus (Camptochironomus) Kieffer. Can. J. Zool. 1998, 76, 361–376. [Google Scholar] [CrossRef]
- Petrova, N.A.; Ilyinskaya, N.B. Inversion polymorphism of natural populations of Camptochironomus tentans (Diptera, Chironomidae) from north-western part of Russia. In Karyosystematics of Invertebrates; MSU Botanical Garden: Moscow, Russia, 1996; pp. 36–57. [Google Scholar]
- Ilyinskaya, N.B.; Petrova, N.A. Karyotype and inversion polymorphism of natural populations of Camptochironomus tentans (Diptera, Chironomidae) from north-western part of Russia. Tsitologiya 1997, 39, 848–856. [Google Scholar]
- Michailova, P. Chromosome Polymorphism of Camptochironomus tentans (Fabricius 1805) (Diptera: Chironomidae) from Łuknajno (Mazurian Lakeland), Poland. Acta Zool. Bulg. 2008, 60, 155–163. [Google Scholar]
- Kiknadze, I.I.; Shobanov, N.A.; Aimanova, K.G.; Andreeva, E.N. Karyotype and morphology of Camptochironomus setivalva (Diptera, chironomidae) larva. Zool. Zhurnal 2000, 79, 695–703. [Google Scholar]
- Shobanov, N.A.; Kiknadze, I.I.; Butler, M.G. Palearctic and Nearctic Chironomus (Camptochironomus) tentans (Fabricius) are different species (Diptera, Chironomidae). Entomol. Scand. 1999, 30, 311–322. [Google Scholar]
- Martin, J.; Guryev, V.; Blinov, A. Population variability in Chironomus (Camptochironomus) species (Diptera, Nematocera) with a Holarctic distribution: Evidence of mitochondrial gene flow. Insect Mol. Biol. 2002, 11, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Polukonova, N.V.; Shaternikov, A.N.; Karmokov, M.K. Inversion polymorphism of non-biting midges Camptochironomus tentans (Fabricius) 1805 (Diptera, Chironomidae) from populations of the Lower Volga region and Central Caucasus. Russ. J. Genet. 2015, 51, 22–32. [Google Scholar] [CrossRef]
- Belyanina, S.I.; Loginova, N.V. A list of chromosomal sequences in chironomus spp. of plumosus group. I. The karyofund of Chironomus balatonicus Devai, Wulker, Scholl, 1983. Tsitologiia 1993, 35, 87–92. [Google Scholar]
- Bolshakov, V.V. Variability of Karyofunds of Chironomus plumosus (Diptera, Chironomidae) from the Rybinsk Reservoir during the Period of Climate Warming. Inland Water Biol. 2021, 14, 256–262. [Google Scholar] [CrossRef]
- Dyomin, S.Y. Variability of the Degree of Condensation of Polytene Chromosomes in the Cells of Different Organs of Chironomus plumosus Larvae from Nature. Ph.D. Thesis, Institute of Cytology of the USSR Academy of Sciences, Leningrad, Russia, 1989. [Google Scholar]
- Ilyinskaya, N.B. The development of 4th instar larvae and diapause. In Chironomus plumosus L. (Diptera, Chironomidae). Systematics, Morphology, Ecology, Production; Sokolova, N.Y., Ed.; Nauka Publishers: Moscow, Russia, 1983; pp. 167–188. [Google Scholar]
- Kiknadze, I.I.; Golygina, V.V.; Filippova, M.A. Evidence of species-specific pericentric inversion in the karyotype of the midge Chironomus balatonicus. Tsitologiia 2002, 44, 97–101. [Google Scholar]
- Keyl, H.G. Chromosomenevolution bei Chironomus. II. Chromosomenumbauten und phylogenetische beziehungen der arten. Chromosoma 1962, 13, 464–514. [Google Scholar] [CrossRef]
- Keyl, H.-G. Chromosomenevolution bei Chironomus I. Strukturabwandlungen an Speicheldrüsen-Chromosomen. Chromosoma 1961, 12, 26–47. [Google Scholar] [CrossRef]
- Petrova, N.A.; Maksimova, F.L. The role of chromosomal rearrangements in the speciation of chironomids (Diptera, chironomidae). Genetica 1978, 14, 1201–1207. (In Russian) [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Karmokov, M.K. Karyotype characteristics, chromosomal polymorphism and gene COI sequences of Chironomus heteropilicornis Wülker, 1996 (Diptera, Chironomidae) from the South Caucasus. Comp. Cytogenet. 2019, 13, 339–357. [Google Scholar] [CrossRef]
- Bolshakov, V.V.; Prokin, A.A. Karyotype and COI sequences of Chironomus sokolovae Istomina, Kiknadze et Karyotype and COI sequences of Chironomus sokolovae Istomina, Kiknadze et Siirin, 1999 (Diptera, Chironomidae) from the bay of Orkhon River, Mongolia. Comp. Cytogenet. 2021, 15, 149–157. [Google Scholar] [CrossRef]
- Bolshakov, V.V.; Movergoz, E.A. Karyotype and COI gene sequences of Chironomus melanotus Keyl, 1961 from the Yaroslavl region, Russia, and the difficulties with its identification using GenBank and BOLD systems. Comp. Cytogenet. 2022, 16, 161–172. [Google Scholar] [CrossRef]
- FigTree v.1.4.3. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 27 September 2022).
- Shobanov, N.A.; Demin, A.G. Chironomus agilis—New species of group plumosus (Diptera, Chironomidae). Zool. Zhurnal 1988, 67, 1489–1497. (In Russian) [Google Scholar]
- Shobanov, N.A. The karyofund of Chironomus plumosus (L.) (Diptera, Chironomidae) I. Standartization of bands according to the Maximova system. Tsitologiia 1994, 36, 117–122. [Google Scholar]
- Golygina, V.V.; Kiknadze, I.I. The revision of chromosome I (AB) mapping in Chironomus plumosus group (Diptera: Chironomidae). Comp. Cytogenet. 2008, 2, 37–55. [Google Scholar]
- Shobanov, N.A. Karyotype and larva morphology of Chironomus oculatus sp.n. (Diptera, chironomidae). Zool. Zhurnal 1996, 75, 1668–1675. (In Russian) [Google Scholar]
- Guryev, V.; Makarevitch, I.; Blinov, A.; Martin, J. Phylogeny of the genus Chironomus (Diptera) inferred from DNA sequences of mitochondrial Cytochrome b and Cytochrome oxidase I. Mol. Phylogenet. Evol. 2001, 19, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Guryev, V.P.; Blinov, A.G. Phylogenetic Relationships among Holarctic Populations of Chironomus entis and Chironomus plumosus in View of Possible Horisontal Transfer of Mitochondrial Genes. Russ. J. Genet. 2002, 38, 239–243. [Google Scholar] [CrossRef]
- Kondo, N.I.; Ueno, R.; Ohbayashi, K.; Golygina, V.V.; Takamura, K. DNA barcoding supports reclassification of Japanese Chironomus species (Diptera: Chironomidae). Entomol. Sci. 2016, 19, 337–350. [Google Scholar] [CrossRef]
- Bolshakov, V.V.; Prokin, A.A.; Artemenko, S.V. Karyotype and COI gene sequence of Chironomus heteropilicornis Wülker, 1996 (Diptera, Chironomidae) from the Gydan Peninsula, Russia. Comp. Cytogenet. 2021, 15, 447–458. [Google Scholar] [CrossRef]
- Karmokov, M.K. Karyotype characteristics and gene COI sequences of Chironomus bonus Shilova et Dzhvarsheishvili, 1974 (Diptera, Chironomidae) from the South Caucasus (Republic of Georgia, Paravani river). Comp. Cytogenet. 2022, 16, 19–38. [Google Scholar] [CrossRef]
- Roslin, T.; Somervuo, P.; Pentinsaari, M.; Hebert, P.D.N.; Agda, J.; Ahlroth, P.; Anttonen, P.; Aspi, J.; Blagoev, G.; Blanco, S.; et al. A molecular-based identification resource for the arthropods of Finland. Mol. Ecol. Resour. 2022, 22, 803–822. [Google Scholar] [CrossRef]
- Demin, A.G.; Polukonova, N.V.; Mugue, N.S. Molecular phylogeny and the time of divergence of minges (Chironomidae, Nematocera, Diptera) inferred from a partial nucleotide sequence of the cytochrome oxidase I gene (COI). Russ. J. Genet. 2011, 47, 1168–1180. [Google Scholar] [CrossRef]
- Song, C.; Wang, X.; Bu, W.; Qi, X. Morphology lies: A case-in-point with a new non-biting midge species from oriental China (Diptera, chironomidae). Zookeys 2020, 2020, 67–77. [Google Scholar] [CrossRef]
- Shobanov, N.A. The Genus Chironomus Meigen (Diptera, Chironomidae) (Systematics, Biology, Evolution). Ph.D. Dissertation, Saint-Petersburg State University, Saint-Petersburg, Russia, 2000. [Google Scholar]
- Golygina, V.V.; Kiknadze, I.I.; Burlak, V.A. Response of an inversion-polymorphic populations of Chironomus plumosus and Chironomus balatonicus to Infection by the entomopathogenic bacterium Bacillus thuringiensis israelensis. Sib. Ecol. J. 2000, 4, 523–528. [Google Scholar]
- Sasa, M.; Kawai, K. Studies on the chironomids midges of Lake Biwa (Diptera, Chironomidae). Lake Biwa Study Monogr. 1987, 3, 1–119. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).