Abstract
Assessing the applicability of theory to major adaptive radiations in deep time represents an extremely difficult problem in evolutionary biology. Neoaves, which includes 95% of living birds, is believed to have undergone a period of rapid diversification roughly coincident with the Cretaceous–Paleogene (K-Pg) boundary. We investigate whether basal neoavian lineages experienced an ecological release in response to ecological opportunity, as evidenced by density compensation. We estimated effective population sizes (Ne) of basal neoavian lineages by combining coalescent branch lengths (CBLs) and the numbers of generations between successive divergences. We used a modified version of Accurate Species TRee Algorithm (ASTRAL) to estimate CBLs directly from insertion–deletion (indel) data, as well as from gene trees using DNA sequence and/or indel data. We found that some divergences near the K-Pg boundary involved unexpectedly high gene tree discordance relative to the estimated number of generations between speciation events. The simplest explanation for this result is an increase in Ne, despite the caveats discussed herein. It appears that at least some early neoavian lineages, similar to the ancestor of the clade comprising doves, mesites, and sandgrouse, experienced ecological release near the time of the K-Pg mass extinction.
1. Introduction
Biologists have speculated about the tempo of evolutionary change since the dawn of evolutionary biology, with Darwin famously embracing gradualism (e.g., [1,2], but see [3,4] for a more nuanced discussion of Darwin’s views). However, early evolutionary biologists held many different views; in fact, Darwin’s half-cousin Francis Galton advocated the idea that evolutionary change occurred in bursts [5]. Ultimately, George Gaylord Simpson brought this topic into the modern synthesis when he published his “Tempo and Mode in Evolution” [6], where he argued that the tempo of evolutionary change was quite variable. Simpson also argued that the facts regarding the fossil record were consistent with the then emerging field of population genetics. Despite those arguments, Simpson was pessimistic regarding the potential for genetics to contribute directly to the study of tempo and mode, observing that geneticists have “…the grave defect that they cannot reproduce the vast and complex horizontal extent of the natural environment and, particularly, the immense span of time in which population changes really occur” [6] (p. xxix). Overall, his view was that geneticists may “…reveal what happens to a hundred rats in the course of 10 years… but not what happened to a billion rats in the course of 10 million years under the fluctuating conditions of earth history.” Much has changed in the 75 years since Simpson expressed these pessimistic views regarding the use genetics to study tempo and mode over deep time; the era of phylogenomics has made it possible to collect large amounts of sequence data that can be combined with information from the fossil record and sophisticated models to yield exactly the types of information the Simpson viewed as solely available from the fossil record.
Arguably, the first efforts to study tempo and mode in a quantitative framework using molecular data began with the realization that the shapes and branch lengths of phylogenetic trees carried substantial information (reviewed by [7,8]). These types of studies led to the inference of major changes in the tempo of vertebrate diversification in deep time [9]. These types of inferences permitted rates of cladogenesis in deep time to be connected with the theory of adaptive radiations, which Schluter [10] defined as, “a proliferation of species within a single clade accompanied by significant interspecific divergence in the kinds of resources exploited and in the morphological and physiological traits used to exploit those resources.” Although there is some disagreement about what constitutes adaptive radiation and how it is recognized (reviewed by Glor [11]), it seems clear that the major diversifications recognized by Alfaro et al. [9] represented short periods of very rapid, even explosive, cladogenesis. While both explosive and adaptive radiations are characterized by a rapid rate of phyletic divergence, adaptive radiations are distinct in that divergent selection is hypothesized to be the driver of rapid speciation and phenotypic divergence.
It has long been argued based on the fossil record that the basal lineages of Neoaves underwent an explosive and possibly adaptive radiation following the Cretaceous–Paleogene (K-Pg) mass extinction (e.g., [12,13,14,15]). The conflicting “long fuse” model of neoavian evolution was based on older timetree studies (e.g., [16,17,18]), some of which used dubious fossil calibrations and/or strong assumptions regarding vicariance phylogeographic hypotheses (e.g., [19,20,21]). There exist some recent paleontological studies that infer the divergence of some taxonomically restrictive clades in the late Cretaceous [14,22]. However, with only a few exceptions [23], recent phylogenomic-scale timetree analyses of birds that have used large, well-vetted fossil databases [24,25,26,27,28,29] have largely reconciled the molecular and paleontological perspectives, and they have corroborated the hypothesis that some basal lineages of neoavian birds experienced an explosive radiation very close in time to the K-Pg boundary, while others such as the speciose Passeriformes did not. Indeed, modern timetrees indicate that early divergences among most supraordinal clades and many lineages of modern orders appear to have occurred within a span of roughly 10 million years. Intervals between some successive divergences occurred in as little as <1 million years (MY). It is worth noting that this rate of speciation is not exceptional by modern standards [30]. Thus, the apparent burst of origin of lineages near the K-Pg boundary could result simply because the lineages that arose during this period did not go extinct, rather than an acceleration of speciation. Although direct evidence for selection is lacking, it can be inferred that the neoavian radiation was adaptive in as much as several distinct and highly diverged ecomorphological apomorphies are recognizable by the mid- to late-Paleocene (e.g., penguins [22], tropicbird-like pelagics [31,32], mousebirds [14,33], and owls [34,35]), and much of the ecological space currently occupied by extant bird families was filled by the early Eocene [36]. Indeed, the ability of species to rapidly diverge and exploit unoccupied niche space may be central to why rapidly produced species did not become extinct.
However, the fossil record and molecular clock analyses have a number of limitations that could impact the inference that an explosive or adaptive radiation occurred at the base of Neoaves near the K-Pg boundary. For example, one could argue that Neoaves is actually characterized by a number of “ghost lineages” that extend into the Cretaceous. Similarly, one could argue that variation in the rate of avian molecular evolution has confounded relaxed clock analyses to the point where the results of those analyses are meaningless. Finally, the fact that fossils are used to calibrate timetrees produced using relaxed clock analyses raises the possibility that these two sources of error could interact. However, these sources of error are not particularly compelling. The dearth of neornithine fossils vis-à-vis abundant archaic non-neornithine birds in uppermost Cretaceous deposits diminishes concerns of preservational bias upon which the ghost lineage hypothesis rests. Likewise, many potential biases in relaxed clock analyses would tend to bias timetrees toward older dates rather than more recent dates [37,38]. Indeed, the inference of a rapid radiation at the base of Neoaves seems relatively clear from the very short branches in multi-locus phylogenetic trees without corrections to make the trees ultrametric [24,25,39,40]. Even some analyses that argued against a radiation near the K-Pg boundary find evidence that branches near the base of Neoaves are quite short [41]. Despite the relatively compelling nature of these arguments, it would be desirable to find ways to test the adaptive (or explosive) radiation hypothesis for Neoaves using data that are independent of the fossil record.
There is indeed evidence of an explosive radiation of Neoaves that is independent of the fossil record or timetrees. The discordance among transposable element (TE) insertions [24,42,43,44], molecular characters for which homoplasy is very limited (but potentially non-zero [45]), provides evidence for extensive conflict among gene trees at the base of Neoaves. That conflict is consistent with the hypothesis that there was extensive incomplete lineage sorting (ILS) near the base of the neoavian tree that has no doubt contributed to the difficulty of confidently resolving branching order among major lineages of Neoaves (e.g., [40,44,46]). Parsimony analyses of indel (insertion–deletion) data also provided evidence for extensive ILS associated with the short branches at the base of the neoavian tree (see Figure 3e,f in Jarvis et al. [24]). Of course, conflicts among gene trees estimated from nucleotide sequences have long been appreciated [24,25,39,40,46,47,48], and this conflict has also been interpreted as evidence for ILS. However, conflict among gene trees can be driven by error in gene trees; in contrast, conflict among low-homoplasy genomic characters (i.e., rare genomic changes) provide much more direct evidence for ILS (reviewed by Braun et al. [49]). Gene tree discordance due to ILS decreases exponentially as branch lengths increase (Figure 1). However, both time and effective population size (Ne) are critical components of coalescent branch lengths (CBLs). Thus, the evidence that the branches at the base of Neoaves in the avian species tree are characterized by large amounts of conflict due to ILS provides direct evidence that those branches represent extremely short periods of time.
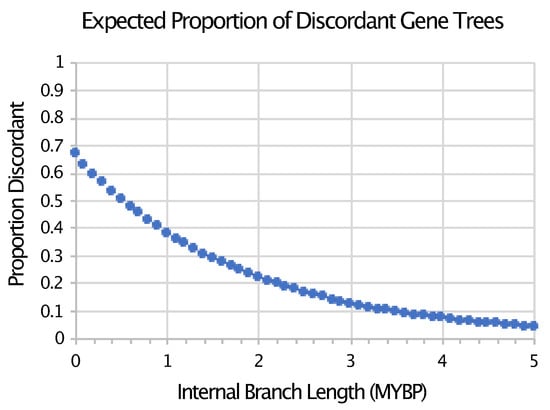
Figure 1.
Expected proportion of discordant gene trees under the neutral multispecies coalescent for a species tree with a specific internal branch length. This figure shows the exponential decay of the expected proportion of discordant gene trees calculated as described by Pamilo and Nei [50] given the simplest case (i.e., sampling individual gene trees from a rooted three taxon tree with a constant population size and generation time). For this figure, we assumed an eight-year generation time (based on [51]; see Methods) and a constant effective population size (Ne) of 112,451 (the median of the recent Ne values reported by Nadachowska-Brzyska et al. [52]).
Despite the now extensive evidence that Neoaves underwent a rapid or explosive radiation near the K-Pg boundary, the question of whether the early diversification of Neoaves represents an adaptive radiation remains open. Corroborating the hypothesis that ancient explosive radiations, such as those highlighted by Alfaro et al. [9], actually involved phenomena similar to the more recent adaptive radiations would seem to require information beyond the rate of cladogenesis. Genomic evidence of a biotic response to ecological opportunity would help to corroborate the hypothesis that Neoaves experienced ecological release near the K-Pg boundary. Such evidence might be found in genomic signatures of ecological release.
Ecological release describes any of a variety of ways in which a species may respond to opportunity, which may or may not in turn lead to adaptive radiation. The prevailing paradigm is that ecological release may follow and result from the exploitation of ecological opportunity that opens an adaptive landscape, whether by evolution of a novel adaptation, the availability of unexploited resources, release from heterospecific antagonists, or some other factor [53,54,55,56,57,58]. Ecological release may be manifested as a flattening of the fitness landscape, broader habitat or resource use, increased trait variation, and density compensation [56], and this at least hypothetically could sometimes lead to adaptive radiation. Studies of the roles and effects of ecological opportunity and release in adaptive radiation have been predominated by experimental systems [59] and oft-cited (and sometimes relatively small) recent radiations (e.g., White Sands lizards [60], Caribbean anoles [61,62], Lake Victoria cichlids [63], Caribbean birds [64], Hawaiian honeycreepers [65], Galapagos finches [66], Madagascan vangas [30,67], white-eyes [30], and auks [68]), and the outcomes of even these are inconsistent. However, there exists a strong difference in the level of subjective inference about the role of ecological opportunity in the radiation of major clades based on the fossil record and in more process-based studies of smaller ‘insular’ recent clades. Studies of large-scale adaptive radiations are necessarily inferential, based on the fossil record (e.g., marine invertebrates [69], ammonites [70], irregular echinoids [71], tracheophyte plants [72], Neotropical cichlids [73], tetanuran dinosaurs [74], or based on comparative phylogenetic birth/death or Lineage Through Time (LTT) analyses of species richness (e.g., various angiosperm clades [75], birds [76], or based on phenotypic diversity (e.g., suboscine ovenbirds and woodcreepers [77], Neotropical cichlids [73], or based on correlation with time-calibrated geophysical and climatic episodes (e.g., passerines [29,78], or some combination of the above.
Density compensation [79,80] is a form of ecological release that has been of particular interest in neontological studies of adaptive radiations, particularly among founding island populations or invasive species. Density compensation is characterized by a transient increase in population size of a founding population in response to an abundance of resources in the absence of heterospecific competitors and/or predators. We reasoned that it might be possible to find evidence for such a transient increase in effective population size in deep time by using rare genomic changes to estimate the amount of ILS.
Herein, we combine estimates of CBL obtained using genome-scale indel data with information from timetrees and reconstructions of ancestral generation times to yield what we believe to be the first empirical estimates of the relative effective population size (Ne of ancestral lineages in very deep time. Our approach bears some similarity to recently described approaches [81,82] and those of Tiley et al. [83], who estimated CBL using binary data (TE insertions and “perfect” transversions, respectively). However, we implemented these analyses in a modified version of the commonly used multispecies coalescent program Accurate Species TRee Algorithm (ASTRAL) [84,85]. We found that some divergences that were likely to have occurred very close to the K-Pg have anomalously short CBL relative to the branch lengths in timetrees; the simplest explanation for that result is a very large Ne for the ancestors of those lineages. Then, we discuss potential sources of bias for the analytical approach we propose and discuss additional data and improvements to timetree analyses that might allow a further corroboration (or refutation) of the inferences we report here.
To the extent that our data show evidence of a transient increase in relative Ne of ancestral lineages, they for the first time provide empirical demographic evidence of a biotic response to hypothesized ecological opportunity that has been independently invoked as a causative agent of adaptive radiation of a major clade.
2. Materials and Methods
We used intronic indels from up to 2515 loci downloaded from the data repository described by Jarvis et al. [86]. Indels were scored using 2matrix [87] using the simple indel coding method [88]. Then, we used the perl programs used in Houde et al. [89] to generate nexus files [90] with charsets corresponding to the following size partitions: (1) all indels (3,918,322 characters); (2) multiresidue indels (≥ 2 base pairs [bp] indels; 702,874 characters); (3) indels with a length ≥10 bp indels (192,851 characters); (4) indels with a length ≥ 50 bp (23,945 characters); and (5) indels with a length ≥100 bp (13,430 characters). Then, indels for each of these size partitions were concatenated to generate nexus files with locus annotation (although these were not analyzed as gene trees), generating five files (one for all intronic indels and four that correspond to the size partitions indicated above). Then, parsimony informative indels were exported to yield relaxed phylip format files used as input by binary-ASTRAL (a variant of the program ASTRAL, available from [91]). Binary-ASTRAL accepts a file with binary characters that are assumed to represent gene tree bipartitions and uses those data to score input trees, outputting information that includes the quartet support, local posterior probabilities for branches, and CBL. We used two trees from Jarvis et al. [24] as input trees: (1) the “total evidence nucleotide tree” (TENT), which was generated by a concatenated maximum likelihood analysis of approximately 42 mega base pairs (Mbp) of sequence data; and (2) the MP-EST* tree, which was generated by applying a multispecies coalescent method (unweighted statistical binning [92] combined with MP-EST* [93]) to the same data.
To complement the direct analyses of indels as binary characters, we also estimated coalescent branch length (CBL) given gene trees estimated using nucleotides, indels, or a combination of nucleotide and indel data. Briefly, gene trees were estimated using IQ-TREE v. 1.6.10 using the best-fitting nucleotide model, the best-fitting binary model, or both (for analyses using a nucleotide partition and an indel partition). The best-fitting models were determined using the -m TEST option. The searches were conducted using the -czb option, which collapses zero-length branches to form polytomies, but we did not collapse any other branches (e.g., based on low bootstrap support). Then, the Jarvis et al. [24] MP-EST* and TENT topologies were scored with ASTRAL 5.6.3 using those gene trees. We conducted the analyses using two different indel weighting schemes: (1) equal weighting of all indels (filenames in the supporting data include “unwt”); and (2) long (≥100 bp) indels upweighted 25x relative to shorter indels and nucleotides (if the latter were included in the analysis) (filenames in the supporting data include “wt25”). The goal of conducting analyses using weighted long indels was to overcome the relatively higher parsimony-measured homoplasy of short indels [89]. The underlying assumption that motivated the wt25 analyses was that observed homoplasy for short indels represents a combination of hemiplasy [94] and true homoplasy, but the observed homoplasy for long indels was essentially limited to hemiplasy. We conducted a total of eight analyses: five analyses (nt, indel unwt, indel wt25, nt + indel unwt, and nt + indel wt25) used gene trees estimated for the 1938 loci that have at least one parsimony informative long (≥100 bp) indel, and the remaining three analyses used gene trees estimated for all 2515 loci with informative indels (nt, indel unwt, and nt + indel unwt).
We used the TENT and MP-EST* timetrees generated by Jarvis et al. [24] for divergence time and branch length estimates. We combined the timetree information with ancestral state reconstructions of generation lengths (Table S1) generated using the fastanc function of phytools v.0.7.16 and R v.3.6.2 [95] and using generation lengths for extant species obtained from IUCN [51] (Table S2). Since CBL for diploid organisms is simply the length of internal branches in generations divided by 2Ne, it is straightforward to calculate an implied Ne as Ne = t/(2 ℓc g), where t is the timetree branch length in years, ℓc is the estimated CBL, and g is the ancestral generation time. Although we speak of Ne estimates herein, we emphasize the potential sources of bias in this analysis and call attention to differences in these estimates using different data partitions (see Discussion). Given the various sources of bias, we believe that the approach described herein should be viewed, at this point, as a method to highlight branches with unusually low CBL given their length in the timetrees; the simplest interpretation of such branches is that they reflect cases where Ne has increased.
We refer to lower taxa by their vernacular English names for ease of reading, but we generally follow the nomenclature used by Jarvis et al. [24] for higher taxa. Note that Pelecaniformes includes the traditionally recognized “Ciconiiformes” [96]. Our use of Aequornithia does not include tropicbirds and Sunbittern, whereas “waterbirds” does. See Table S3 for a full account of species representing higher taxa in this study.
3. Results
The normalized Quartet Score (QS) reported by ASTRAL [84] indicates the proportion of indels or gene trees that are congruent with a reference species tree in quartet analysis; thus, QS provides a criterion that can be used to evaluate both data and tree topologies. QS was consistently higher (albeit to a modest degree) for the MP-EST* tree than for the TENT (Table 1). Arguably, this is not surprising for the analyses of gene trees. After all, the MP-EST* tree was generated using a multispecies coalescent method with some conceptual similarities to ASTRAL. In contrast, the TENT was generated by an analysis of concatenated data. However, conducting direct analyses of indels provides a different way of examining phylogeny, and these results indicate that the distribution of indels, including large (and possibly homoplasy-free) indels, provides support for the MP-EST* tree when it is interpreted in the framework of the multispecies coalescent. These results suggest that the MP-EST* tree may be closer to the true avian species tree, so we report results based on the MP-EST* tree henceforth (see Supplementary Materials for all results of analyses using the TENT).

Table 1.
Normalized quartet scores.
The QS of long indels was considerably higher than those of indel partitions that included short indels (Table 1). The QS of gene trees, which are the source of information in used in the majority of multispecies coalescent analyses, was quite similar to the QS for short indels. In quartet analysis of gene trees, 25x weighting of long indels yielded lower QS than unweighted indels. In addition, within the gene tree analyses, QS was higher using nucleotides, whether alone or in combination with indels. Perhaps surprisingly, the QS for gene trees was always much lower than the QS for long indels; this was even the case with wt25 combined evidence trees that should be highly consistent with the long indel data. This probably reflects the fact that there are very few long indels per locus, so most branches in those gene trees are based on the nucleotide and short indel data. These results emphasize the potential value of long indels (and possibly other types of rare genomic changes) for phylogenetics (see Braun et al. [49] for additional discussion). Based on these results, we emphasize analyses of long indels in this study. We represent ≥50 bp indels as the best compromise between sample size and QS (see Supplementary Materials for analyses using all indel size classes and gene trees).
The highest estimated Ne values ranged from 7,577,755 to 44,060,394 across indel size partitions, with higher estimates in more size inclusive partitions (Figure 2 and Table 2). The lowest estimated Ne values ranged from 118,644 to 321,754 across indel size classes, with higher estimates in more size inclusive partitions. The highest Ne estimates common to both trees and across all indel size partitions was the Columbimorphae branch.
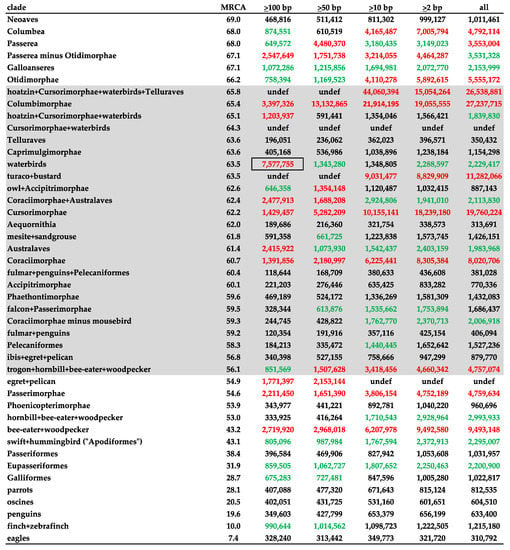
Figure 2.
Estimated Ne of all indel size classes using the MP-EST* tree. The shaded area corresponds to the Paleocene epoch. Red values are equal or greater than the upper quartile; green values are equal of greater than the median. The outlined value is a deviation from the observed trend of increasing Ne estimates for more inclusive sets of indels. This apparently spurious value may be due to the paucity of long indels tracking very short branches. MRCA = most recent common ancestor, undef = cannot be calculated because coalescent branch length is zero. See text for additional details.

Table 2.
Estimated Ne median and upper quartiles for the MP-EST* tree.
Bee-eater + woodpecker and Passerimorphae fall within the upper quartile of Ne estimates more recently than 55 MYBP. Egret + pelican fall within the upper quartile of Ne estimates more recently than 55 MYBP where Ne could be calculated. Preceding 66 MYBP on the MP-EST* timetree, Passerea, upper quartile Columbea, Otidimorphae, and Passerea minus Otidimorphae are within the upper quartile in two to four of the indel size partitions.
All indel size partitions revealed a distinct spike in Ne estimates near the K-Pg boundary (Figure 3 and Figure 4, Figures S1 and S2). All gene tree analyses showed a similar distinct spike in estimated Ne near the K-Pg boundary (see Supplementary Materials; Figures S3–S8; Tables S4, S10 and S11). Based on the median and upper quartile for the Ne estimates, nucleotide gene trees produced much lower Ne estimates than indel gene trees but similar values to those based on combined nucleotide + indel gene trees (Table 2). Only ≥ 100 bp ≥ 50 bp indels produced values comparable to those of nucleotide gene trees.
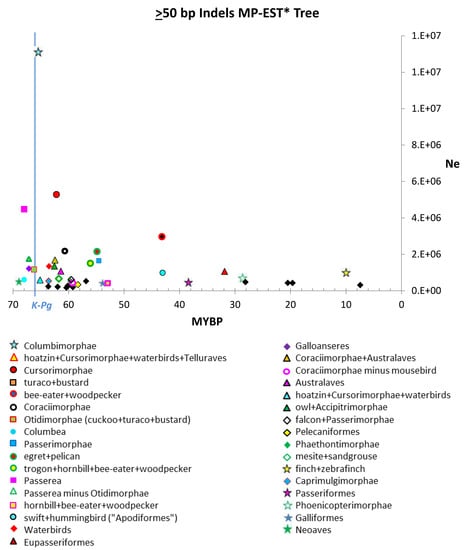
Figure 3.
Estimated Ne of ≥ 50 bp indels size partition plotted on MP-EST* timetree time scale. There is a spike in some Ne estimates near the Cretaceous–Paleogene (K-Pg) boundary. Ne could not be estimated for the hoatzin + Cursorimorphae + waterbirds + Telluraves branch and the turaco + bustard branch because coalescent branch length (CBL) was equal to zero. MYBP = million years before present.
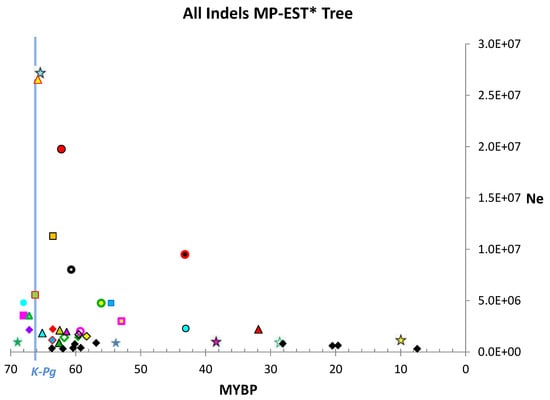
Figure 4.
Estimated Ne of “all indels” size partition plotted on MP-EST* timetree time scale. There is a spike in some Ne estimates near the K-Pg boundary, similar to that observed in analysis of only long indels, which are less homoplasious but much less numerous. Symbols as in Figure 3. MYBP = million years before present.
We calculated the ancestral generation length that would be required to reduce our Ne estimates for each branch into the median and upper quartile of each size partition (Table S21). Longer generation lengths were typically implied using short indels, which are characterized by shorter CBL. Of course, the recalculated ancestral generation lengths were negligible for clades whose Ne estimates were originally below the median and upper quartile. Among the clades with the highest estimated Ne, Columbimorphae required a generation length of 22.1–60.1 years (yrs) for the upper quartile and 41.2–167.5 yrs for the median. Values for hoatzin + Cursorimorphae + waterbirds + Telluraves ranged from 31.5 to 118.9 yrs for the upper quartile and 76.5–272.8 yrs for the median. Waterbirds also had a particularly higher implied generation lengths of 4.0–59.5 for the upper quartile and 9.1–110.9 for the median. Although turaco + bustard had among the highest estimated Ne in many size partitions, its implied generation lengths were lower: 16.6–24.7 yrs for the upper quartile and 40.3–50.2 yrs for the median.
4. Discussion
4.1. Estimates of Ne
The availability of large genomic datasets has for the first time opened the possibility for estimating effective population size of common ancestral lineages in deep time using a multispecies coalescent approach. Hemiplasy, i.e., the conflict in allelic versus population phylogeny specifically due to ILS [94], was modeled to be a function of the time between successive divergence events as measured in coalescent units, measured in generations and influenced by Ne [50]. We postulate that estimates of hemiplasy and time measured in coalescent units might be used to solve for Ne in the basal lineages of Neoaves.
In order to accomplish this, it is foremost necessary to have accurate measurements of hemiplasy that are not inflated either by allelic homoplasy or by apparent gene tree conflict that is not adequately supported by the data. To this end, we employ insertion/deletion (indel) data in comparison and in combination with nucleotide data. Indels have been shown to be less homoplasious than nucleotides as measured character-by-character by parsimony [24,89]. Jarvis et al. [24] found that internal branch length was found to explain 87% of the variation in measured “apparent” indel homoplasy and by extension inferred to closely estimate hemiplasy, since this is the only source of homoplasy expected to be positively correlated with branch length. We caution that this degree of correlation should not be accepted at face value as a direct measure of hemiplasy or of ILS. The Pamilo and Nei model [50] describes the relationship of branch length (in coalescence units) to the percentage of gene trees that are congruent versus incongruent with that branch. However, Jarvis et al. [24] considered only indels with character state transitions that mapped by parsimony to each branch of the tree, rather than all the indels that were either consistent or inconsistent with each branch. Nevertheless, in addition to their relatively low homoplasy, indel data are appealing in that they represent pangenomic binary alleles that may be particularly amenable to estimating CBL. The addition of indels to nucleotide data improved the ability to reject some hard polytomies among basal lineages of Neoaves in coalescent analysis [89].
4.2. Caveats in the Estimation of Ne
4.2.1. Accuracy of Ne Estimates
There are several potential sources of error in our Ne estimation, including incorrect tree topology and concomitant errors in branch length estimation, incorrectly estimated gene trees in the coalescent analysis (because our dataset consisted primarily of a single representative species for each order of extant birds, some of the quartets in our analyses may fall into the “Felsenstein zone” of short internal and two long terminal branches [97]), incorrectly estimated coalescence (i.e., misassigned to branch), incorrectly estimated generation lengths, homoplasy in the data and/or poorly supported gene trees that can lead to artificially high estimates of gene tree discordance, insufficient sampling of indels and/or gene trees, and insufficient sampling of lineages. Evidence that sample size or homoplasy may confound these estimates comes from the observation that more size-inclusive indel partitions (i.e., those that include short indels) produced consistently larger Ne estimates as well as some differences in rank-order (Tables S5–S9); still, a comparable spike in Ne was observed at the K-Pg boundary in much the same lineages). On the one hand, we examined datasets of long indels because they exhibit the lowest measured levels of homoplasy. On the other hand, we examined more size-inclusive partitions including short indels as well, because they comprise orders of magnitude more indels, which are still more parsimony consistent than nucleotides [89].
Most Ne estimates based on the MP-EST* tree and on the TENT are in generally good agreement with one another (Tables S5–S9). This suggests that differences in tree topology do not have a large effect on our estimates. Of course, differences in tree topology necessarily preclude comparison of those specific branches that are not recovered in both trees, and some of the highest estimated Ne were among branches not shared by the two trees. That differences in tree topology seems to otherwise have had little effect on Ne estimation in this case is fortuitous, because no large phylogeny is likely to be historically accurate in its entirety. Moreover, the early divergences among Neoaves are particularly uncertain, and some may even represent a hard polytomy [44,46,86]. Incorrect tree topology necessarily results in zero or negative CBL (“undef” in Tables S4–S11, S13–S22), and it is a likely reason for why we were unable to calculate Ne for as many as two branches on the MP-EST* tree and as many as 6 on the TENT, or perhaps why we may have obtained anomalously high Ne estimates for others. However, we note that differences in tree topology between the MP-EST* tree and TENT as well as the trees of other authors (e.g., [25]) mostly involve the rearrangement of only very short branches. Since very short branches cannot vary much in length, these differences are unlikely to have introduced much error in our estimates of Ne. Moreover, branch length was not correlated with CBL or Ne (see Section 4.2.6).
However, similarity of Ne estimates on the MP-EST* tree and TENT may not in fact reflect accuracy. It also could be interpreted that the data behave similarly in response to biases shared by both trees, or that the faults of the trees pale in comparison to other biases that make the datasets behave similarly.
4.2.2. Homoplasy and Sequence Alignment
Pamilo and Nei [50] introduced a model by which the probability of concordant versus discordant gene trees is related to the process of lineage sorting of neutral ancestral polymorphisms due to genetic drift. This process is quantifiably related to elapsed time as measured in coalescent units, i.e., generations and effective population size (Ne). The extent to which any data can be used to evaluate hemiplasy or gene tree-species tree discordance depends on how much homoplasy is in the data, in as much as it is difficult if not impossible to distinguish homoplasy from hemiplasy. Indels might seem particularly well suited to estimate levels of hemiplasy for the purpose of estimating Ne because they are measurably more homoplasy-free than nucleotides [24,89], are independent of the nucleotide-based phylogenies to which they may be compared, and as binary characters, they produce simple bipartitions in quartet analysis. Indeed, the strong correlation of internode length (i.e., time elapsed between lineage divergences) and the percent of parsimony-consistent (%Retention Index = 1.0) indels was interpreted as evidence of ILS [24]. However, the reliability of indels in phylogenetic analysis has been questioned at times due to their dependence on sequence alignment [98,99], and this is certainly one of the major sources of homoplasy, especially in regions of low sequence complexity and repeats. This may be of particular concern in genomic datasets that rely heavily on high-throughput genome sequencing technologies that may systematically introduce alignment errors. Tandem repeats and mononucleotide repeats may also introduce significant error during genome assembly and multispecies alignment. There are even some known genome assembly errors with the potential to create artifactual long indels; specifically, some allelic differences can be assembled incorrectly as tandem repeats [100]. We expect improved genome assembly methods to resolve many of those issues. Perhaps as importantly as questions of alignment and genome assembly, indels lag far behind nucleotides in terms of ‘substitution’ models that faithfully describe differences in rates of insertion versus deletion, as well as differences in rate, frequency, and homoplasy associated with differences in indel length and mechanism of gain or loss. We are only beginning to learn their behavior in multispecies coalescent analysis [82,89].
We compared the performance of nucleotide and indel data, separately and combined, in quartet analysis of gene trees. Gene tree concordance generated from nucleotides are the standard form of data used to relate Ne to time in coalescent units [50]. Indeed, QS were higher using nucleotide data than indel data (Table 1), suggesting that nucleotide data perform better than indels at recovering gene trees despite the lesser homoplasy of indels. This paradox might accrue from reduced variance in the vastly larger number of nucleotide characters in any given locus. In addition, counter-intuitively, while estimated Ne quartiles of nucleotide gene trees were lower than those based on indels (Table 2 and Table S12), the very highest Ne estimates that included nucleotide data were generally higher, in some cases seemingly unrealistically higher (e.g., turaco + bustard), than those using indels alone in the gene tree analysis (Tables S10 and S11). It is all the more curious that Columbimorphae wasn’t among the very highest, since it was recovered as such in the size partitioned indel analyses.
We also compared the nucleotide and indel gene tree results to those of indels treated as individual characters. QS of indel ≥2 bp, and “all” the size partitions were somewhat lower but similar to those of gene trees that included nucleotide data (Table 1). However, the QS of size partitions of longer indels were much higher than those of all the gene tree QS. That longer indels yield higher QS is intuitive, since they exhibit lower homoplasy; yet paradoxically, gene trees using weighted indels (nt + wt25, wt25) yielded lower QS than those with unweighted indels, whether or not combined with nucleotides (nt + unwt, unwt).
Springer et al. [81,82] chose to utilize TE insertions because of their alleged negligible homoplasy, neutrality, linkage disequilibrium, and lack of intralocus recombination. Treated as separate loci, TEs could be used to define simple bipartitions using parsimony. However, we note that any individual indel, including TEs, may be reconstructed as coalescing above or below a given node depending either on whether character state transformations are treated as accelerated or decelerated in parsimony analysis (i.e., ACCTRAN versus DELTRAN in PAUP [101]) or depending on the particular taxa sampled in quartet analysis. We, too, are confident that the 2515 loci we studied are physically unlinked, define bipartitions in gene trees, and that the history of recombination events has little or no effect on their evolution. Deletions are sequences that do not exist and are therefore immune to recombination. Insertions are sequences that do exist that hypothetically can be cleaved by recombination when in a heterozygous state. However, the method of simple indel coding scores indels as homologous only if they have identical 5’ and 3’ beginnings and endings. Thus, a recombinant indel could be lost and appear homoplastic indel at worst. While introns are undoubtedly not entirely neutral, the degree to which TEs are neutral remains an open question. A significant advantage to not restrict our analyses to TEs is that there are many more length-inclusive indels, of which TEs are of course a type. With larger datasets, there is less variance. There are very few of the largest indels that coalesce on the shortest branches of the neoavian tree (data not shown), which could lead to sampling artifacts and spurious results. This is the most likely explanation for a few discrepancies between estimates of CBL and Ne based on long indels and those of larger datasets of more size-inclusive indels (outlined values in Figure 2 and Table S4) because timetree branch lengths and generation lengths are invariant across the size class comparisons on the reference tree. It is more difficult to rationalize deviation in intermediate size partitions from the expected uniform trend of increased estimated Ne from least inclusive to most inclusive across all size partitions.
We selected the inclusiveness of indel size partitions on the basis of observed incremental levels of parsimony informativeness that we surmise correspond roughly to different forms of indels (e.g., replication slippage, unequal crossing over, TEs; see Table 2, Figure 3 in [86]). On average, longer indels are generally more parsimony consistent than shorter ones; however, single bp indels are more parsimony consistent than 2–6 bp indels.
We focus on the results produced by estimating CBL and Ne on the MP-EST* tree because normalized quartet scores (Table 1) on it are marginally yet without exception higher than those for the TENT (Table 1). In other words, the indel data are more consistent with the MP-EST* tree than with the TENT. This provides some support for the hypothesis that the MP-EST* tree is a better estimate of deep avian phylogeny than the TENT, albeit within the limits of our taxon sample and indel dataset (i.e., introns of Jarvis et al. [24,86] represent only approximately 2% of the orthologous single-copy portion of the avian genome).
4.2.3. Biases Associated with Relaxed Clock Analyses
We used the published MP-EST* and TENT timetrees of Jarvis et al. [24] for the divergence times in our analyses. Thus, the sequence data we used to score indels were extracted from the same genome assemblies as those used to produce the timetrees, and they consist of the exact same broad sampling of taxa. The timetree analysis of Jarvis et al. was particularly well justified and carefully executed in its choice of fossil calibrations, clocklike loci, and relaxed clock methods used [24,26]. Credence to these divergence dates is afforded to the extent that they have been corroborated by other studies [25,27]. Some analyses that argued for earlier divergences among Neoaves found evidence that branches near the base of Neoaves are quite short. These could be readily reconciled with the K-Pg explosion simply on issues of timetree calibration. For example, some ancient estimates of divergence times for basal Neoaves resulted only from the difference in choice of prior maximum age constraint [23,26]. Others [18,41] depended on calibrations derived from calculated averages that included obsolete literature. We are ultimately interpreting any branch in the species tree for which the CBL is especially short relative to the timetree branch length as evidence for increased Ne. The observation that there are branches near the base of Neoaves where Ne appears elevated (given the criterion we used) indicates that there was a transient spike in Ne as the base of Neoaves, regardless of when the radiation actually occurred. If we assume this transient spike in Ne is evidence of ecological release (see below, Section 4.3) but choose to believe a much earlier origin of Neoaves, it would simply mean that the inferred ecological release occurred during the Cretaceous. Of course, the basis for that ecological release is unclear for those timetrees that place the deepest divergences for Neoaves substantially earlier than the K-Pg boundary; in principle, it could reflect the impact of the “Cretaceous Terrestrial Revolution” [102,103], but that event corresponds to a relatively long period of time and is therefore unlikely to drive an explosive radiation. The simplest hypothesis that explains the Ne spike we observe, if it is evidence of ecological release, is that the K-Pg boundary represents an event that somehow created ecological opportunity and that those timetrees suggesting more recent divergences are in fact accurate.
Much has been written about the sources of error in timetree estimation using relaxed molecular clocks. For example, it has long been appreciated that divergence times themselves are not identifiable; only the product of rate and time is identifiable [104]. In the absence of a perfect fossil record where all extinct taxa are identified correctly and stratigraphic ages have been established with a high degree of precision, it is simply impossible to elicit dates with absolute precision [105]. However, identifiability is not necessary to obtain useful parameter estimates and the situation for standard Bayesian relaxed clock analyses is far from hopeless; the combination genome-scale datasets and improved analytical methods are clearly improving estimates of divergence times (reviewed by Ho [106]). Moreover, newer models that use a total evidence approach (i.e., combined molecular and morphological datasets) to accomplish divergence time estimation have the potential to offer a fundamentally different way to approach relaxed clock analyses [107,108]. An early attempt to use morphological clocks [109] to examine deep divergences within Neornithes produced somewhat older divergence times that more recent molecular studies [24,25,26,27,28] (with the exception of Mitchell et al. [23]). It seems reasonable to expect these more sophisticated relaxed clock analyses to yield more accurate avian timetrees. However, even if the improvements to methods for timetree estimation are relatively modest, it would have a limited impact on the approach, since the relative internal branch lengths are the most important values for our approach
The multispecies coalescent can lead to a problem with the potential to be more pernicious than the standard problems that apply to all relaxed clock analyses (Figure 5); this issue could have an impact on the analyses we have undertaken in this study. Specifically, we would like to assess the lengths of internal branches that reflect the times between speciation events (labeled τ1 and τ2 in the left part of Figure 5), but molecular divergence times actually predate the species divergence times [18,110]. When nuclear loci are used for dating, it reflects a mixture of trees with different divergence times that predate the actual speciation time [111]; herein, we use T1 and T2 to indicate the mean coalescent times for gene trees that are concordant with the species tree (exemplified by the red gene tree in the left part of Figure 5). This issue may not be that problematic when Ne is constant and internal branches are relatively long because differences between the means of the molecular divergence times (T2–T1) should be relatively close to the desired parameter (i.e., the difference between speciation times; τ2–τ1). However, this illustration should also make it clear that changes in Ne can alter estimates of T2–T1) precisely because those changes will have an impact on the average coalescent time. For example, t2 (T2–τ2) would be larger than t1 (T1–τ1) if Ne for the common ancestor of all three hypothetical taxa was larger than Ne for the common ancestor of taxa A and B (and vice versa). Thus, variation in Ne has the potential to distort branch lengths. In general, increased Ne on the internal branch (i.e., the branch uniting species A and B in the tree to the left of Figure 5) will reduce the estimated branch length, whereas reduced Ne will increase the estimated branch length.
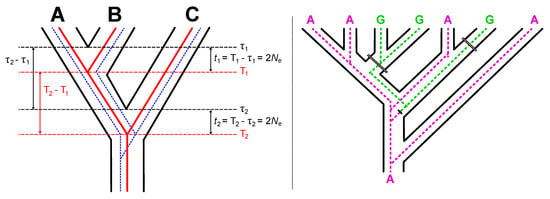
Figure 5.
The impact of the multispecies coalescent on divergence time estimates. Black lines define the species trees and colored lines depict gene trees. The solid red line in the three-taxon tree to the left provides an example of a concordant gene tree. To illustration the difference between speciation times (τ1 and τ2) and molecular divergence times, we have drawn the divergences in for lineages in the red gene tree to correspond to the mean coalescent times (T1 and T2). The dashed blue line is an example of a discordant gene tree; the proportion of discordant gene trees increases as the time between speciation events (τ2–τ1) decreases. The species tree to the right illustrates the impact of a discordant gene tree on the expected proportion of each site pattern. The gene tree shown within that species tree actually underwent a single substitution that changed the ancestral state A (purple) to the derived state G (green). However, the gene tree-species tree discordance results in a site pattern that appears homoplastic when it is actually hemiplastic. Note that maximum parsimony reconstructs two A→G substitutions on the branches, which is indicated using the wide double lines on the species tree. However, there is actually a single substitution deeper in time (the narrower double line on the gene tree). Although likelihood calculations are more complex than the simple parsimony mapping, analyses that do not consider discordance among gene trees will inflate the length of certain branches in a manner similar to this illustration. See text for additional details.
The fact that analyses of data from multiple nuclear genes actually reflects a mixture of trees, some of which are concordant with the species tree but differing in branch lengths and others of which are topologically discordant, makes the estimation of branch lengths even more difficult. As the time between speciation events decreases, the number of trees with a topology identical to the species tree (i.e., the red gene tree in the left of Figure 5) will decrease, whereas the number of discordant trees will increase (e.g., the dashed blue gene tree in the left of Figure 5). Most relaxed molecular clock analyses are actually concatenated analyses, so they should inherit a disturbing behavior of concatenated maximum likelihood phylogenetic analyses [112]. Specifically, the fact that concatenated analyses assume a single underlying tree can cause hemiplastic site patterns to be misinterpreted as multiple substitutions and, in doing so, inflate internal branch lengths. Mendes and Hahn [112] named the phenomenon “SPILS” (substitutions produced by ILS). We expect the SPILS phenomenon to have the potential to distort the estimates of branch lengths in relaxed clock analyses based data from many loci (as in Jarvis et al. [24]).
The multispecies coalescent may distort estimates of internal branch lengths obtained using relaxed clock analyses that rely on concatenated data, but we do not believe that they represent fundamental problems for the approach we used. We say this because a naïve expectation regarding branches associated with elevated Ne is that SPILS would create a bias that yields longer branches in the timetree. However, almost all phylogenetic analyses of Neoaves [24,25,39,40] have very short branches at the base of Neoaves. Thus, any overestimation of branch lengths must be relatively modest. Nevertheless, we believe that this is a potential source of bias that could have an impact on the precise values of Ne. After all, any overestimation of internal branch lengths for the timetree is expected to lead to higher estimates of Ne in absolute terms for a given estimate of the CBL.
Ultimately, our analyses will highlight branches as being associated with an elevated Ne whenever the CBL is especially short relative to the timetree branch lengths. Although there could be some non-linear biases in the precise values of Ne, it should not change the large-scales patterns we infer. We would still highlight cases where Ne underwent an increase. Nevertheless, it would be desirable to find ways to improve our basic approach. An obvious improvement would be the use of a relaxed clock method that directly incorporates the multispecies coalescent; such methods do exist (e.g., StarBEAST [113]), but they are unlikely to be practical for phylogenomic-scale data. However, it might be reasonable to reduce the dataset size for the timetree estimation and fix the topology based on the results of phylogenomic analyses. Alternatively, one might estimate the timetree using loci selected from parts of the genome where Ne is reduced locally. Tiley et al. [83] reported that conserved non-exonic elements (from Edwards et al. [114]) had shorter internal branch lengths than any other data type they examined. They proposed that this reflects a reduction in SPILS due to local reductions in Ne driven by Hill–Robertson interference [115]. If true, standard relaxed molecular clock methods might produce nearly unbiased timetrees if conserved non-exonic element data are used. We believe that exploring these approaches and combining their results with CBL estimates obtained using indels will be interesting.
4.2.4. Generation Length
The ability to accurately estimate coalescent units may be limited by the impossibility of knowing generation lengths of long-extinct ancestral lineages and the uncertainty of timetree dating. Inaccurately estimated generation lengths are a potential source of error in Ne estimation, but they are unlikely to have major effects. With a few notable exceptions such as Procellariiformes, generation length does not vary a great deal among living birds. Reported generation lengths for the extant species used in our study range from 2.7 (Speckled mousebird) to 30.7 years (Northern fulmar) (Table S2), with a mean of 8.6 years and a median of 6.7 years. Despite these shortcomings, reasonable constraints on generation lengths can be inferred by ancestral state analysis (see, e.g., [116]) to estimate relative differences in Ne among lineages and through time.
We used a maximum likelihood model of ancestral state reconstruction of generation length in our analyses (Table S1; [95]). To assess the plausibility that inaccuracies could have resulted in spuriously high estimated Ne, we calculated the generation lengths that would be required to reduce our highest Ne estimates to the levels of the first quartile and median on our timetree (Tables S21 and S22). The resulting generation lengths of up to 119 years to achieve the upper quartile and 273 years for the median are patently untenable. Thus, it does not appear that inaccurately reconstructed generation lengths could solely account for what might be inflated Ne estimates.
4.2.5. Taxon Sampling Bias
Several uncertainties and questions remain even if our Ne estimates are robust. Our observation of elevated Ne near the K-Pg boundary might result from taxon sampling bias. With few exceptions, we studied only singular representatives of extant orders. The divergences among most extant avian orders and supraordinal lineages are estimated to have occurred “within 10–15 million years … during the K-Pg transition, with many interordinal divergences within the 1- to 3-million-year range” [24]. Since the species we studied were mostly single representatives of each order, our sampling was densest and therefore more likely to have been able to detect outliers in that time interval (Figure 2). Our study included a similar number of data the Paleocene (56–66 MY interval) and the collective time before and after that epoch, but the number of data per equivalent 3 MY intervals was higher in the Paleocene (n = 3–11) than before and afterwards (n = 1–6). The percentages of Ne estimates greater or equal to the upper quartile and median within each indel size class did not differ substantially between the Paleocene and the collective time before and after that epoch. Even so, the extent to which some Ne estimates within the Paleocene exceed those before and afterwards is remarkable. Ultimately, the role of sample size and sampling bias will only become clearer from denser and more uniform taxon sampling through time.
It is possible that our Ne estimates are within the range of estimated Ne for current or recent populations, but that our sample of branches later than 56 MY is insufficient to detect a representative range of Ne. Nadachowska-Brzyska et al. [52] reported Pairwise Sequentially Markovian Coalescent (PSMC)-estimated Ne of current populations with a median of 112,451 and upper quartile of 305,674, but with much higher estimates for Pleistocene than current populations for most species and up >4 million for some. Foremost, we note that both of our studies examined relatively few species (the same 48), which themselves may not fully represent the variation of Ne in either extant or extinct birds. We recovered Ne estimates an order of magnitude higher on four branches (i.e., Columbimorphae, Cursorimorphae, Cursorimorphae + waterbirds + Telluraves, and hoatzin + Cursorimorphae + waterbirds + Telluraves on the MP-EST* tree). The extent to which a transient spike in Ne is compelling evidence of density compensation in response to ecological release is if it is observed in lineages that experienced radiation. This is not to say that ecological release necessarily results in radiation or that a given radiation was necessarily adaptive. Ornithologists have identified the neoavian clade as a radiation so explosive that some divergences may represent hard polytomies [44,46]. Thus, it is notable that the Neoaves branch did not yield unusually high Ne estimates, although all six of the first divergences within Neoaves were within the upper quartile based on one or more indel size class datasets (Figure 2, Table 2, Tables S4 and S12). Since the most recent common ancestor (MRCA) of Neoaves somewhat predates the K-Pg [24,25,27], it therefore may not be expected to bear the signature of ecological release due to that event. Ecological opportunity would appear to have presented itself to descendent lineages instead.
Estimates of Ne were >1 million in four of the “magnificent seven” lineages (i.e., Telluraves, Aequornithia, Phaethontimorphae, Otidimorphae, Caprimulgimorphae, Columbimorphae, and Phoenicopterimorphae [40]). The magnificent seven correspond to the major lineages implicated in a potential hard neoavian polytomy near the K-Pg boundary (see below, Section 4.3). Columbimorphae was most consistently the highest in indel analyses. Dismissing the ≥100 bp indels Ne estimate as an outlier, the highest Ne estimates in the MP-EST* tree included Columbimorphae, hoatzin + Cursorimorphae + waterbirds + Telluraves, turaco-bustard, and Cursorimorphae (Figure 2, Tables S5–S9). Cursorimorphae + waterbirds was undefinable. Thus, it is perhaps not coincidental that select waterbirds (i.e., Sphenisciformes [22] and Phaethontiformes [31,32]), presumed cursorimorphs (e.g., “Graculavidae” [117]), owls (Strigiformes [34,35]), mousebirds (Coliiformes sensu lato [14,33]), and caprimulgimorphs [118] have among the richest and oldest fossil records among Neoaves [36,119]. In contrast and notably, Columbimorphae was among our highest Ne estimates (Figure 2, Tables S5–S9), but it has no early Cenozoic fossil record whatsoever.
The long terminal branches of our taxon sample are in many cases an artifact of taxon sampling that betray an untold number of potentially rapid divergences along a lineage. Just as the ancestral character state reconstruction of internal branches on a full reference tree improves with increased taxon sampling and the dissection of long branches, greater taxon sampling should improve the accuracy of CBL inference. We expect that increased lower-taxon sampling will not only better reflect the range of variation in Ne estimates, but it also may improve the accuracy of the estimates themselves.
4.2.6. Other Potential Biases
We regressed both CBL and Ne as a function of timetree internal branch length to test whether our Ne estimates could represent an artifact related to internal branch length. CBL is highly correlated with branch length (R2 >0.85 for all size partitions on MP-EST* tree). This is expected because, by definition, long branches provide more time for new alleles to arise. In contrast, Ne is not at all correlated with internal branch length (R2 < 0.08 for all size partitions on MP-EST* tree, although long indel size partitions had the lowest). Thus, high Ne estimates do not necessarily result from correlation with the very short intervals divergences of the explosive neoavian radiation. That being said, unexpectedly higher Ne estimates due to lower or even zero CBL in longer indel size partitions than in more inclusive indel partitions may be attributable to the paucity of long indels that coalesce on short branches, i.e., a high variance in small samples. This is most likely the case for the ≥100 bp indel partition for waterbirds, which are unquestionably monophyletic.
It may legitimately be asked whether ancient signatures of Ne can persist in the face of successive bouts of ILS along a phylogeny. There is no theoretical reason that Ne should not be temporally stable in neutrally evolving populations that exceed the “50/500” criterion [120] (our estimates do vastly). The popular expectation that Ne is not stable may be rooted in the fact that most studies of Ne in natural populations are biased in focus toward species of conservation concern that have experienced population bottlenecks. Signatures of ILS in recently diverged species are well documented (e.g., [83,121]) and are calculated to persist as long as 10 million years [122]. One important consideration for indels as a marker of ILS is that they are not subject to recombination. If we focus on gene trees, the length of segments with a very ancient coalescent time will be limited by recombination. Overall, recombination is expected to make it nearly impossible to find long regions that have persisted in polymorphic state over long periods of time [123]. In contrast, a reasonably large number of indels might persist as segregating polymorphisms for a large period of time precisely because recombination within an indel is not possible. It is important to recognize the long tail of the distribution of coalescent times (see Figure 1); the probability that any individual indel will fall on the far end of that distribution may be low, but we still expect a large number of indels to fall in that part of the distribution when they are surveyed at the whole-genome level.
Despite the many caveats described above, we believe that we have demonstrated proof in principle of a method to estimate relative differences in effective population size among lineages of common ancestors in deep time, using genomic data taken from single individuals of multiple species. Many questions remain concerning the accuracy of the absolute values of our estimates, the potential causes of estimation error, whether the relatively high Ne estimates near the K-Pg we observe simply reflect sampling bias of ordinal representatives that mostly diverged at that time, and even if the K-Pg Ne spike is real, whether it is evidence of ecological release that can be specifically linked with adaptive radiation in neoavian birds. We expect that the ongoing family-level taxon sampling of birds more uniformly through time (B10K Phase 2 [124]) will do much to resolve these questions.
4.3. Ecological Theory of Adaptive Radiation
Of adaptive radiation, Schluter wrote, “It is regarded as the hallmark of adaptive evolution and may well be the most common syndrome in the origin and proliferation of taxa” [54] (p. 1). Although as stated in the introduction, there remains some disagreement about what exactly constitutes adaptive radiation and its pervasiveness [11]. As noted, any distinction between explosive and adaptive radiations is couched in the primacy of divergent natural selection in accelerating speciation. For instance, an increase in phyletic diversity could result from reduced rates of extinction or from habitat fragmentation and reproductive isolation or from drift along ridges of the adaptive landscape [54]. However, the mere act of inferring rates of diversification in deep time is problematic [125]. Regardless, the prevailing paradigm is that adaptive radiation may follow and result from the exploitation of ecological opportunity, whether by the evolution of a novel adaptation, the availability of unexploited resources, release from heterospecific antagonists—be they competitors or predators, climatic change, etc., that opens an adaptive landscape [53,54,55,56,57,58].
It has long been argued based on the fossil record that neoavian birds experienced an explosive, if not adaptive, radiation following the K-Pg mass extinction (e.g., [12,13,14,15]). The appeal of the hypothesis that K-Pg caused the adaptive radiation of neoavian birds was propelled by widespread acceptance of the global effects of the Chicxulub meteor impact [126,127] (but see [27]) coupled with the known fossil record and recent timetree analyses [36,128].
Ecological theory of adaptive radiation holds that ecological opportunity in the form of increased habitat or resources, the absence of heterospecific competitors or predators [129,130], and/or relaxation of directional or stabilizing selection or other factors [54,56]) may lead to ecological release. Ecological release describes any of a variety of ways in which a species may respond to opportunity. Hypothesized signatures of ecological release include reduced extinction, increased intraspecific competition, a flattening of the fitness landscape, increased phenotypic variance, and, most importantly for our purposes, a transient increase in population size due to density compensation [79,80]. Ecological release, it is theorized, may or may not then result in adaptive radiation (reviewed by Yoder et al. [56]).
Empirical studies of the relationships between ecological opportunity and release and adaptive radiation have thus far been limited to relatively recent small radiations in which information on population demographics is readily available. Most methods for estimating past Ne (e.g., [131,132,133,134,135,136]) cannot be used to address population size in the basal lineages of Neoaves, because it is not possible to sample multiple individuals of hypothetical ancestral lineages. PSMC [116] is exceptional, because it can trace ancient Ne from a single genome based on the coalescence of linked units of heterozygosity. Unfortunately, PSMC loses resolution beyond 3 MY [115], so it is insufficient to evaluate divergences as old as the hypothesized neoavian radiation.
If it were possible to assess ancient population size in the basal lineages of Neoaves, then it might be possible to find evidence of density compensation associated with the ecological release and adaptive radiation. When plotted on a time scale, our Ne estimates reveal a pronounced spike near the K-Pg boundary. On first appearances, these results are consistent with density compensation in response to ecological release at the K-Pg boundary. Many potential heterospecific antagonists of modern birds, be they competitors and/or predators, became extinct by the K-Pg boundary. Potential antagonists included the abundant, diverse, and volant Enantiornithes [15,137]. The Chicxulub meteor impact is hypothesized to have decimated a large part of the terrestrial landscape, which may have provided a vast unexploited habitat concomitant with limited floral and faunal succession, especially for highly vagile species such as birds [138]. Accordingly, we observe a pronounced transient spike in estimated Ne near the K-Pg boundary. Of course, the “spike” we observe is on individual branches among lineages. A subsequent reduction in Ne in subordinate branches is necessary in order to demonstrate that this is indeed a transient spike in Ne in any given lineage. Indeed, we observed reduced Ne on numerous branches of subordinate clades later in time. One biological inference of process according to the theory of adaptive radiation is that increased intraspecific competition resulting from increased abundance may promote phenotypic differentiation and speciation and increasingly complex biotic and trophic interactions. The population size of descendant lineages is expected to return (i.e., decrease) to stable levels as ecological space is filled.
Columbimorphae consistently yielded among the highest Ne estimates in all manner of our analyses, yet apart from Columbiformes, its extant species diversity is low. Thus, the presumed abundance of the stem columbimorph does not appear to have manifested itself in speciose mesite or sandgrouse clades. It is important to remember that ecological release and density compensation do not always result in radiation [54,57]. It is also possible that once-diverse groups have since undergone extensive extinction leaving few descendants [9]. It would be impossible to detect ancient explosive radiations in these clades by sampling extant taxa [139]. In addition, seemingly inconsistent with the high estimated Ne of Columbimorphae is that there is currently no known early Cenozoic fossil record of Columbimorphae whatsoever. One might expect that more abundant species would be more likely to be known from the fossil record, all other things being equal. However, it is conceivable that fossil members of groups such as Columbimorphae may be known but not be recognized as such by paleontologists [128], or that the fossil record of this primarily southern clade has been inadequately sampled [27,140]. High Ne estimates for Columbimorphae suggests that paleontologists might profitably ‘be on the lookout’ both for deposits conducive to their preservation as well as for described paleospecies that may not be recognized as members of this osteologically diverse clade.
4.4. Comparison to Extant Birds
Of course, it would be desirable to make direct comparisons of our Ne estimates to those obtained using other techniques as an independent check of accuracy. Although methods such as PSMC permit estimation of Ne in lineages deeper than divergence events, they unfortunately do not permit Ne estimation in the very deep time on which we focused. Conversely, we cannot make direct comparisons with Ne estimates on extant species or recent lineages, because our dataset does not include alleles that can coalesce on terminal or even sufficiently recent branches. Such data would require samples of populations or a sufficient number of species with very recent ancestry. Moreover, our measurements fundamentally contrast with PSMC, because PSMC tracks Ne continuously through time in units of linked heterozygosity. Instead, we interpret our Ne estimates as the harmonic mean of a branch. Neutral ancestral polymorphisms are lost predictably over time through lineage sorting, and Ne may be reduced by population bottlenecks that may have occurred at any time along a branch. New mutations subsequent to a bottleneck restore heterozygosity, but they do not restore ancestral polymorphisms [52]. That being said, the upper quartile and median values of our Ne estimates for all size partitions of indels for deep lineages are considerably higher than those reported as current based on PSMC for the same 48 species we studied. It is notable, though, that most of these were inferred to have had effective population sizes an order of magnitude higher during the Pleistocene than they currently are [52].
Furthermore, the relevance of Ne estimates in extant taxa for judging the accuracy of those near the K-Pg boundary is questionable if the latter document an extraordinary episode of avian demography (see above, Section 4.3). We take some comfort that all but the highest of our estimates of Ne are plausible in comparison to those estimated for living species, notwithstanding wildly variable estimates of Ne in extant taxa that may be due in part to different meanings of Ne and methods of calculating it [141,142,143]. It is worth recalling that Ne is a measure that is based on genetic diversity that may differ significantly from census population size. Measurements of Ne may also be affected by population structure [144,145,146,147,148], selection [149,150], range expansion [151,152], introgressive hybridization, and intralocus recombination [131], even in the context of the same meaning of Ne and when measured by the same method. The scale at which these effects are measured is also relevant. For example, it is often assumed that the Ne of founding populations will be small and selection will be strong in novel environments. However, Ne is expected to increase with range expansion, as has been observed for many invasive species and as would be expected in a scenario of range expansion following the Chicxulub meteor impact. However, there may be a significant difference between the reduced Ne of marginal populations that are not well adapted to local conditions (which have been the conservation focus of most studies of existing small populations) and the increased Ne of the metapopulation, depending on the level of population structure and gene flow [152]. Similarly, the ecological theory of adaptive radiation predicts that selection will vary through time, beginning with a reduction in normalizing selection and flattening of the selection landscape, followed by local increases in selection driving speciation either in response to local conditions or due to intraspecific competition [56]. Selection may vary considerably among loci used in any particular genetic analysis of Ne [152], and again among sub-populations (whereas indels are assumed to be neutral markers). Most contemporary studies amplify these disparities, since they have overwhelmingly focused on demic populations of conservation concern. These and other well-known factors such as biased sex ratio, and sex- or age-biased reproductive variance, could contribute to differences between censused and effective population sizes [145]. For all these reasons, our results may have limited value for direct comparison with studies of extant species that have explored the role and magnitude of density compensation in response to ecological opportunity using either censused or effective population size.
The intervals of time between successive divergences among many basal lineages of Neoaves was estimated to be as little as <1 MY [24]. This is well within the timeframe when introgressive hybridization persists among extant birds [153]. Introgressive hybridization has much the same effect as ILS in as much as ancestral polymorphisms may be reintroduced into the gene pool, which hypothetically would result in shortened apparent CBL and elevated Ne. Introgressive hybridization can result in asymmetry of incongruent quartet gene trees, so it is perhaps worth noting that Houde et al. [89] found an asymmetry of incongruent quartet gene trees among the deep branches of Neoaves. Unfortunately, they did not perform a symmetry test nor did they investigate where the incongruent trees mapped in the genome, as recommended by Springer et al. [81].
We emphasize that we do not advocate the accuracy of our Ne estimates in terms of their absolute values, particularly because we generally obtained higher Ne estimates using more size-inclusive indel datasets than with the longest indels, especially the highest estimates. Either may have its benefits or biases. Short indels are several orders of magnitude more numerous and exhibit much lower variance, but they are measurably more parsimony-homoplasious than long indels and yield shorter CBL. Despite the uncertainty of the magnitude of our Ne estimates and the causes of differences of estimates from long and short indels, both produced a distinct spike in estimated Ne near the K-Pg boundary (Figure 3 and Figure 4, Figures S1 and S2). The fact that gene tree analyses comprising nucleotide and/or indel data produced a similar spike (Figures S3–S8) suggests that indels are providing meaningful relative measures of Ne among lineages and through time in any individual analysis.
5. Conclusions and Future Directions
We estimated Ne in basal lineages of birds using indels and gene trees generated from nucleotides, indels, or both in the context of well-understood relationships between gene tree concordance, time, and demography couched in coalescent theory. All datasets produced somewhat different absolute estimates of Ne, but qualitatively similar estimates relative to others among the lineages within each analysis. Long indels generated more credible estimates than gene trees based on QS, regardless of whether gene trees comprised nucleotides, indels, or both. Although the similarity of absolute value and rank order of Ne estimates calculated on the multispecies coalescent MP-EST* tree and concatenation TENT of Jarvis et al. [24] suggest that long indels may be somewhat less susceptible than shorter indels to whatever factors may complicate Ne estimation, their relative paucity may under-sample coalescent events associated with very short branches characteristic of rapid divergences and occasionally lead to spurious results. We discuss numerous caveats and sources of potential error in our estimates related to homoplasy, generation length, taxon and temporal sampling, tree topology, and divergence time estimates.
We observed a spike in estimated Ne among some of the basal-most lineages of Neoaves beginning at the K-Pg boundary. This observation is consistent with density compensation in response to ecological opportunity. It is also consistent with the hypothesis that the K-Pg mass extinction spurned an explosive radiation of neoavian birds, whether that was adaptive or non-adaptive. To the best of our knowledge, such demographic data have never previously been available for the ancestral lineages of major clades, largely because it generally requires population sampling.
The proximate tests of the validity of our results from the perspective of their relevance to ecological theory will accrue from ongoing improved taxon sampling that will provide more uniform sampling of divergences through time. The relevance of our results to the ecological theory of adaptive radiation will ultimately depend on complementary studies of rates of speciation and of ecological specialization near the K-Pg boundary. Nevertheless, we view our result as yet another piece in an ever-accruing collection of data that are consistent with and therefore afford confidence to the hypothesis that Neoaves underwent an adaptive radiation in response to ecological release following the K-Pg mass extinction. Perhaps more importantly, we believe that our study represents an, albeit preliminary and imperfect, proof-in-principle for a novel approach for estimating population demography in deep time.
Supplementary Materials
The following is available online at https://www.mdpi.com/1424-2818/12/4/164/s1, Supplementary Materials.zip, including S1: Ne estimates from Indels on TENT, with comparisons to MP-EST* tree, S2: Ne estimates from Gene Trees, S3: Generation Length Sensitivity Analysis, Figure S1: Estimated Ne of size partitioned indels plotted on MP-EST* timetree time scale, Figure S2: Estimated Ne of size partitioned indels plotted on TENT timetree time scale, Figure S3: Estimated Ne of nucleotide gene trees plotted on MP-EST* timetree time scale, Figure S4: Estimated Ne of indel gene trees plotted on MP-EST* timetree time scale, Figure S5: Estimated Ne of nt+indel gene trees plotted on MP-EST* timetree time scale, Figure S6: Estimated Ne of nucleotide gene trees plotted on TENT timetree time scale, Figure S7: Estimated Ne of indel gene trees plotted on TENT timetree time scale, Figure S8: Estimated Ne of nt+indel gene trees plotted on TENT timetree time scale, Table S1: Ancestral state reconstruction of generation length, Table S2: Generation lengths of species studied, Table S3: Species sampled with clade names, Table S4: Estimated Ne on TENT, Table S5: Rank ordered Ne estimates ≥100 bp indels, Table S6: Rank ordered Ne estimates ≥50 bp indels, Table S7: Rank ordered Ne estimates ≥10 bp indels, Table S8: Rank ordered Ne estimates ≥2 bp indels, Table S9: Rank ordered Ne estimates all indels, Table S10: Gene tree estimated Ne of nucleotide and indel data using MP-EST* tree, Table S11: Gene tree estimated Ne of nucleotide and indel data using TENT, Table S12: Upper quartile and median Ne estimates of TENT indel and gene trees, Table S13: Rank ordered gene tree Ne estimates 1938 loci nucleotides MP-EST* vs TENT, Table S14: Rank ordered gene tree Ne estimates 1938 loci unwt indels MP-EST* vs TENT, Table S15: Rank ordered gene tree Ne estimates 1938 loci wt25 indels MP-EST* vs TENT, Table S16: Rank ordered gene tree Ne estimates 1938 loci nts+unwt Indels MP-EST* vs TENT, Table S17: Rank ordered gene tree Ne estimates 1938 loci nts+wt25 Indels MP-EST* vs TENT, Table S18: Rank ordered gene tree Ne estimates 2515 loci nucleotides MP-EST* vs TENT, Table S19: Rank ordered gene tree Ne estimates 2515 loci unwt indels MP-EST* vs TENT, Table S20: Rank ordered gene tree Ne estimates 2515 loci nts+unwt Indels MP-EST* vs TENT, Table S21: Sensitivity analysis of generation length on MP-EST* tree, Table S22: Sensitivity analysis of generation length on TENT. Data files are available from Zenodo at doi.org/10.5281/zenodo.3757429
Author Contributions
Conceptualization, P.H. and E.L.B.; methodology, P.H. and E.L.B.; software, E.L.B.; validation, P.H., E.L.B. and L.Z.; formal analysis, P.H., E.L.B. and L.Z.; investigation, P.H. and E.L.B.; resources, P.H. and E.L.B.; data curation, P.H. and E.L.B.; writing—original draft preparation, P.H. and E.L.B.; writing—review and editing, P.H., E.L.B. and L.Z.; visualization, P.H. and E.L.B.; supervision, P.H.; project administration, P.H.; funding acquisition, E.L.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by US National Science Foundation grant DEB-1655683 to E.L.B.
Acknowledgments
We thank Siavash Mirarab for his substantial contributions to the conceptual development and execution of this study. We also thank the Bird 10,000 Genomes Project (B10K) for providing the genomic data upon which this study was based.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Eldredge, N. Punctuated equilibria: An alternative to phyletic gradualism. In Models in Paleobiology; Schopf, T.J., Ed.; Freeman, Cooper and Company: San Fransisco, IL, USA, 1972; pp. 82–115. [Google Scholar]
- Gould, S.J. Tempo and mode in the macroevolutionary reconstruction of Darwinism. Proc. Natl. Acad. Sci. USA 1994, 91, 6764–6771. [Google Scholar] [CrossRef] [PubMed]
- Penny, D. Charles Darwin, Gradualism and Punctuated Equilibria. Syst. Zool. 1983, 32, 72. [Google Scholar] [CrossRef]
- Rhodes, F.H.T. Darwinian gradualism and its limits: The development of Darwin’s views on the rate and pattern of evolutionary change. J. Hist. Biol. 1987, 20, 139–157. [Google Scholar] [CrossRef]
- Gillham, N.W. Evolution by jumps: Francis Galton and William Bateson and the mechanism of evolutionary change. Genetics 2001, 159, 1383–1392. [Google Scholar] [PubMed]
- Simpson, G.G. Tempo and Mode in Evolution; Columbia Biological Series, No. 15; Columbia University Press: New York, NY, USA, 1944. [Google Scholar]
- Mooers, A.Ø.; Heard, S.B. Inferring Evolutionary Process from Phylogenetic Tree Shape. Q. Rev. Boil. 1997, 72, 31–54. [Google Scholar] [CrossRef]
- Ricklefs, R.E. Estimating diversification rates from phylogenetic information. Trends Ecol. Evol. 2007, 22, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Alfaro, M.E.; Santini, F.; Brock, C.; Alamillo, H.; Dornburg, A.; Rabosky, D.L.; Carnevale, G.; Harmon, L.J. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc. Natl. Acad. Sci. USA 2009, 106, 13410–13414. [Google Scholar] [CrossRef] [PubMed]
- Schluter, L. Ecological Causes of Adaptive Radiation. Am. Nat. 1996, 148, S40–S64. [Google Scholar] [CrossRef]
- Glor, R.E. Phylogenetic Insights on Adaptive Radiation. Annu. Rev. Ecol. Evol. Syst. 2010, 41, 251–270. [Google Scholar] [CrossRef]
- Feduccia, A. ‘Big bang’ for tertiary birds? Trends Ecol. Evol. 2003, 18, 172–176. [Google Scholar] [CrossRef]
- Feduccia, A. Avian extinction at the end of the Cretaceous: Assessing the magnitude and subsequent explosive radiation. Cretac. Res. 2014, 50, 1–15. [Google Scholar] [CrossRef]
- Ksepka, D.; Stidham, T.A.; Williamson, T.E. Early Paleocene landbird supports rapid phylogenetic and morphological diversification of crown birds after the K–Pg mass extinction. Proc. Natl. Acad. Sci. USA 2017, 114, 8047–8052. [Google Scholar] [CrossRef] [PubMed]
- Field, D.J.; Bercovici, A.; Berv, J.S.; Dunn, R.; Fastovsky, D.E.; Lyson, T.R.; Vajda, V.; Gauthier, J.A. Early Evolution of Modern Birds Structured by Global Forest Collapse at the End-Cretaceous Mass Extinction. Curr. Boil. 2018, 28, 1825–1831.e2. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.; Penny, D. Mass Survival of Birds Across the Cretaceous-Tertiary Boundary: Molecular Evidence. Science 1997, 275, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Van Tuinen, M.; Hedges, S.B. Calibration of avian molecular clocks. Mol. Boil. Evol. 2001, 18, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.W.; Rest, J.; García-Moreno, J.; Sorenson, M.D.; Mindell, D.P. Strong mitochondrial DNA support for a Cretaceous origin of modern avian lineages. BMC Boil. 2008, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Cracraft, J. Avian evolution, Gondwana biogeography and the Cretaceous–Tertiary mass extinction event. Proc. R. Soc. B Boil. Sci. 2001, 268, 459–469. [Google Scholar] [CrossRef]
- Paton, T.; Haddrath, O.; Baker, A.J. Complete mitochondrial DNA genome sequences show that modern birds are not descended from transitional shorebirds. Proc. R. Soc. B Boil. Sci. 2002, 269, 839–846. [Google Scholar] [CrossRef]
- Wright, T.; Schirtzinger, E.E.; Matsumoto, T.; Eberhard, J.R.; Graves, G.R.; Sanchez, J.J.; Capelli, S.; Müller, H.; Scharpegge, J.; Chambers, G.; et al. A Multilocus Molecular Phylogeny of the Parrots (Psittaciformes): Support for a Gondwanan Origin during the Cretaceous. Mol. Boil. Evol. 2008, 25, 2141–2156. [Google Scholar] [CrossRef]
- Blokland, J.C.; Reid, C.M.; Worthy, T.H.; Tennyson, A.; Clarke, J.A.; Scofield, R.P. Chatham Island Paleocene fossils provide insight into the palaeobiology, evolution, and diversity of early penguins (Aves, Sphenisciformes). Palaeontol. Electron. 2019, 22, 1–92. [Google Scholar] [CrossRef]
- Mitchell, K.J.; Cooper, A.; Phillips, M.J. Comment on “Whole-genome analyses resolve early branches in the tree of life of modern birds”. Science 2015, 349, 1460. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, E.; Mirarab, S.; Aberer, A.J.; Li, B.; Houde, P.; Li, C.; Ho, S.Y.W.; Faircloth, B.C.; Nabholz, B.; Howard, J.T.; et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 2014, 346, 1320–1331. [Google Scholar] [CrossRef] [PubMed]
- Prum, R.O.; Berv, J.S.; Dornburg, A.; Field, D.J.; Townsend, J.P.; Lemmon, E.M.; Lemmon, A.R. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 2015, 526, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Cracraft, J.; Houde, P.; Ho, S.Y.W.; Mindell, D.P.; Fjeldså, J.; Lindow, B.; Edwards, S.V.; Rahbek, C.; Mirarab, S.; Warnow, T.; et al. Response to Comment on “Whole-genome analyses resolve early branches in the tree of life of modern birds”. Science 2015, 349, 1460. [Google Scholar] [CrossRef] [PubMed]
- Claramunt, S.; Cracraft, J. A new time tree reveals Earth history’s imprint on the evolution of modern birds. Sci. Adv. 2015, 1, e1501005. [Google Scholar] [CrossRef]
- Kimball, R.T.; Oliveros, C.H.; Wang, N.; White, N.; Barker, F.K.; Field, D.J.; Ksepka, D.; Chesser, R.T.; Moyle, R.G.; Braun, M.J.; et al. A Phylogenomic Supertree of Birds. Diversity 2019, 11, 109. [Google Scholar] [CrossRef]
- Oliveros, C.H.; Field, D.J.; Ksepka, D.T.; Barker, K.; Aleixo, A.; Andersen, M.J.; Alström, P.; Benz, B.W.; Braun, E.L.; Braun, M.J.; et al. Earth history and the passerine superradiation. Proc. Natl. Acad. Sci. USA 2019, 116, 7916–7925. [Google Scholar] [CrossRef]
- Moyle, R.G.; Filardi, C.E.; Smith, C.E.; Diamond, J. Explosive Pleistocene diversification and hemispheric expansion of a “great speciator”. Proc. Natl. Acad. Sci. USA 2009, 106, 1863–1868. [Google Scholar] [CrossRef]
- Bourdon, E.; Bouya, B.; Iarochène, M. Earliest African neornithine bird: A new species of Prophaethontidae (Aves) from the Paleocene of Morocco. J. Vertebr. Paléontol. 2005, 25, 157–170. [Google Scholar] [CrossRef]
- Bourdon, E.; Mourer-Chauviré, C.; Amaghzaz, M.; Bouya, B. New specimens of Lithoptila abdounensis (Aves, Prophaethontidae) from the lower Paleogene of Morocco. J. Vertebr. Paléontol. 2008, 28, 751–761. [Google Scholar] [CrossRef]
- Zelenkov, N.V.; Dyke, G. The Fossil Record and Evolution of Mousebirds (Aves: Coliiformes). Palaeontol. 2008, 51, 1403–1418. [Google Scholar] [CrossRef]
- Rich, P.V.; Bohaska, D.J. The world’s oldest owl: A new strigiform from the Paleocene of southwestern Colorado. Smithson. Contrib. Paleobiol. 1976, 27, 87–94. [Google Scholar]
- Kurochkin, E.N.; Dyke, G. The first fossil owls (Aves: Strigiformes) from the Paleogene of Asia and a review of the fossil record of Strigiformes. Paléontol. J. 2011, 45, 445–458. [Google Scholar] [CrossRef]
- Mayr, G. Paleogene Fossil Birds; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Berv, J.S.; Field, D.J. Genomic Signature of an Avian Lilliput Effect across the K-Pg Extinction. Syst. Boil. 2017, 67, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Field, D.J.; Berv, J.S.; Hsiang, A.Y.; Lanfear, R.; Landis, M.J.; Dornburg, A. Timing the extant avian radiation: The rise of modern birds, and the importance of modeling molecular rate variation. PeerJ Prepr. 2019. [Google Scholar] [CrossRef]
- Hackett, S.J.; Kimball, R.T.; Reddy, S.; Bowie, R.C.K.; Braun, E.L.; Braun, M.J.; Chojnowski, J.L.; Cox, W.; Han, K.-L.; Harshman, J.; et al. A Phylogenomic Study of Birds Reveals Their Evolutionary History. Science 2008, 320, 1763–1768. [Google Scholar] [CrossRef]
- Reddy, S.; Kimball, R.T.; Pandey, A.; Hosner, P.A.; Braun, M.J.; Hackett, S.J.; Han, K.-L.; Harshman, J.; Huddleston, C.J.; Kingston, S.; et al. Why Do Phylogenomic Data Sets Yield Conflicting Trees? Data Type Influences the Avian Tree of Life more than Taxon Sampling. Syst. Boil. 2017, 66, 857–879. [Google Scholar] [CrossRef]
- Hedges, S.B.; Marin, J.; Suleski, M.; Paymer, M.; Kumar, S. Tree of life reveals clock-like speciation and diversification. Mol. Boil. Evol. 2015, 32, 835–845. [Google Scholar] [CrossRef]
- Matzke, A.; Churakov, G.; Berkes, P.; Arms, E.M.; Kelsey, D.; Brosius, J.; Kriegs, J.O.; Schmitz, J. Retroposon Insertion Patterns of Neoavian Birds: Strong Evidence for an Extensive Incomplete Lineage Sorting Era. Mol. Boil. Evol. 2012, 29, 1497–1501. [Google Scholar] [CrossRef]
- Suh, A.; Smeds, L.; Ellegren, H. The Dynamics of Incomplete Lineage Sorting across the Ancient Adaptive Radiation of Neoavian Birds. PLoS Boil. 2015, 13, e1002224. [Google Scholar] [CrossRef]
- Suh, A. The phylogenomic forest of bird trees contains a hard polytomy at the root of Neoaves. Zool. Scr. 2016, 45, 50–62. [Google Scholar] [CrossRef]
- Han, K.-L.; Braun, E.L.; Kimball, R.T.; Reddy, S.; Bowie, R.C.K.; Braun, M.J.; Chojnowski, J.L.; Hackett, S.J.; Harshman, J.; Huddleston, C.J.; et al. Are Transposable Element Insertions Homoplasy Free: An Examination Using the Avian Tree of Life. Syst. Boil. 2011, 60, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Poe, S.; Chubb, A.L. Birds in A Bush: Five Genes Indicate Explosive Evolution of Avian Orders. Evolution 2004, 58, 404–415. [Google Scholar] [CrossRef]
- Chojnowski, J.L.; Kimball, R.T.; Braun, E.L. Introns outperform exons in analyses of basal avian phylogeny using clathrin heavy chain genes. Gene 2008, 410, 89–96. [Google Scholar] [CrossRef]
- Kimball, R.T.; Wang, N.; Heimer-McGinn, V.; Ferguson, C.; Braun, E.L. Identifying localized biases in large datasets: A case study using the avian tree of life. Mol. Phylogenet. Evol. 2013, 69, 1021–1032. [Google Scholar] [CrossRef]
- Braun, E.L.; Cracraft, J.; Houde, P. Resolving the Avian Tree of Life from Top to Bottom: The Promise and Potential Boundaries of the Phylogenomic Era. In Avian Genomics in Ecology and Evolution; Springer: Cham, Switzerland, 2019; pp. 151–210. ISBN 978-3-030-16476-8. [Google Scholar]
- Pamilo, P.; Nei, M. Relationships between gene trees and species trees. Mol. Boil. Evol. 1988, 5, 568–583. [Google Scholar] [CrossRef]
- International Union for Conservation of Nature. 2006 IUCN Red List of Threatened Species, 2006. Available online: http://www.iucnredlist.org/ (accessed on 1 March 2008).
- Nadachowska-Brzyska, K.; Li, C.; Smeds, L.; Zhang, G.; Ellegren, H. Temporal Dynamics of Avian Populations during Pleistocene Revealed by Whole-Genome Sequences. Curr. Boil. 2015, 25, 1375–1380. [Google Scholar] [CrossRef]
- Simpson, G.G. The Major Features of Evolution; Columbia University Press: New York, NY, USA, 1953. [Google Scholar]
- Schluter, D. The Ecology of Adaptive Radiation; Oxford University Press: Oxford UK, 2000; ISBN 9780198505228. [Google Scholar]
- Gavrilets, S.; Losos, J.B.; Rowland, J.M.; Emlen, D.J. Adaptive Radiation: Contrasting Theory with Data. Science 2009, 323, 732–737. [Google Scholar] [CrossRef]
- Yoder, J.; Clancey, E.; Roches, S.D.; Eastman, J.M.; Gentry, L.; Godsoe, W.; Hagey, T.J.; Jochimsen, D.; Oswald, B.P.; Robertson, J.; et al. Ecological opportunity and the origin of adaptive radiations. J. Evol. Boil. 2010, 23, 1581–1596. [Google Scholar] [CrossRef]
- Stroud, J.T.; Losos, J.B. Ecological Opportunity and Adaptive Radiation. Annu. Rev. Ecol. Evol. Syst. 2016, 47, 507–532. [Google Scholar] [CrossRef]
- Gillespie, R.; Bennett, G.M.; De Meester, L.; Feder, J.L.; Fleischer, R.C.; Harmon, L.J.; Hendry, A.P.; Knope, M.L.; Mallet, J.; Martin, C.H.; et al. Comparing Adaptive Radiations Across Space, Time, and Taxa. J. Hered. 2020, 111, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Kassen, R. Toward a General Theory of Adaptive Radiation. Ann. N. Y. Acad. Sci. 2009, 1168, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Roches, S.D.; Robertson, J.M.; Harmon, L.J.; Rosenblum, E.B. Ecological release in White Sands lizards. Ecol. Evol. 2011, 1, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Losos, J.B.; De Queiroz, K. Evolutionary consequences of ecological release in Caribbean Anolis lizards. Boil. J. Linn. Soc. 1997, 61, 459–483. [Google Scholar] [CrossRef]
- Mahler, D.L.; Revell, L.J.; Glor, R.E.; Losos, J.B. Ecological Opportunity and The Rate of Morphological Evolution in the Diversification of Greater Antillean Anoles. Evolution 2010, 64, 2731–2745. [Google Scholar] [CrossRef]
- Meyer, A.; Kocher, T.D.; Basasibwaki, P.; Wilson, A.C. Monophyletic origin of Lake Victoria cichlid fishes suggested by mitochondrial DNA sequences. Nature 1990, 347, 550–553. [Google Scholar] [CrossRef]
- Cox, G.W.; Ricklefs, R.E. Species Diversity and Ecological Release in Caribbean Land Bird Faunas. Oikos 1977, 28, 113. [Google Scholar] [CrossRef]
- Lovette, I.J.; Bermingham, E.; Ricklefs, R.E. Clade-specific morphological diversification and adaptive radiation in Hawaiian songbirds. Proc. R. Soc. B Boil. Sci. 2002, 269, 37–42. [Google Scholar] [CrossRef]
- Grant, P.R.; Grant, B.R. How and Why Species Multiply: The Radiation of Darwin’s Finches (princeton Series In Evolutionary Biology), 1st ed.; Princeton University Press: Princeton, NJ, USA, 2008; ISBN 978-0-691-13360-7. [Google Scholar]
- Jønsson, K.A.; Fabre, P.-H.; Fritz, S.A.; Etienne, R.S.; Ricklefs, R.E.; Jørgensen, T.B.; Fjeldså, J.; Rahbek, C.; Ericson, P.G.P.; Woog, F.; et al. Ecological and evolutionary determinants for the adaptive radiation of the Madagascan vangas. Proc. Natl. Acad. Sci. USA 2012, 109, 6620–6625. [Google Scholar] [CrossRef]
- Weir, J.T.; Mursleen, S. Diversity-Dependent Cladogenesis and Trait Evolution in the Adaptive Radiation of The Auks (Aves: Alcidae). Evolution 2012, 67, 403–416. [Google Scholar] [CrossRef]
- Sepkoski, J.J. A factor analytic description of the Phanerozoic marine fossil record. Paleobiology 1981, 7, 36–53. [Google Scholar] [CrossRef]
- Neige, P.; Dera, G.; Dommergues, J.-L. Adaptive radiation in the fossil record: A case study among Jurassic ammonoids. Palaeontology 2013, 56, 1247–1261. [Google Scholar] [CrossRef]
- Boivin, S.; Saucede, T.; Laffont, R.; Steimetz, E.; Neige, P. Diversification rates indicate an early role of adaptive radiations at the origin of modern echinoid fauna. PLoS ONE 2018, 13, e0194575. [Google Scholar] [CrossRef]
- Niklas, K.J.; Tiffney, B.H.; Knoll, A.H. Patterns in vascular land plant diversification. Nature 1983, 303, 614–616. [Google Scholar] [CrossRef]
- López-Fernández, H.; Arbour, J.H.; Winemiller, K.O.; Honeycutt, R.L. Testing for Ancient Adaptive Radiations in Neotropical Cichlid Fishes. Evolution 2013, 67, 1321–1337. [Google Scholar] [CrossRef] [PubMed]
- Rauhut, O.W.M.; Pol, D. Probable basal allosauroid from the early Middle Jurassic Cañadón Asfalto Formation of Argentina highlights phylogenetic uncertainty in tetanuran theropod dinosaurs. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Magallón, S.; Sanderson, M.J. Absolute Diversification Rates in Angiosperm Clades. Evolution 2001, 55, 1762–1780. [Google Scholar] [CrossRef]
- Jetz, W.; Thomas, G.; Joy, J.B.; Hartmann, K.; Mooers, A.O. The global diversity of birds in space and time. Nature 2012, 491, 444–448. [Google Scholar] [CrossRef]
- Claramunt, S. Discovering Exceptional Diversifications at Continental Scales: The Case of the Endemic Families of Neotropical Suboscine Passerines. Evolution 2010, 64, 2004–2019. [Google Scholar] [CrossRef]
- Moyle, R.G.; Oliveros, C.H.; Andersen, M.J.; Hosner, P.A.; Benz, B.W.; Manthey, J.; Travers, S.L.; Brown, R.M.; Faircloth, B.C. Tectonic collision and uplift of Wallacea triggered the global songbird radiation. Nat. Commun. 2016, 7, 12709. [Google Scholar] [CrossRef]
- MacArthur, R.H.; Diamond, J.M.; Karr, J.R. Density Compensation in Island Faunas. Ecology 1972, 53, 330–342. [Google Scholar] [CrossRef]
- Wright, S.J. Density compensation in island avifaunas. Oecologia 1980, 45, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Springer, M.S.; Molloy, E.K.; Sloan, D.B.; Simmons, M.P.; Gatesy, J. ILS-Aware Analysis of Low-Homoplasy Retroelement Insertions: Inference of Species Trees and Introgression Using Quartets. J. Hered. 2019, 111, 147–168. [Google Scholar] [CrossRef]
- Springer, M.S.; Gatesy, J. Retroposon Insertions within a Multispecies Coalescent Framework Suggest that Ratite Phylogeny is not in the ‘Anomaly Zone’. BioRxiv 2019, 643296. [Google Scholar] [CrossRef]
- Tiley, G.P.; Pandey, A.; Kimball, R.T.; Braun, E.L.; Burleigh, J.G. Whole genome phylogeny of Gallus: Introgression and data-type effects. Avian Res. 2020, 11, 7. [Google Scholar] [CrossRef]
- Zhang, C.; Rabiee, M.; Sayyari, E.; Mirarab, S. ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 2018, 19, 153. [Google Scholar] [CrossRef] [PubMed]
- Mirarab, S. Species Tree Estimation Using ASTRAL: Practical Considerations. arXiv 2019, arXiv:1904.03826. [Google Scholar]
- Jarvis, E.; Mirarab, S.; Aberer, A.J.; Li, B.; Houde, P.; Li, C.; Ho, S.Y.W.; Faircloth, B.C.; Nabholz, B.; Howard, J.T.; et al. Phylogenomic analyses data of the avian phylogenomics project. GigaScience 2015, 4, 4–9. [Google Scholar] [CrossRef]
- Salinas, N.R.; Little, D.P. 2matrix: A utility for indel coding and phylogenetic matrix concatenation1. Appl. Plant Sci. 2014, 2, 1300083. [Google Scholar] [CrossRef]
- Simmons, M.P.; Ochoterena, H. Gaps as characters in sequence-based phylogenetic analyses. Syst. Boil. 2000, 49, 369–381. [Google Scholar] [CrossRef]
- Houde, P.; Braun, E.L.; Narula, N.; Minjares, U.; Mirarab, S. Phylogenetic Signal of Indels and the Neoavian Radiation. Diversity 2019, 11, 108. [Google Scholar] [CrossRef]
- Maddison, D.R.; Swofford, D.L.; Maddison, W.P.; Cannatella, D. Nexus: An Extensible File Format for Systematic Information. Syst. Boil. 1997, 46, 590–621. [Google Scholar] [CrossRef] [PubMed]
- GitHub—Smirarab/ASTRAL: Accurate Species TRee Algorithm. Available online: https://github.com/smirarab/ASTRAL (accessed on 21 February 2020).
- Mirarab, S.; Bayzid, M.S.; Boussau, B.; Warnow, T. Statistical binning enables an accurate coalescent-based estimation of the avian tree. Science 2014, 346, 1250463. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yu, L.; Edwards, S.V. A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evol. Boil. 2010, 10, 302. [Google Scholar] [CrossRef]
- Avise, J.C.; Robinson, T. Hemiplasy: A New Term in the Lexicon of Phylogenetics. Syst. Boil. 2008, 57, 503–507. [Google Scholar] [CrossRef]
- Revell, L.J. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2011, 3, 217–223. [Google Scholar] [CrossRef]
- Cracraft, J.C. Avian higher-level relationships and classification: Nonpasseriforms. In The Howard and Moore Complete Checklist of the Birds of the World, 4th ed.; Dickinson, E.C., Remsen, J.V., Jr., Eds.; Aves Press: Northhampton, UK, 2013; Volume 1. [Google Scholar]
- Felsenstein, J. Cases in which Parsimony or Compatibility Methods will be Positively Misleading. Syst. Boil. 1978, 27, 401–410. [Google Scholar] [CrossRef]
- Morgan-Richards, M.; Trewick, S.A.; Bartosch-Härlid, A.; Kardailsky, O.; Phillips, M.J.; McLenachan, P.A.; Penny, D. Bird evolution: Testing the Metaves clade with six new mitochondrial genomes. BMC Evol. Boil. 2008, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Saurabh, K.; Holland, B.; Gibb, G.; Penny, D. Gaps: An Elusive Source of Phylogenetic Information. Syst. Boil. 2012, 61, 1075–1082. [Google Scholar] [CrossRef]
- Korlach, J.; Gedman, G.; Kingan, S.B.; Chin, C.-S.; Howard, J.T.; Audet, J.-N.; Cantin, L.; Jarvis, E.D. De novo PacBio long-read and phased avian genome assemblies correct and add to reference genes generated with intermediate and short reads. GigaScience 2017, 6, 1–16. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods); Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Lloyd, G.T.; Davis, K.E.; Pisani, D.; Tarver, J.E.; Ruta, M.; Sakamoto, M.; Hone, D.W.E.; Jennings, R.; Benton, M.J. Dinosaurs and the cretaceous terrestrial revolution. Proc. Biol. Sci. 2008, 275, 2483–2490. [Google Scholar] [CrossRef] [PubMed]
- Barba-Montoya, J.; Dos Reis, M.; Schneider, H.; Donoghue, P.C.J.; Yang, Z. Constraining uncertainty in the timescale of angiosperm evolution and the veracity of a Cretaceous Terrestrial Revolution. New Phytol. 2018, 218, 819–834. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Rannala, B. Bayesian Estimation of Species Divergence Times Under a Molecular Clock Using Multiple Fossil Calibrations with Soft Bounds. Mol. Boil. Evol. 2005, 23, 212–226. [Google Scholar] [CrossRef] [PubMed]
- Rannala, B. Conceptual issues in Bayesian divergence time estimation. Philos. Trans. R. Soc. B Boil. Sci. 2016, 371, 20150134. [Google Scholar] [CrossRef]
- Ho, S.Y.W. The changing face of the molecular evolutionary clock. Trends Ecol. Evol. 2014, 29, 496–503. [Google Scholar] [CrossRef]
- Ronquist, F.; Lartillot, N.; Phillips, M.J. Closing the gap between rocks and clocks using total-evidence dating. Philos. Trans. R. Soc. B Boil. Sci. 2016, 371, 20150136. [Google Scholar] [CrossRef]
- Zhang, C.; Stadler, T.; Klopfstein, S.; Heath, T.A.; Ronquist, F. Total-Evidence Dating under the Fossilized Birth-Death Process. Syst. Boil. 2015, 65, 228–249. [Google Scholar] [CrossRef]
- Lee, M.S.; Cau, A.; Naish, D.; Dyke, G. Morphological Clocks in Paleontology, and a Mid-Cretaceous Origin of Crown Aves. Syst. Boil. 2014, 63, 442–449. [Google Scholar] [CrossRef]
- Arbogast, B.S.; Edwards, S.V.; Wakeley, J.; Beerli, P.; Slowinski, J.B. Estimating divergence times from molecular data on phylogenetic and population genetic timescales. Annu. Rev. Ecol. Syst. 2002, 33, 707–740. [Google Scholar] [CrossRef]
- Edwards, S.V.; Beerli, P. Perspective: Gene Divergence, Population Divergence, And the Variance in Coalescence Time in Phylogeographic Studies. Evolution 2000, 54, 1839–1854. [Google Scholar] [CrossRef]
- Mendes, F.K.; Hahn, M.W. Gene Tree Discordance Causes Apparent Substitution Rate Variation. Syst. Boil. 2016, 65, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Ogilvie, H.A.; Bouckaert, R.R.; Drummond, A.J. Starbeast2 brings faster species tree inference and accurate estimates of substitution rates. Mol. Biol. Evol. 2017, 34, 2101–2114. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.V.; Cloutier, A.; Baker, A.J. Conserved nonexonic elements: A novel class of marker for phylogenomics. Syst. Biol. 2017, 66, 1028–1044. [Google Scholar] [CrossRef]
- Hill, W.G.; Robertson, A. The effect of linkage on limits to artificial selection. Genet. Res. 1966, 8, 269. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Inference of human population history from individual whole-genome sequences. Nature 2011, 475, 493–496. [Google Scholar] [CrossRef]
- Olson, S.L.; Parris, D.C. The Cretaceous Birds of New Jersey. Smithson. Contrib. Paleobiol. 1987, 63, 1–22. [Google Scholar] [CrossRef]
- Chen, A.; White, N.; Benson, R.B.J.; Braun, M.J.; Field, D.J. Total-Evidence Framework Reveals Complex Morphological Evolution in Nightbirds (Strisores). Diversity 2019, 11, 143. [Google Scholar] [CrossRef]
- Ksepka, D.; Clarke, J.A. Phylogenetically vetted and stratigraphically constrained fossil calibrations within Aves. Palaeontol. Electron. 2015. [Google Scholar] [CrossRef]
- Harmon, L.J.; Braude, S. Conservation of Small Populations: In An Introduction to Methods and Models in Ecology, Evolution, and Conservation Biology; JSTOR: New York, NY, USA, 2010; pp. 125–138. [Google Scholar]
- Ebersberger, I.; Galgoczy, P.; Taudien, S.; Taenzer, S.; Von Haeseler, A.; Platzer, M. Mapping Human Genetic Ancestry. Mol. Boil. Evol. 2007, 24, 2266–2276. [Google Scholar] [CrossRef]
- Hudson, R.R.; Coyne, J.A. Mathematical Consequences of the Genealogical Species Concept. Evolution 2002, 56, 1557–1565. [Google Scholar] [CrossRef]
- Springer, M.S.; Gatesy, J. The gene tree delusion. Mol. Phylogenet. Evol. 2016, 94, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Stiller, J.; Zhang, G. Comparative Phylogenomics, a Stepping Stone for Bird Biodiversity Studies. Diversity 2019, 11, 115. [Google Scholar] [CrossRef]
- Louca, S.; Pennell, M.W. Extant timetrees are consistent with a myriad of diversification histories. Nature 2020, 1–4. [Google Scholar] [CrossRef]
- Alvarez, L.W.; Asaro, F.; Michel, H.V.; Alvarez, W. Extraterrestrial Cause for the Cretaceous-Tertiary Extinction. Science 1980, 208, 1095–1108. [Google Scholar] [CrossRef]
- Schulte, P.; Alegret, L.; Arenillas, I.; Arz, J.A.; Barton, P.J.; Bown, P.R.; Bralower, T.J.; Christeson, G.L.; Claeys, P.; Cockell, C.S.; et al. The Chicxulub Asteroid Impact and Mass Extinction at the Cretaceous-Paleogene Boundary. Science 2010, 327, 1214–1218. [Google Scholar] [CrossRef]
- Mayr, G. The origins of crown group birds: Molecules and fossils. Palaeontology 2014, 57, 231–242. [Google Scholar] [CrossRef]
- Wright, S.J. Extinction-Mediated Competition: The Anolis Lizards and Insectivorous Birds of the West Indies. Am. Nat. 1981, 117, 181–192. [Google Scholar] [CrossRef]
- Buckley, L.B.; Jetz, W. Insularity and the determinants of lizard population density. Ecol. Lett. 2007, 10, 481–489. [Google Scholar] [CrossRef]
- Gattepaille, L.; Günther, T.; Jakobsson, M. Inferring Past Effective Population Size from Distributions of Coalescent Times. Genetics 2016, 204, 1191–1206. [Google Scholar] [CrossRef]
- Ho, S.Y.W.; Shapiro, B. Skyline-plot methods for estimating demographic history from nucleotide sequences. Mol. Ecol. Resour. 2011, 11, 423–434. [Google Scholar] [CrossRef]
- Schiffels, S.; Durbin, R. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 2014, 46, 919–925. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, S.; Harris, K.; Song, Y.S. Estimating Variable Effective Population Sizes from Multiple Genomes: A Sequentially Markov Conditional Sampling Distribution Approach. Genetics 2013, 194, 647–662. [Google Scholar] [CrossRef] [PubMed]
- Barbato, M.; Orozco-Terwengel, P.; Tapio, M.; Bruford, M.W. SNeP: A tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front. Genet. 2015, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Palacios, J.A.; Wakeley, J.; Ramachandran, S. Bayesian Nonparametric Inference of Population Size Changes from Sequential Genealogies. Genetics 2015, 201, 281–304. [Google Scholar] [CrossRef]
- Longrich, N.R.; Tokaryk, T.; Field, D.J. Mass extinction of birds at the Cretaceous–Paleogene (K–Pg) boundary. Proc. Natl. Acad. Sci. USA 2011, 108, 15253–15257. [Google Scholar] [CrossRef]
- Janzen, D.H. Who Survived the Cretaceous? Science 1995, 268, 785. [Google Scholar] [CrossRef]
- Rabosky, D.L.; Lovette, I.J. Explosive Evolutionary Radiations: Decreasing Speciation or Increasing Extinction Through Time? Evolution 2008, 62, 1866–1875. [Google Scholar] [CrossRef]
- Cracraft, J.; Claramunt, S. Conceptual and analytical worldviews shape differences about global avian biogeography. J. Biogeogr. 2017, 44, 958–960. [Google Scholar] [CrossRef]
- Wang, J. Estimation of effective population sizes from data on genetic markers. Philos. Trans. R. Soc. B Boil. Sci. 2005, 360, 1395–1409. [Google Scholar] [CrossRef]
- Charlesworth, B. Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 2009, 10, 195–205. [Google Scholar] [CrossRef]
- Wang, J.; Santiago-Rubio, E.; Caballero, A. Prediction and estimation of effective population size. Heredity 2016, 117, 193–206. [Google Scholar] [CrossRef]
- Alò, D.; Turner, T.F. Effects of Habitat Fragmentation on Effective Population Size in the Endangered Rio Grande Silvery Minnow. Conserv. Boil. 2005, 19, 1138–1148. [Google Scholar] [CrossRef]
- Turner, T.F.; Osborne, M.J.; Moyer, G.R.; Benavides, M.A.; Alò, D. Life history and environmental variation interact to determine effective population to census size ratio. Proc. R. Soc. B Boil. Sci. 2006, 273, 3065–3073. [Google Scholar] [CrossRef]
- Nielsen, R.; Beaumont, M. Statistical inferences in phylogeography. Mol. Ecol. 2009, 18, 1034–1047. [Google Scholar] [CrossRef]
- Mazet, O.; Rodríguez, W.; Chikhi, L. Demographic inference using genetic data from a single individual: Separating population size variation from population structure. Theor. Popul. Boil. 2015, 104, 46–58. [Google Scholar] [CrossRef]
- Mazet, O.; Rodríguez, W.; Grusea, S.; Boitard, S.; Chikhi, L. On the importance of being structured: Instantaneous coalescence rates and human evolution—lessons for ancestral population size inference? Heredity 2015, 116, 362–371. [Google Scholar] [CrossRef]
- Moyer, G.R.; Winemeller, K.O.; McPhee, M.V.; Turner, T.F. Historical Demography, Selection, And Coalescence of Mitochondrial and Nuclear Genes in Prochilodus Species of Northern South America. Evolution 2005, 59, 599–610. [Google Scholar] [CrossRef]
- Khatri, B.; Burt, A. Robust Estimation of Recent Effective Population Size from Number of Independent Origins in Soft Sweeps. Mol. Boil. Evol. 2019, 36, 2040–2052. [Google Scholar] [CrossRef]
- Lessa, E.P.; Cook, J.A.; Patton, J.L. Genetic footprints of demographic expansion in North America, but not Amazonia, during the Late Quaternary. Proc. Natl. Acad. Sci. USA 2003, 100, 10331–10334. [Google Scholar] [CrossRef] [PubMed]
- Braasch, J.; Barker, B.S.; Dlugosch, K.M. Expansion history and environmental suitability shape effective population size in a plant invasion. Mol. Ecol. 2019, 28, 2546–2558. [Google Scholar] [CrossRef] [PubMed]
- Kraus, R.; Kerstens, H.H.D.; Van Hooft, P.; Megens, H.-J.; Elmberg, J.; Tsvey, A.; Sartakov, D.; Soloviev, S.; Crooijmans, R.P.M.A.; Groenen, M.; et al. Widespread horizontal genomic exchange does not erode species barriers among sympatric ducks. BMC Evol. Boil. 2012, 12, 45. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).