Abstract
Gut microbiota, through both its species composition and its metabolites, impacts expression and activity of intestinal drug transporters. This phenomenon directly affects absorption process of orally administered drugs and contributes to the observed inter-individual variability in pharmacotherapeutic responses. This review summarizes mechanistic evidence from in vitro and animal studies and integrates clinical observations in which alterations in gut microbiota are associated with changes in oral drug exposure, consistent with potential regulation of key intestinal drug transporters—such as P-glycoprotein (P-gp, ABCB1), Breast Cancer Resistance Protein (BCRP, ABCG2), MRP2/3 proteins (ABCC2/3), and selected Organic Anion-Transporting Polypeptides (OATPs, e.g., SLCO1A2, SLCO2B1)—by major bacterial metabolites including short-chain fatty acids (SCFAs), secondary bile acids, and tryptophan-derived indoles. The molecular mechanisms involved include activation of nuclear and membrane receptors (PXR, FXR, AhR, TGR5), modulation of transcriptional and stress-response pathways (Nrf2, AP-1) with simultaneous suppression of pro-inflammatory pathways (NF-κB), and post-translational modifications (e.g., direct inhibition of P-gp ATPase activity by Eggerthella lenta metabolites). The review also highlights the pharmacokinetic implications of, e.g., tacrolimus, digoxin, and metformin. In conclusion, the significance of “drug–transporter–microbiome” interactions for personalized medicine is discussed. Potential therapeutic interventions are also covered (diet, pre-/probiotics, fecal microbiota transplantation, modulation of PXR/FXR/AhR pathways). Considering the microbiota as a “second genome” enables more accurate prediction of drug exposure, reduction in toxicity, and optimization of dosing for orally administered preparations.
1. Introduction
The human gastrointestinal tract harbors a dense and diverse community of microbes (the gut microbiota) that plays a pivotal role in host physiology [1]. Beyond involvement in digestion and immunity modulation, gut bacteria and their metabolites can profoundly influence drug pharmacokinetics. However, clinical pharmacokinetic studies rarely allow direct attribution of altered drug exposure to transporter regulation. Observed differences in bioavailability may instead reflect other microbiota-dependent mechanisms, such as luminal drug metabolism, microbial degradation, changes in intestinal permeability, or modifications in enterohepatic cycling. Therefore, clinical observations should be interpreted cautiously and viewed as consistent with, rather than confirmatory of, transporter-mediated effects. Two major families of membrane transport proteins participate in regulating the intestinal handling of xenobiotics: solute carrier (SLC) influx transporters and ATP-binding cassette (ABC) efflux transporters. SLC transporters mediate the uptake of structurally diverse drugs and endogenous substrates into enterocytes, whereas ABC proteins primarily function as ATP-driven efflux pumps that limit intestinal drug accumulation and shape first-pass pharmacokinetics [2]. In the intestine, key SLC representatives include OATP1A2 and OATP2B1 (SLCO1A2/SLCO2B1), PEPT1 (SLC15A1), OCTs and OATs (SLC22 family), and MATEs (SLC47), while the major ABC transporters involved in drug disposition are ABCB1 (P-gp), ABCG2 (BCRP) and ABCC2 (MRP2). Together, these coordinated influx–efflux systems determine the extent of oral drug absorption and contribute significantly to inter-individual variability in systemic drug exposure. Given that gut microbiota and their metabolites can modulate the expression and activity of both SLC and ABC transporters, understanding their baseline physiological roles is crucial for interpreting the microbiota–drug transport interactions discussed in this review. In particular, emerging evidence indicates that the gut microbiota regulates the expression and activity of intestinal drug transporters, thereby affecting oral drug absorption and disposition [3,4].
An overview of the major intestinal drug transporters, including their localization and functional orientation, is presented in Figure 1.
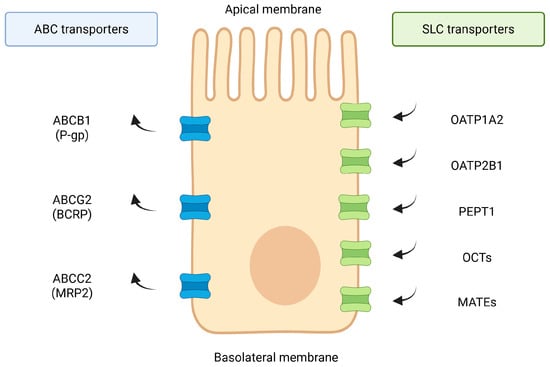
Figure 1.
Overview of major intestinal drug transporters. Schematic localization of key apical and basolateral intestinal transporters. ABC efflux pumps (ABCB1/P-gp, ABCG2/BCRP, ABCC-family) mediate ATP-dependent export of xenobiotics toward the lumen, whereas SLC influx systems (OATP1A2, OATP2B1, PEPT1, OCTs/OATs, MATEs) facilitate uptake of clinically relevant drugs. Transporter families and orientations summarized based on established data [1,2,5,6,7,8,9,10,11,12,13,14] and additional reference sources (Zhou et al., 2017 [2]; Zhou & Shu, 2022 [14]). Created in BioRender. Plust, M. (2025) https://BioRender.com/610jab7.
Intestinal transport proteins such as P-glycoprotein (P-gp, ABCB1), breast cancer resistance protein (BCRP, ABCG2), organic anion-transporting polypeptides (OATPs), and multidrug resistance-associated proteins (MRPs) are expressed in the intestinal epithelium/enterocytes, where they govern uptake and efflux of xenobiotics [2,5,6,15]. Alterations in the function of these transporters can lead to significant changes in drug bioavailability and inter-individual variability in drug responses [16]. Notably, conventional pharmacogenomic factors alone often fail to explain observed variability in drug disposition, pointing to the microbiome as a “second genome” influencing drug transport and metabolism. This review provides an in-depth analysis of how specific gut microbial species and their metabolites regulate key intestinal drug transporters at the molecular level [17,18]. We discuss cellular mechanisms involved, including receptor-activated transcriptional pathways and post-translational modifications, and highlight examples of drug–microbiota–transporter interactions that impact pharmacokinetics. We further integrate findings from in vitro studies, animal models, and clinical observations and consider implications for personalized medicine and drug development.
2. Gut Microbiota and Intestinal Drug Transporters
The level of expression of intestinal drug transporters is dynamic and can be shaped by gut microbial signals [19]. Germ-free animals or antibiotic-treated models often exhibit altered transporter levels compared to animals with a normal microbiota, underscoring a microbiota-dependent regulation [20,21]. Figure 2 summarizes baseline alterations in intestinal transporter expression observed in germ-free and microbiota-depleted models.

Figure 2.
Baseline alterations in intestinal drug transporters under germ-free and microbiota-depleted conditions. Germ-free mice exhibit reduced apical expression of key drug transporters, including ABCB1/P-gp, ABCG2/BCRP, MRP2, and OATP2B1, whereas conventional microbiota restore physiological transporter levels. Antibiotic treatment produces the opposite pattern for some transporters, with increased P-gp expression and variable effects on BCRP, MRP2, and OATP2B1. These baseline differences highlight the essential role of the gut microbiota in maintaining intestinal transporter homeostasis [20,21]. Created in BioRender. Plust, M. (2025) https://BioRender.com/610jab7.
For example, colonization of germ-free mice restores normal intestinal transporter expression patterns, while broad-spectrum antibiotic depletion of the microbiota can dysregulate transporter genes [22]. Specific commensal bacteria have been linked to the modulation of transporters. Notably, Foley et al. identified a “functional core” microbiome (enriched in Clostridia and Bacilli classes) in mice that was necessary and sufficient for inducing P-glycoprotein expression in the intestinal epithelium [1,23]. Metagenomic analysis of this core community revealed an enhanced capacity for producing short-chain fatty acids (SCFAs) and secondary bile acids, metabolites which positively correlated with P-gp levels. In contrast, dysbiosis characterized by the loss of such beneficial microbes can lead to transporter downregulation [24,25,26].
In ulcerative colitis (UC), a condition marked by reduced Firmicutes (including butyrate-producing Clostridia) and lower luminal SCFA/bile acid levels, intestinal P-gp expression is significantly diminished [27,28,29]. This reduction in P-gp in UC patients was found to coincide with a loss of microbiota-derived metabolites capable of inducing P-gp, suggesting that a healthy microbiome tonically sustains certain transporter levels [1,30].
Together, these findings establish that the gut microbiota exerts a bidirectional control over intestinal transporters: commensal-derived signals can upregulate transporter expression contributing to mucosal barrier function, whereas microbial imbalances or pathogenic signals may downregulate transporters and compromise drug handling [31,32,33]. The ATP-binding cassette (ABC) family—notably P-gp (ABCB1), BCRP (ABCG2), and MRP2/3 (ABCC2/3) as well as uptake transporters such as OATP2B1 (SLCO2B1) and OATP1A2 on the apical enterocyte membrane are the key intestinal drug transporters regulated by microbiota [1,2,5,6,7,8,9,10,11,12,13,16,17]. From a structural and functional standpoint, intestinal transporters comprise both ATP-dependent efflux pumps of the ABC family and numerous influx systems belonging to the solute carrier (SLC) superfamily. Key SLC drug transporters expressed in enterocytes include PEPT1 (SLC15A1), OATP1A2 and OATP2B1 (SLCO1A2/2B1), as well as members of the SLC22 family such as OCT1/3 and OATs. In addition, transporters involved in luminal cation handling, including PMAT (SLC29A4) and MATE1/2-K (SLC47A1/2), contribute to intestinal uptake and retention of several therapeutics. These SLC transporters play essential roles in governing the absorption of peptide-like drugs, statins, antihistamines, metformin, and other cationic or amphipathic agents [2,14]. This transporter repertoire works in concert to determine the fraction of an orally ingested drug that reaches systemic circulation.
The gut microbiota can impact both transcriptional regulation of the transporter genes and post-translational modulation of transporter proteins, as discussed below. It is important to note that changes in transporter activity due to microbial factors may either increase drug exposures (by reducing efflux or enhancing uptake) or decrease drug levels (by inducing efflux or reducing uptake), depending on the specific context [14,34,35].
3. Microbial Metabolites and Transporter Regulation: SCFAs, Bile Acids, and Tryptophan Derivatives
Gut microbes produce metabolites that can act as signaling molecules to the host [36]. Short-chain fatty acids, secondary bile acids, and tryptophan catabolites have emerged as key mediators of microbiota–transporter interactions [37,38]. Those metabolites engage host receptors and signaling pathways (e.g., G-protein coupled receptors and nuclear receptors) to modulate the expression of drug transporter genes in the intestinal epithelium [39]. Figure 3 illustrates the major classes of microbiota-derived metabolites and their associated signaling pathways regulating intestinal drug transporters.
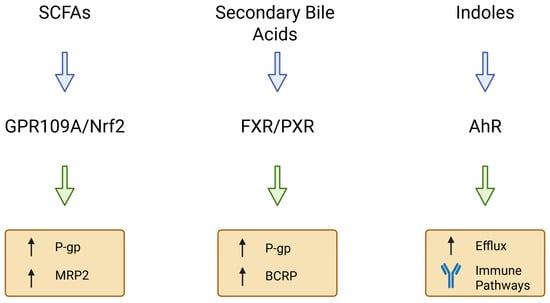
Figure 3.
Microbial metabolites and signaling pathways regulating intestinal drug transporters. Short-chain fatty acids (SCFAs) regulate P-gp and MRP2 primarily via GPR109A and Nrf2-dependent signaling [40,41]. Secondary bile acids activate FXR and PXR, modulating ABCB1/P-gp, ABCG2/BCRP and OATP transporters [42,43]. Indole derivatives signal through AhR, influencing efflux transporter expression and immune-associated regulatory pathways [44]. Together, these metabolite groups represent major microbiota-derived modulators of intestinal transporter homeostasis. Created in BioRender. Plust, M. (2025) https://BioRender.com/610jab7.
3.1. Short-Chain Fatty Acids Upregulating Drug Transporters
SCFAs (acetate, propionate, and notably butyrate) are produced by fermentation of dietary fibers by anaerobic bacteria (primarily Firmicutes like Faecalibacterium prausnitzii and Roseburia spp.) [45].
Butyrate has long been recognized as a key signaling metabolite in the gut; it serves as the preferred energy source for colonocytes and modulates gene expression via epigenetic and receptor-mediated mechanisms [46,47]. Butyrate can also robustly induce P-glycoprotein expression and activity in intestinal cells. Mechanistically, butyrate is an inhibitor of histone deacetylases (HDAC), leading to hyperacetylation of histones and altered transcription of target gene [48], including promoter regions of ABCB1 gene (encoding P-gp), thereby enhancing transcription. In addition, butyrate and other SCFAs activate certain G-protein coupled receptors on intestinal epithelial cells (such as FFAR2/GPR43, FFAR3/GPR41, and GPR109A) [1,49,50,51]. GPCR activation triggers intracellular signaling cascades (e.g., via PKC or MAPKs) that can converge on transcription factors regulating transporter genes. Butyrate has also been shown to activate Nrf2 (nuclear factor erythroid 2-related factor 2), a redox-sensitive transcription factor, which activation leads to increased expression of various cytoprotective genes, including certain transporters [52,53]. In colon carcinoma cell models, pharmacologic activation of Nrf2 upregulated P-gp expression, consistent with this pathway’s involvement [54,55].
Notably, the upregulation of P-gp by butyrate has been demonstrated in vitro and in vivo in the context of not only xenobiotic handling but also in mitigating intestinal inflammation (by promoting the efflux of anti-inflammatory endogenous substrates) [23,56,57]. For example, butyrate-producing commensals are depleted in ulcerative colitis, and this correlates with reduced colonic P-gp and a pro-inflammatory state. Replenishing butyrate (through fiber diet or probiotics) in animal models restores P-gp levels and improves colitis, underscoring SCFAs’ regulatory role [58]. Furthermore, probiotics such as Lactobacillus acidophilus that increase luminal butyrate or otherwise stimulate epithelial cells can enhance P-gp expression. Studies have shown that Lactobacilli treatment of intestinal epithelial monolayers leads to higher P-gp levels and function via activation of AP-1 transcription factors (c-Fos/c-Jun). This AP-1 activation may be downstream of SCFA signaling or other microbe-associated molecular patterns, illustrating how commensal bacteria orchestrate host transporter defenses [59,60,61,62].
Beyond P-gp, SCFAs may also influence other transporters. There is evidence that butyrate can modulate the expression of MRP family transporters and uptake carriers, although data are less extensive than for P-gp. In the kidney, SCFAs have been reported to enhance OAT (organic anion transporter) expression and facilitate toxin excretion [63,64,65]. By analogy, in the gut, SCFA signaling might upregulate efflux pumps like MRP2 (which exports organic anions and conjugated drug metabolites) via Nrf2 or other pathways, since Nrf2 is a known inducer of MRP2 in response to oxidative stress. Indeed, treatment of colitic rats with SCFA-producing fiber (inulin) prevented the downregulation of Mrp2 that otherwise occurs in colitis, suggesting SCFAs help maintain MRP2 expression under inflammatory conditions [66,67,68]. Thus, SCFAs broadly act as beneficial modulators, promoting a transporter expression profile that enhances mucosal barrier function and xenobiotic clearance [44]. SCFAs also regulate specific SLC transporters. In a conventional-vs-germ-free mouse model, colonic expression of the butyrate transporter SLC5A8 and the SCFA receptor GPR109A was markedly reduced under germ-free conditions and restored by microbial colonization, indicating that SCFA-producing taxa directly shape epithelial uptake capacity [40]. Furthermore, human biopsy data demonstrate that individuals with higher fecal SCFA levels exhibit increased colonic expression of the organic cation transporter OCT3 (SLC22A3), suggesting a link between microbial SCFA availability and intestinal uptake of cationic drugs such as metformin [41].
3.2. Secondary Bile Acids and Nuclear Receptor Signaling
Bile acids are another class of microbiota-dependent molecules that regulate drug transporter genes. Primary bile acids (e.g., cholic acid, chenodeoxycholic acid) are synthesized from cholesterol in the liver, secreted in bile, and can be reabsorbed in the intestine [69]. The gut microbiota enzymatically converts primary bile acids into secondary bile acids (such as deoxycholic acid, lithocholic acid, and ursodeoxycholic acid) via deconjugation and dihydroxylation reactions [70]. These secondary bile acids not only facilitate lipid absorption but also serve as potent signaling ligands for host nuclear receptors, notably the farnesoid X receptor (FXR) and pregnane X receptor (PXR), as well as membrane G-protein coupled receptors like TGR5. Through these receptors, bile acids can modulate the transcription of numerous genes including those encoding drug transporters [42]. Beyond ABC transporters, bile-acid-dependent FXR and PXR signaling also regulates several intestinal SLC systems, including the apical bile acid transporter ASBT (SLC10A2), the basolateral OSTα/β complex, and selected OATP and OAT family members, integrating microbiota-driven bile acid remodeling with coordinated control of both drug uptake and efflux [43,71].
Certain secondary bile acids, such as lithocholic acid (LCA) and deoxycholic acid (DCA), are known agonists of PXR. PXR is a xenobiotic-sensing nuclear receptor highly expressed in the intestine (and liver) that, upon activation, forms a heterodimer with RXR and binds to response elements in the promoter regions of target genes [1,72]. PXR activation typically induces the expression of genes involved in drug metabolism and transport, including CYP3A4 (a major drug-metabolizing enzyme) and multiple drug transporters. Notably, activation of PXR has been shown to increase the transcription of P-glycoprotein (MDR1/ABCB1) and BCRP (ABCG2) in intestinal models [1,73]. Foley et al. demonstrated that LCA and DCA at physiologically relevant concentrations could induce P-gp expression in colonic epithelial cells, especially when combined with butyrate. This synergistic effect aligns with a model where butyrate and secondary bile acids activate parallel pathways (HDAC inhibition/GPCRs and PXR, respectively) that converge on boosting MDR1 gene transcription [1,23]. Thus, a microbiota rich in bile acid-metabolizing bacteria (e.g., certain Clostridium clusters that carry bile salt hydrolases and 7α-dehydroxylation enzymes) is able to generate higher levels of secondary bile acids, which in turn activate PXR in the intestine and upregulate protective transporters like P-gp and MRP2 [74,75].
FXR is another bile acid-activated nuclear receptor expressed in the ileum and liver. While FXR primarily governs bile acid homeostasis (controlling synthesis and enterohepatic cycling), it can indirectly influence drug transporter expression. In the intestine, FXR activation (for instance by chenodeoxycholate or microbial metabolites like certain secondary bile acids) induces the production of fibroblast growth factor 19 (FGF19), which signals to the liver to reduce bile acid synthesis [76,77]. Additionally, intestinal FXR activation upregulates genes involved in bile acid export such as organic solute transporter α and β (OSTα/β, the basolateral bile acid efflux transporter) and may downregulate the apical bile acid uptake transporter sodium-dependent bile acid transporter (ASBT), to protect against bile acid overload [78]. Regarding drug transporters, FXR agonism has been reported to increase MRP2 expression in liver, aiding the biliary excretion of bile acids. In enterocytes, FXR’s effect on classical drug transporters is less direct, but by altering the bile acid pool and local inflammation, FXR can modulate transporter regulation [79,80]. For example, FXR activation tends to have anti-inflammatory effects in the gut. Reduced inflammation can relieve NF-κB-mediated suppression of certain transporters (NF-κB, a pro-inflammatory transcription factor, often represses transporter gene expression during inflammation). Thus, via maintaining anti-inflammatory tone, FXR-active bile acids from the microbiota might indirectly preserve higher P-gp/BCRP levels [81,82]. Moreover, FXR shares some target gene overlap with PXR and CAR; studies have indicated FXR ligands can cross-activate PXR target genes to a degree. Overall, the microbiota–bile acid–FXR/PXR axis is a crucial pathway wherein microbial metabolism of bile acids leads to activation of host receptors that transcriptionally induce drug transporters like P-gp and MRPs [83,84].
Beyond P-gp, MRP2 (ABCC2), an apical efflux pump exporting organic anions and phase II conjugates [85] in the intestine, can also be upregulated by bile-acid-activated receptors. Its expression in enterocytes could be enhanced by PXR (which is known to induce hepatic MRP2) and by FXR (to facilitate extrusion of glucuronidated bile acids and bilirubin) [71].
In a healthy microbiome setting, ample secondary bile acids and luminal butyrate likely maintain not only P-gp but also an array of efflux transporters that function coordinately to protect the mucosa from both toxins and inflammatory mediators [22,86]. Indeed, an observed phenomenon in ulcerative colitis is an imbalance in the P-gp/MRP2 axis: UC patients typically show reduced P-gp expression but paradoxically increased MRP2 expression in inflamed colon [87]. One hypothesis suggests that dysbiosis in UC (low SCFA, low secondary bile acids) fails to sustain P-gp (which has anti-inflammatory roles), while inflammatory signals (or residual FXR activation by accumulated primary bile acids) selectively induce MRP2 as a stress response [88].
3.3. Tryptophan Metabolites and Aryl Hydrocarbon Receptor (AhR) Activation
Dietary tryptophan is another substrate for gut microbial metabolism, leading to various indole and indole-derivative compounds [89,90]. The microbial tryptophan metabolites (e.g., indole, indole-3-acetic acid, indole-3-propionic acid, tryptamine, and others) are important modulators of gut epithelial biology and immune homeostasis [91,92]. A key mechanism by which they signal to the host is through activation of the aryl hydrocarbon receptor (AhR), a ligand-activated transcription factor expressed in intestinal epithelial and immune cells. AhR is traditionally known for sensing environmental toxins (like dioxins), but endogenous and microbial indoles are natural AhR ligands in the gut lumen [93,94]. Activation of AhR has been shown to transcriptionally upregulate the gene encoding BCRP (ABCG2), a major drug efflux transporter [95,96]. Studies in human intestinal cells and other models demonstrate that AhR acts as a direct transcriptional activator of ABCG2 [97]. For instance, treating cells with known AhR agonists (such as TCDD or indole-3-carbinol) increases BCRP mRNA and protein levels, whereas knocking down AhR reduces baseline BCRP expression [98]. A xenobiotic response element (XRE) has been identified in the ABCG2 promoter, through which ligand-activated AhR can enhance BCRP transcription [99]. Therefore, gut microbial metabolites that engage AhR can booster the intestinal efflux of substrates via BCRP [100,101]. Indole and certain indole derivatives from commensal bacteria (e.g., Clostridium sporogenes produces indole-3-propionic acid, a potent AhR agonist) likely contribute to baseline BCRP expression in the gut [102]. This is supported by observations that colonization with AhR ligand-producing microbiota increases epithelial expression of detoxifying enzymes and transporters, reinforcing the intestinal barrier [103].
In addition to BCRP, AhR activation may influence other transporters and metabolizing enzymes that contain XREs in their regulatory regions (such as certain phase I/II enzymes and possibly ABCB1). However, BCRP is the clearest example of an efflux pump directly upregulated by AhR [104,105]. By effluxing dietary carcinogens and microbial metabolites, BCRP induction via AhR is considered a key cytoprotective response [106,107].
It should be noted that tryptophan metabolites also modulate immune pathways (e.g., IL-22 production via AhR in innate lymphoid cells) which can secondarily affect transporter expression by altering the inflammatory milieu [108,109].
In the context of drug transport, its activation by microbiota-derived indoles enhances the gut’s capability to pump out potentially harmful compounds via transporters like BCRP and possibly P-gp. Some probiotics (e.g., certain Lactobacillus strains) produce indole-3-aldehyde, activating AhR and promoting mucosal barrier integrity, this could translate to maintained or elevated transporter levels as part of the barrier function [110,111]. Overall, tryptophan metabolite signaling through AhR constitutes a crucial link between microbiota metabolism and the upregulation of intestinal transporters involved in xenobiotic defense [112].
3.4. Other Microbial Metabolites and Factors
While SCFAs, bile acids, and indoles are among the most studied, other microbial products can potentially impact transporter functions as well:
- Phenolic Metabolites: Gut bacteria metabolize polyphenols and aromatic compounds into phenolic acids that may activate receptors like PXR or Nrf2 [40]. For example, urolithins (derived from polyphenols by gut microbes) have been noted to induce phase II enzymes and could affect transporters via Nrf2 activation, though direct evidence on transporter genes is still emerging [113].
- Trimethylamine N-oxide (TMAO): Produced from dietary choline/carnitine by a two-step microbial-host process, TMAO has been implicated in modulating bile acid and cholesterol transport. Some studies suggest TMAO may downregulate hepatic transporters (like OATP and NTCP) via FXR signaling. In the intestine, a high-TMAO environment (as in some dysbiosis) could conceivably alter FXR and hence transporter expression, although details remain to be clarified [114].
- Microbial Enzymes and Postbiotics: Microbes secrete enzymes or peptide signaling molecules that interact with host cells. Probiotics can release soluble proteins that activate MAPK pathways in epithelial cells, potentially influencing transporter gene expression similar to how Lactobacillus activated AP-1 to increase P-gp. Additionally, microbial fermentation gases (H2S, methane) or small molecules (like spermine, polyamines) might impact cellular signaling pathways [115,116,117,118], that modulate transporter post-translational modification (e.g., phosphorylation status affecting transporter activity) [119].
- Pathogen-associated Molecular Patterns (PAMPs): Components of microbial cells, such as lipopolysaccharide (LPS) from Gram-negative bacteria, can also affect transporter function. LPS triggers Toll-like receptor 4 (TLR4) and downstream NF-κB inflammatory signaling in the gut [120,121]. Acute exposure to LPS or a pro-inflammatory state has been shown to downregulate P-glycoprotein expression and inhibit its function in various tissues [122]. For example, LPS administration in rodents impaired P-gp-mediated efflux at blood–tissue barriers. In the intestine, inflammation due to pathogenic bacteria or microbial imbalance could similarly suppress P-gp and other transporter levels as part of the broader NF-κB driven response [122,123]. Infections like Citrobacter rodentium in mice reduce colonic P-gp expression, an effect linked to increased epithelial permeability and inflammation [124]. Such findings highlight that not all microbial signals induce transporters—some, particularly from pathogens or dysbiosis, can inhibit transporter expression and activity, diminishing the gut’s drug efflux capability [125,126]. The summary is provided in Table 1 below.
 Table 1. Major microbiota-derived factors and their known effects on intestinal drug transporters and the underlying host pathways involved.
Table 1. Major microbiota-derived factors and their known effects on intestinal drug transporters and the underlying host pathways involved.
4. Molecular Pathways of Microbiota-Driven Transporter Regulation
It should be emphasized that most of the mechanistic pathways described in this section are supported predominantly by preclinical in vitro and animal studies. Direct confirmation of these mechanisms in the human intestinal epithelium remains limited, and therefore they should be interpreted as biologically plausible rather than definitively established. Understanding which host molecular pathways are involved provides mechanistic insight into how microbes influence transporter expression. Several key pathways have emerged: nuclear receptors (e.g., PXR, FXR, CAR, VDR), transcription factors (e.g., Nrf2, NF-κB, AP-1), and kinase signaling cascades (affecting post-translational modifications of transporters). These pathways can be activated or inhibited by microbial metabolites, leading to up- or down-regulation of transporter proteins [128]. Although ABC efflux pumps such as P-gp, BCRP, and MRPs have been most extensively characterized, these microbiota-responsive pathways also regulate clinically important SLC transporters. Mechanistic studies indicate that microbial metabolites influence the transcription and membrane abundance of various SLCO and SLC22 carriers, suggesting that the microbiota orchestrates a coordinated modulation of both uptake and efflux systems (Zhou i in., 2017 [2]; Zhou i Shu, 2022 [14]).
As shown in Figure 4, representative microbial metabolites—including butyrate, lithocholic acid, indole-3-propionic acid and LPS—activate distinct host receptors (HDAC, PXR, AhR, TLR4), leading to differential regulation of intestinal efflux transporters such as P-gp and BCRP.

Figure 4.
Microbial metabolite–receptor interactions regulating intestinal efflux transporters. Butyrate activates HDAC-dependent signaling, inducing P-glycoprotein (P-gp). Lithocholic acid activates PXR, upregulating P-gp and BCRP. Indole-3-propionic acid signals through AhR to increase BCRP expression. LPS activates TLR4, modulating inflammatory signaling that influences P-gp and BCRP. Together, these pathways represent key nodes linking microbiota-derived metabolites to transporter regulation. Created in BioRender. Plust, M. (2025) https://BioRender.com/zl2fzz8.
4.1. Nuclear Receptors as Xenobiotic and Metabolite Sensors
Nuclear receptors (NRs) are ligand-activated transcription factors that play central roles in xenobiotic sensing and metabolic gene regulation. In the context of gut microbiota and drug transporters, the most relevant NRs include:
- Pregnane X Receptor (PXR): PXR is activated by various microbial metabolites (secondary bile acids like LCA, certain dietary compounds possibly modified by microbes, etc.). When activated in intestinal enterocytes, PXR binds to response elements on genes to induce a suite of xenobiotic-handling proteins—like ABCB1 (P-gp), ABCC2 (MRP2), ABCG2 (BCRP), and CYP3A4 [3,43,129,130,131]. In addition to ABC transporters, PXR activation has been shown to regulate multiple SLC genes, including members of the SLCO (OATP) and SLC22 (OAT/OCT) families, indicating that microbiota-derived PXR ligands may reshape both intestinal drug uptake and efflux (Zhou i in., 2017 [2,58]; Zhou i Shu, 2022 [14]). PXR thus serves as a crucial mediator by which microbiota can enhance efflux transporter expression to handle increased luminal loads of foreign chemicals. Interestingly, gut microbes themselves can modulate PXR signaling not only by providing ligands but also by influencing PXR expression levels [132].
- Constitutive Androstane Receptor (CAR): CAR is another xenobiotic-sensing NR which often overlaps with PXR in target genes but is usually active basally and further induced by certain ligands (e.g., phenobarbital-type inducers) [133]. They pinpointed CAR as a likely transcription factor mediating microbiota-induced changes in Abcb1 (P-gp) expression [134]. It appears that some microbiota metabolites may repress CAR activity under normal conditions, and removing them (with antibiotics) triggers CAR, which then boosts P-gp expression. CAR’s exact endogenous ligands from microbiota are not well-characterized; indirect modulation via altered bile acid pools or cytokine signaling is possible [135]. CAR activation is generally associated with induction of certain efflux pumps and phase II enzymes, so it aligns with an increase in transporter expression [136].
- Farnesoid X Receptor (FXR): FXR, activated by bile acids, influences primarily bile acid transporters. Microbiota-controlled bile acid profiles will determine FXR activation. Intestinal FXR activation tends to maintain barrier integrity and can have varying effects on drug transporters [137]. Some studies in FXR-knockout mice show alterations in P-gp and BCRP levels during cholestatic conditions, implying FXR may contribute to their regulation. Notably, FXR agonists (e.g., obeticholic acid) given to mice shift the gut microbiota and also change expression of some ABC transporters, though disentangling cause/effect is complex [138].
- Vitamin D Receptor (VDR): VDR can be activated by lithocholic acid (a secondary bile acid) which is a VDR ligand. Activation of VDR has been shown to induce P-gp expression in the gut as well, since vitamin D/VDR signaling upregulates ABCB1 in various tissues. Microbiota that produce LCA could activate VDR locally, potentially contributing to P-gp regulation [139,140].
4.2. Transcription Factors and Signaling Pathways
- Nrf2 (Nuclear factor E2-related factor 2): Nrf2 is a master regulator of cellular antioxidant and defensive responses. SCFAs like butyrate can activate Nrf2. When Nrf2 translocates to the nucleus, it binds antioxidant response elements (AREs) in gene promoters. Nrf2 drives expression of many phase II metabolism enzymes (UGTs, GSTs) and certain transporters including MRP2 and MRP3. Butyrate-mediated Nrf2 activation has been linked to increased P-gp levels as well. For example, berberine (a plant alkaloid that also modulates gut microbes) was shown to upregulate P-gp via Nrf2-dependent mechanisms in colitic rats [141,142].
- NF-κB (Nuclear Factor kappa B): NF-κB is a key inflammatory transcription factor activated by microbial PAMPs (LPS, flagellin) and cytokines [143]. Activation of NF-κB generally leads to production of pro-inflammatory mediators, but it can also repress certain genes. In inflammatory states of the gut, NF-κB activation correlates with decreased expression of transporters like P-gp and BCRP [144,145]. The mechanism may involve NF-κB interfering with the binding of positive factors (like PXR or constitutive transcription factors) on transporter gene promoters [146]. Additionally, NF-κB induces nitric oxide and oxidative stress that can impair transporter function [147]. Overall, chronic NF-κB activation (e.g., in IBD or infection) is associated with transporter downregulation, contributing to barrier compromise [148].
- AP-1 (Activator Protein 1): AP-1 refers to dimeric transcription factors composed of Fos and Jun proteins that respond to MAPK signaling [149]. The MDR1 (P-gp) gene promoter contains AP-1 binding sites known to enhance its transcription [150,151]. Beneficial microbes can activate AP-1 in epithelial cells, e.g., L. acidophilus was shown to induce c-Fos/c-Jun, resulting in higher P-gp expression [152]. On the other hand, certain bacterial toxins or stress may activate JNK pathways leading to AP-1, but in contexts like oxidative stress AP-1 might also be repressive [153].
- PPARs (Peroxisome Proliferator-Activated Receptors): These lipid-sensing NRs (PPARα/δ/γ) can be activated by microbial metabolites (e.g., certain fatty acids) [154]. While not classic drug transporter regulators, PPARγ activation by microbiota (as in fermentation products) has anti-inflammatory effects that indirectly preserve transporter function [155]. Some studies indicate PPARα agonists can increase ABCB1 expression in the liver; whether similar occurs in gut is under investigation [156,157].
- HIF-1 (Hypoxia Inducible Factor 1): The gut mucosa experiences an altered oxygen gradient in dysbiosis, possibly activating HIF-1. HIF-1 is known to induce certain transporters (like ABCB1 in hypoxic tumors) [158,159]. A fiber-rich, microbiota-driven increase in butyrate actually consumes oxygen in the colon and can stabilize HIF-1, which has been linked to enhanced barrier function [160]. Microbiota-induced HIF-1 helps upregulate P-gp or other transporters as part of adaptation to low oxygen, though direct evidence in vivo is limited [161].
4.3. Post-Translational Modifications of Transporters
One remarkable example is the post-translational inhibition of P-glycoprotein by a bacterial metabolite, as described earlier, Eggerthella lenta secretes a factor (a family of isoflavonoid molecules) that directly interacts with P-gp and inhibits its ATPase activity [16,162,163,164]. This prevents P-gp from cycling and pumping substrates, effectively reducing efflux function without changing P-gp expression [162,165]. Such inhibition was observed in cell culture and translated to increased drug absorption in mice colonized with E. lenta [16]. The inhibitor is structurally similar to plant-derived P-gp inhibitors, highlighting how microbial metabolites can mimic known drug-transporter inhibitors [166,167].
Other PTMs include phosphorylation, glycosylation, ubiquitination of transporters:
- Kinase signaling: Transporter proteins like BCRP and P-gp can be phosphorylated by kinases (e.g., PKA, PKC, JNK) which may alter their localization or activity [6,168]. The gut microbiota modulates host kinase signaling (for example, microbial secondary bile acids can activate PKC or Src kinases) [169]. There is evidence that intestinal BCRP requires phosphorylation by JAK2/3 (Janus kinases) for full activity. In inflammatory states, cytokines activate JAK/STAT pathways which could modify BCRP [170,171]. Microbiota that reduce inflammation might maintain proper JAK-mediated BCRP activation, whereas dysbiosis could lead to aberrant phosphorylation and diminished function [172]. Similarly, P-gp function can be modulated by PKC-mediated phosphorylation; some bacterial toxins activate PKC and might internalize P-gp, reducing efflux at the membrane [173].
- Glycosylation: N-glycosylation of P-gp and BCRP is needed for stability and trafficking to the plasma membrane [174]. Microbiota affect the gut epithelial glycosylation patterns (e.g., via influencing nutrient availability like monosaccharides or modulating endoplasmic reticulum stress). If microbial products interfere with normal glycosylation (for instance, some bacterial infections cause ER stress), transporters could misfold or be targeted for degradation, lowering their surface expression [175].
- Ubiquitin-proteasome degradation: Inflammatory signals triggered by microbes can promote ubiquitin tagging of certain proteins [176,177]. Commensals that activate pathways like AMPK may actually enhance transporter stability by preventing misfolded protein accumulation [178]. It has been noted that butyrate can increase expression of chaperone proteins and stabilize tight junction and transporter proteins in the membrane, though more specific data on transporter PTMs are needed [179].
- Membrane microenvironment: Microbial metabolites can also alter the membrane lipid composition [180]. P-gp activity is known to depend on membrane lipid environment. Secondary bile acids can incorporate into membranes and might modulate how P-gp interacts with the bilayer, potentially changing its conformation and drug affinity [181].
- Although these post-translational mechanisms are biologically plausible and supported by selected in vitro studies, the evidence linking microbiota-derived metabolites directly to transporter PTMs remains fragmentary. Additional work is required to determine the extent to which these modifications occur in vivo and whether they meaningfully influence human drug disposition.
- Figure 5 summarizes the integrated relationships between microbiota-derived metabolites, host signaling pathways, regulated intestinal drug transporters, and representative clinical drugs affected by these interactions.
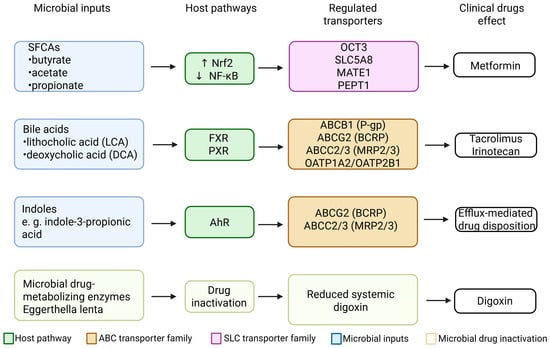 Figure 5. Comprehensive summary of microbial inputs, host signaling pathways, regulated intestinal drug transporters, and affected clinical drugs. Short-chain fatty acids (SCFAs) regulate several SLC transporters, including OCT3, SLC5A8,TE1/2-K, and PEPT1, primarily through activation of Nrf2 and inhibition of NF-κB, influencing the disposition of metformin. Secondary bile acids such as lithocholic acid (LCA) and deoxycholic acid (DCA) activate FXR and PXR, modulating efflux transporters (ABCB1/P-gp, ABCG2/BCRP, ABCC2/3) and uptake transporters (OATP1A2 and OATP2B1), with clinical implications for tacrolimus and irinotecan/SN-38. Indole derivatives activate AhR, upregulating BCRP and MRP2/3 and affecting efflux-mediated drug disposition. Eggerthella lenta reduces digoxin to dihydrodigoxigenin, decreasing the systemic fraction of active digoxin independently of host transport mechanisms. Collectively, these pathways illustrate the coordinated influence of gut microbiota on intestinal drug transport and pharmacokinetics. Created in BioRender. Plust, M. (2025) https://BioRender.com/610jab7.
Figure 5. Comprehensive summary of microbial inputs, host signaling pathways, regulated intestinal drug transporters, and affected clinical drugs. Short-chain fatty acids (SCFAs) regulate several SLC transporters, including OCT3, SLC5A8,TE1/2-K, and PEPT1, primarily through activation of Nrf2 and inhibition of NF-κB, influencing the disposition of metformin. Secondary bile acids such as lithocholic acid (LCA) and deoxycholic acid (DCA) activate FXR and PXR, modulating efflux transporters (ABCB1/P-gp, ABCG2/BCRP, ABCC2/3) and uptake transporters (OATP1A2 and OATP2B1), with clinical implications for tacrolimus and irinotecan/SN-38. Indole derivatives activate AhR, upregulating BCRP and MRP2/3 and affecting efflux-mediated drug disposition. Eggerthella lenta reduces digoxin to dihydrodigoxigenin, decreasing the systemic fraction of active digoxin independently of host transport mechanisms. Collectively, these pathways illustrate the coordinated influence of gut microbiota on intestinal drug transport and pharmacokinetics. Created in BioRender. Plust, M. (2025) https://BioRender.com/610jab7.
5. Clinical Observations and Human Studies
Direct studies in humans are more limited but growing, given the complexity of human microbiome variability and ethical considerations. Importantly, because intestinal transporter expression or activity is rarely measured directly in human studies, changes in drug levels associated with microbiota alterations cannot be assumed to result solely from transporter modulation. Microbiota-driven effects on luminal metabolism, drug degradation, intestinal permeability, or enterohepatic circulation may equally contribute to these clinical observations. Still, several lines of evidence in clinical or pharmacological studies point to microbiota–transporter interactions:
- Association studies: Analyses of intestinal biopsies have shown correlations between microbiota composition and transporter gene expression. As one example, in a cohort of UC patients versus healthy controls, P-gp expression in colon biopsies was lower in UC and directly correlated with the abundance of butyrate-producing Firmicutes in the patients’ gut microbiota. Those with more Roseburia/Faecalibacterium had higher mucosal P-gp, whereas those over-colonized by Proteobacteria (like E. coli) had lower P-gp. This is consistent with mechanistic data and suggests microbiome profiling might predict transporter expression in individuals [182,183].
- Fecal metabolite associations: Similarly, levels of fecal SCFAs and secondary bile acids in patients have been linked to transporter modulation. Patients on high-fiber diets (with presumably higher SCFAs) showed increased expression of genes like ABCB1 and SLC22A3 (OCT3) in colon biopsies in one small study (potentially explaining better drug tolerance) [41]. Conversely, individuals on broad-spectrum antibiotics (which wipe out SCFA producers) had transient reductions in fecal butyrate and coincident drops in P-gp expression (noted in one study examining loperamide response, though more data are needed) [184].
- Drug pharmacokinetics and microbiome status: There are reported instances where changes in a patient’s microbiota (due to antibiotics or illness) altered drug levels in ways consistent with transporter effects:
- Tacrolimus: Clinical anecdotal reports and pilot studies have noted that patients on tacrolimus (an immunosuppressant) experienced elevated drug levels and toxicity after courses of broad-spectrum antibiotics [185]. While some of this is due to loss of bacterial drug metabolism, the study by Degraeve et al. suggests a mechanistic basis: antibiotics increase intestinal P-gp which lowers tacrolimus absorption [17]. In a controlled setting, one study co-administered the P-gp inhibitor zosuquidar to kidney transplant patients and found that normally, P-gp limits tacrolimus absorption, but when gut bacteria are suppressed (as in patients on multiple antibiotics), tacrolimus AUC variance increased markedly until P-gp was blocked. This indicates that microbiota differences could be part of why some patients need higher tacrolimus doses—those with certain microbiomes might have higher baseline P-gp activity [186,187].
- Digoxin: A classic case is digoxin, a cardiac glycoside. About 10% of patients harbor Eggerthella lenta strains that metabolize digoxin, reducing its plasma levels. Interestingly, those same strains secrete P-gp inhibitors [188]. A recent trial measured digoxin pharmacokinetics in subjects before and after altering their microbiota (through diet) [189]. In those where E. lenta increased, early-phase digoxin blood levels were higher (consistent with P-gp inhibition increasing absorption rate), although total exposure (AUC) did not change much because metabolic degradation counteracted it [190].
- Metformin: Emerging data link metformin response to the microbiome. Metformin’s absorption occurs mainly in the upper intestine via transporters like OCT1 and SERT, and it exerts glucose-lowering effects partly through actions on the gut. Some studies found that microbiome composition (abundance of Akkermansia and SCFA producers) correlates with better metformin response [191]. Mechanistically, metformin itself alters bile acid pools and gut microbes, which could upregulate intestinal apical bile acid transporters and limit metformin diarrhea side-effects [192,193].
- Cancer chemotherapeutics: The irinotecan example is notable clinically. Cancer patients on irinotecan sometimes receive antibiotics like neomycin to mitigate diarrhea [194]. Neomycin kills β-glucuronidase-producing bacteria, preventing reactivation of toxic SN-38 in the colon [195]. Additionally, neomycin might modulate transporter expression: small studies hint that neomycin co-treatment leads to higher expression of MRP2 in colon (so that SN-38 glucuronide is effluxed more, reducing exposure of colon mucosa) [196]. Probiotic trials in cancer patients also showed reduced irinotecan diarrhea, presumably by microbiota modulation affecting both metabolism and transport [197]. More broadly, cancer patients often have altered microbiomes due to diet/antibiotics, which may influence oral bioavailability of drugs like tyrosine kinase inhibitors (many of which are P-gp/BCRP substrates) [198].
Direct clinical approaches employing microbiota modifiers are being explored as potential ways to influence drug transport and toxicity [199]. For example, fecal microbiota transplantation (FMT) in UC can restore some butyrate-producing bacteria; one might predict it could also restore colonic P-gp expression, contributing to mucosal healing by exporting inflammatory mediators [200]. Measuring transporter expression pre- and post-FMT could be an interesting clinical research avenue. Probiotics are another form of microbiota modification: administering Lactobacillus or Bifidobacterium strains to patients might increase intestinal P-gp and tighten the barrier—potentially useful in IBD or in reducing drug-induced gut toxicity. However, it could also decrease the absorption of certain oral drugs (a consideration for co-administration timing) [1,17,201,202].
In summary, human data, while still emerging, align with the pattern established in experimental models: a microbiome abundant in SCFA and secondary bile acid producers (often associated with healthy diets) tends to promote a high-expression, high-function state of intestinal efflux transporters, which can reduce the absorption (and toxicity) of certain drugs but also protect the host. On the other hand, a disrupted or inflammatory microbiome can reduce these transporters, possibly increasing drug absorption unpredictably or compromising epithelial defense. This variability underscores the need for personalized approaches in medicine [203,204].
Case Examples of Drug–Microbiota–Transporter Interactions
Examples where microbiota-mediated changes in transporters have been shown to alter drug pharmacokinetics or response (Table 2).

Table 2.
Selected Drugs Affected by Microbiota–Transporter Interactions.
6. Implications for Personalized Medicine and Drug Development
The intricate interplay between gut microbiota and intestinal transporters has several important implications:
Personalized Medicine and Microbiome Profiling
Inter-individual differences in drug absorption and response (even among those with the same genetic features for drug-metabolizing enzymes and transporters) can be partially explained by differences in gut microbiome composition [205]. As such, analyzing a patient’s microbiome (sometimes termed “pharmacomicrobiomics”) could help personalize drug therapy. For drugs with a narrow therapeutic index or known transporter-mediated variability (e.g., tacrolimus, digoxin, certain anticancer agents), knowing if a patient’s microbiota composition is skewed towards high SCFA producers vs. dysbiosis might guide dose selection [1,16,17,188,206]. For instance, a patient with low butyrate-producing bacteria (perhaps detectable by low fecal butyrate or specific 16S rRNA sequencing) might have lower baseline P-gp expression, thus absorbing more of a P-gp substrate drug, and could be started at a lower dose to avoid toxicity [207]. Conversely, a patient on chronic antibiotics or with microbiome depletion might need higher doses to achieve efficacy due to upregulated efflux transporters. In the future, clinical assays might measure microbial metabolites (like fecal LCA or butyrate levels) as biomarkers for transporter activity in the gut [208].
Interventions to modify the microbiome can be personalized. If a patient is a poor absorber of a life-saving drug due to high efflux, one could consider co-administering a specific probiotic that competes with or reduces the efflux-enhancing bacteria, or even providing an enzyme/pro-drug that the microbiome will convert to a transporter inhibitor in situ (a novel therapeutic strategy hinted by the E. lenta digoxin case) [209]. On the other hand, if a patient suffers toxicity from a drug because their microbiome inactivates a transporter (as E. lenta does to P-gp), one might use targeted antibiotics or dietary changes to transiently remove that effect (e.g., avoid foods or supplements high in certain flavonoids that feed those bacteria during drug therapy) [188,189,190,210]. The concept of a “microbial pharmacotype” has been proposed—classifying individuals by their microbiome’s capacity to modulate drugs. Incorporating this into precision medicine could improve drug safety and efficacy, especially for drugs where transporter polymorphisms alone do not fully predict outcomes [18,163].
7. Drug–Drug–Microbiota Interactions
Classically, drug–drug interactions (DDIs) consider how one drug affects the metabolism or transport of another (often via enzyme or transporter inhibition/induction). We now should extend this model to drug–drug–microbiota interactions. For example, consider a patient taking Drug A that alters the microbiome (like a long-term antibiotic, or a proton pump inhibitor which is known to shift gut flora) and Drug B that is a transporter substrate. Drug A could change microbial metabolite profiles and thus the expression of transporters that handle Drug B [211,212].
A concrete example, a patient on rifampin (a PXR-activating antibiotic) will not only directly induce P-gp via PXR but also markedly alter the gut microbiota composition (rifampin has antibacterial effects) [213]. The microbiome change might reduce SCFA levels (since rifampin can kill commensals), which paradoxically could counteract some induction or alter other transporters [214]. The net effect on Drug B (say a P-gp substrate like dabigatran) could be complex, rifampin’s direct PXR induction increases P-gp, reducing dabigatran levels, but microbiome-mediated effects might further modulate this outcome (perhaps enhancing it if SCFAs drop, since SCFAs were also inducing P-gp normally) [215]. Understanding these three-way interactions is important in polypharmacy.
Another example is metformin and antibiotics: metformin’s efficacy partly depends on microbiota; co-prescription of antibiotics for an infection can cause blood sugar control to fluctuate in diabetic patients on metformin [216]. This is likely due to microbiota disruption affecting both drug metabolism and transporter expression (like OCTs in gut or bile acid transport via FXR) [217]. Thus, clinicians should be aware that adding or removing an antibiotic, or any microbiome-impacting drug (certain psychiatric medications, NSAIDs, etc., also change gut flora), can have knock-on effects on transporter-mediated drug handling [218].
8. Pharmacokinetic Modeling and Drug Development
In drug development, physiologically based pharmacokinetic (PBPK) models are used to predict drug absorption and disposition. Traditionally, these models include terms for gut transporter expression and activity that are fixed or vary by genetics. Incorporating the microbiome as a dynamic compartment is the new frontier [219,220]. Some researchers have begun developing PBPK models that include intestinal compartments with microbial drug metabolism activity [221]. This can be expanded to microbial effects on transporters: e.g., models can simulate how varying butyrate concentrations in colon affect P-gp function and thus the fraction of drug absorbed at different gut segments [222]. By plugging in patient-specific microbiome data (obtained via sequencing or metabolomics), one could predict individualized drug AUCs and Cmax with better accuracy than using a one-size transporter expression value [223].
Another implication is in toxicity testing, as animal models for drug toxicity might need to consider microbiota differences. Often, lab animals have different microbiota than humans, which could lead to discrepancies in transporter expression and hence drug exposure [224]. A drug causing intestinal toxicity in rats might do so because those rats lack a certain microbiome protection (like producing mucosal P-gp); gnotobiotic models colonized with human flora or co-housed animals might be useful to bridge this gap [225].
Microbiota Modifiers Affecting Microbiota–Transporter Pathways
Recognizing these pathways opens the possibility of using microbiota modifiers to influence them for patient benefit:
- Probiotics or Prebiotics: Administering specific strains known to induce transporters (like Lactobacillus for P-gp) could be a strategy to reduce drug absorption when desirable, such as preventing systemic uptake of a toxin or modulating local drug delivery [226]. In inflammatory conditions, probiotic-induced P-gp might help export inflammatory mediators (as P-gp exports endocannabinoids that suppress neutrophils) [227].
- Inhibitors of negative pathways: Blocking the effect of LPS/TLR4 with drugs (e.g., TLR4 antagonists) might prevent the inflammatory downregulation of transporters during infections or sepsis, potentially protecting the barrier and preventing unpredictable drug absorption in critical illness [228].
- Microbiome engineering: Fecal transplants or engineered bacterial consortia could be deployed to manage chronic diseases where transporter regulation is a factor. For example, in IBD, a consortium that produces high butyrate and secondary bile acids might be given to maintain P-gp and promote remission (as low P-gp is implicated in colitis severity) [229]. In metabolic disease, modulating bile-acid metabolizing bacteria might alter FXR signaling and intestinal nutrient transporters to treat obesity/diabetes [230].
- Adjunct therapy in chemotherapy: Given the role of microbial β-glucuronidases and transporters in irinotecan toxicity, combining chemotherapy with microbiome-targeted interventions (antibiotics, enzymatic inhibitors like pPB—a bacterial β-glucuronidase inhibitor, or transporter inducers) is being investigated. Early trials with such adjuncts show promise in reducing side effects without compromising efficacy [231].
In all, the convergence of microbiology and pharmacology heralds a more holistic approach to treatment—where one not only prescribes a drug but also can manipulate the patient’s microbiota or use its signals to optimize therapy.
9. Conclusions
The regulation of intestinal drug transporters by the gut microbiota represents a sophisticated multi-tiered interaction between our microbial symbionts and our own physiology. Specific gut microbes and their metabolites (short-chain fatty acids, secondary bile acids, indoles, etc.) can modulate key transport proteins like P-glycoprotein, BCRP, OATPs, and MRPs through well-defined molecular pathways, engaging nuclear receptors (PXR, FXR, AhR, CAR), transcription factors (Nrf2, NF-κB, AP-1), and even direct chemical inhibition of transporter function. These interactions have tangible outcomes on drug pharmacokinetics, influencing how much of an orally administered drug is absorbed, how it is distributed, and the intensity of its effects. They also affect the intestinal barrier’s ability to defend against toxins and maintain immune homeostasis.
Importantly, the microbiota’s influence on drug transporters can be both helpful and harmful. In some cases, it supports the body by speeding up drug elimination, protecting against toxins, and reducing inflammation. In others, it can be problematic, making a drug less effective or even causing toxicity. Therefore, treatment should be personalized: for one patient, strengthening the microbiota might boost transporter activity and lower side effects, while for another, reducing its influence could help improve drug absorption.
As research progresses, a more integrative pharmacology is emerging, combining microbiomics, genomics, and traditional PK/PD. Clinical management of drugs may soon routinely include microbiome assessments, and interventions like diet or probiotics could become standard adjuncts to modulate drug transport and response. For drug developers and regulatory sciences, considering the microbiome as an active “organ” influencing drug disposition will be important in designing trials and dosage guidelines (Figure 6).

Figure 6.
Pharmacomicrobiomics and personalized therapy.
In conclusion, the gut microbiota is a key regulator of intestinal drug transporters through advanced molecular and cellular mechanisms that we are just beginning to fully elucidate. Embracing this knowledge will enhance our ability to predict drug responses, avoid adverse interactions, and personalize therapy. As this field advances, we move closer to a future of precision medicine where not only the patient’s genome but also their microbiome is factored into optimal drug treatment strategies. The old adage “we are what we eat” might be amended to “we absorb what our gut microbes let through,” highlighting the integral role of our microbial partners in pharmacology.
Author Contributions
Conceptualization, P.R. and M.D., methodology, P.R., O.P., M.P. and M.D.; formal analysis, P.R., O.P., M.P. and M.D.; investigation, P.R., O.P., M.P. and M.D.; resources, P.R. and M.D.; data curation, P.R., O.P., M.P. and M.D.; writing—original draft preparation, P.R., O.P. and M.P.; writing—review and editing, P.R., O.P., M.P. and M.D.; supervision, M.D.; project administration, P.R.; funding acquisition, M.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed thus study. Data sharing is not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Foley, S.E.; Tuohy, C.; Dunford, M.; Grey, M.J.; De Luca, H.; Cawley, C.; Szabady, R.L.; Maldonado-Contreras, A.; Houghton, J.M.; Ward, D.V.; et al. Gut microbiota regulation of P-glycoprotein in the intestinal epithelium in maintenance of homeostasis. Microbiome 2021, 9, 183. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Zhu, L.; Wang, K.; Murray, M. Recent advance in the pharmacogenomics of human solute carrier transporters (SLCs) in drug disposition. Adv. Drug Deliv. Rev. 2017, 116, 21–36. [Google Scholar] [CrossRef]
- Drozdzik, M.; Czekawy, I.; Oswald, S.; Drozdzik, A. Intestinal drug transporters in pathological states: An overview. Pharmacol. Rep. 2020, 72, 1173–1194. [Google Scholar] [CrossRef] [PubMed]
- Nigam, S.K.; Granados, J.C. OAT, OATP, and MRP Drug Transporters and the Remote Sensing and Signaling Theory. Annu. Rev. Pharmacol. Toxicol. 2023, 63, 637–660. [Google Scholar] [CrossRef] [PubMed]
- Oostendorp, R.L.; Beijnen, J.H.; Schellens, J.H. The biological and clinical role of drug transporters at the intestinal barrier. Cancer Treat. Rev. 2009, 35, 137–147. [Google Scholar] [CrossRef]
- Brouwer, K.L.R.; Evers, R.; Hayden, E.; Hu, S.; Li, C.Y.; Meyer Zu Schwabedissen, H.E.; Neuhoff, S.; Oswald, S.; Piquette-Miller, M.; Saran, C.; et al. Regulation of drug transport proteins—From mechanisms to clinical impact: A white paper on behalf of the International Transporter Consortium. Clin. Pharmacol. Ther. 2022, 112, 461–484. [Google Scholar] [CrossRef]
- Suominen, L.; Stenberg, E.; Sjöstedt, N.; Kidron, H. Food additives inhibit intestinal drug transporters but have limited effect on in vitro drug permeability. Mol. Pharm. 2025, 22, 5627–5637. [Google Scholar] [CrossRef]
- Murakami, T.; Takano, M. Intestinal efflux transporters and drug absorption. Expert Opin. Drug Metab. Toxicol. 2008, 4, 923–939. [Google Scholar] [CrossRef]
- Mercado-Lubo, R.; McCormick, B.A. The interaction of gut microbes with host ABC transporters. Gut Microbes 2010, 1, 301–306. [Google Scholar] [CrossRef]
- Montagnani, M.; Bottalico, L.; Potenza, M.A.; Charitos, I.A.; Topi, S.; Colella, M.; Santacroce, L. The crosstalk between gut microbiota and nervous system: A bidirectional interaction between microorganisms and metabolome. Int. J. Mol. Sci. 2023, 24, 10322. [Google Scholar] [CrossRef]
- Mao, Q.; Unadkat, J.D. Role of the breast cancer resistance protein (BCRP/ABCG2) in drug transport—An update. AAPS J. 2015, 17, 65–82. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Duong, V.A.; Maeng, H.J. Pharmaceutical formulations with P-glycoprotein inhibitory effect as promising approaches for enhancing oral drug absorption and bioavailability. Pharmaceutics 2021, 13, 110. [Google Scholar] [CrossRef] [PubMed]
- Cvetkovic, M.; Leake, B.; Fromm, M.F.; Wilkinson, G.R.; Kim, R.B. OATP and P-glycoprotein transporters mediate the cellular uptake and excretion of fexofenadine. Drug Metab. Dispos. 1999, 27, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Shu, Y. Transcriptional regulation of solute carrier (SLC) drug transporters. Drug Metab. Dispos. 2022, 50, 1238–1250. [Google Scholar] [CrossRef]
- Kyaw, K.S.; Adegoke, S.C.; Ajani, C.K.; Nwabor, O.F.; Onyeaka, H. Toward in-process technology-aided automation for enhanced microbial food safety and quality assurance in milk and beverages processing. Crit. Rev. Food Sci. Nutr. 2022, 64, 1715–1735. [Google Scholar] [CrossRef]
- Kyaw, T.S.; Zhang, C.; Sandy, M.; Trepka, K.; Zhang, S.; Ramirez Hernandez, L.A.; Ramirez, L.; Goh, J.J.N.; Yu, K.; Dimassa, V.; et al. Human gut Actinobacteria boost drug absorption by secreting P-glycoprotein ATPase inhibitors. iScience 2024, 27, 110122. [Google Scholar] [CrossRef]
- Degraeve, A.L.; Haufroid, V.; Loriot, A.; Gatto, L.; Andries, V.; Vereecke, L.; Elens, L.; Bindels, L.B. Gut microbiome modulates tacrolimus pharmacokinetics through the transcriptional regulation of ABCB1. Microbiome 2023, 11, 138. [Google Scholar] [CrossRef]
- Trepka, K.R.; Olson, C.A.; Upadhyay, V.; Zhang, C.; Turnbaugh, P.J. Pharma[e]cology: How the gut microbiome contributes to variations in drug response. Annu. Rev. Pharmacol. Toxicol. 2025, 65, 355–373. [Google Scholar] [CrossRef]
- Sun, C.; Chen, L.; Shen, Z. Mechanisms of gastrointestinal microflora on drug metabolism in clinical practice. Saudi Pharm. J. 2019, 27, 1146–1156. [Google Scholar] [CrossRef]
- Swanson, H.I. Drug metabolism by the host and gut microbiota: A partnership or rivalry? Drug Metab. Dispos. 2015, 43, 1499–1504. [Google Scholar] [CrossRef]
- Lundberg, R.; Toft, M.F.; August, B.; Hansen, A.K.; Hansen, C.H. Antibiotic-treated versus germ-free rodents for microbiota transplantation studies. Gut Microbes 2016, 7, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Cresci, G.A.; Thangaraju, M.; Mellinger, J.D.; Liu, K.; Ganapathy, V. Colonic gene expression in conventional and germ-free mice with a focus on the butyrate receptor GPR109A and the butyrate transporter SLC5A8. J. Gastrointest. Surg. 2010, 14, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Foley, S.E.; Dente, M.J.; Lei, X.; Sallis, B.F.; Loew, E.B.; Meza-Segura, M.; Fitzgerald, K.A.; McCormick, B.A. Microbial metabolites orchestrate a distinct multi-tiered regulatory network in the intestinal epithelium that directs P-glycoprotein expression. mBio 2022, 13, e01993-22. [Google Scholar] [CrossRef] [PubMed]
- Vendramin, R.; Litchfield, K.; Swanton, C. Cancer evolution: Darwin and beyond. EMBO J. 2021, 40, e108389. [Google Scholar] [CrossRef]
- Portincasa, P.; Bonfrate, L.; Vacca, M.; De Angelis, M.; Farella, I.; Lanza, E.; Khalil, M.; Wang, D.Q.; Sperandio, M.; Di Ciaula, A. Gut microbiota and short chain fatty acids: Implications in glucose homeostasis. Int. J. Mol. Sci. 2022, 23, 1105. [Google Scholar] [CrossRef]
- Theis, B.F.; Park, J.S.; Kim, J.S.A.; Zeydabadinejad, S.; Vijay-Kumar, M.; Yeoh, B.S.; Saha, P. Gut feelings: How microbes, diet, and host immunity shape disease. Biomedicines 2025, 13, 1357. [Google Scholar] [CrossRef]
- Ufer, M.; Häsler, R.; Jacobs, G.; Haenisch, S.; Lächelt, S.; Faltraco, F.; Sina, C.; Rosenstiel, P.; Nikolaus, S.; Schreiber, S.; et al. Decreased sigmoidal ABCB1 (P-glycoprotein) expression in ulcerative colitis is associated with disease activity. Pharmacogenomics 2009, 10, 1941–1953. [Google Scholar] [CrossRef]
- Kumari, R.; Ahuja, V.; Paul, J. Fluctuations in butyrate-producing bacteria in ulcerative colitis patients of North India. World J. Gastroenterol. 2013, 19, 3404–3414. [Google Scholar] [CrossRef]
- Sultan, S.; El-Mowafy, M.; Elgaml, A.; Ahmed, T.A.E.; Hassan, H.; Mottawea, W. Metabolic influences of gut microbiota dysbiosis on inflammatory bowel disease. Front. Physiol. 2021, 12, 715506. [Google Scholar] [CrossRef]
- Zhu, S.; Han, M.; Liu, S.; Fan, L.; Shi, H.; Li, P. Composition and diverse differences of intestinal microbiota in ulcerative colitis patients. Front. Cell Infect. Microbiol. 2022, 12, 953962. [Google Scholar] [CrossRef]
- Prame Kumar, K.; Ooi, J.D.; Goldberg, R. The interplay between the microbiota, diet and T regulatory cells in the preservation of the gut barrier in inflammatory bowel disease. Front. Microbiol. 2023, 14, 1291724. [Google Scholar] [CrossRef] [PubMed]
- Takiishi, T.; Fenero, C.I.M.; Câmara, N.O.S. Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers 2017, 5, e1373208. [Google Scholar] [CrossRef] [PubMed]
- Mogoş, G.F.R.; Manciulea, M.; Enache, R.-M.; Pavelescu, L.A.; Popescu, O.A.; Cretoiu, S.M.; Marinescu, I. Intestinal microbiota in early life: Latest findings regarding the role of probiotics as a treatment approach for dysbiosis. Nutrients 2025, 17, 2071. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Zhao, L.; Li, C.; Xiao, J.; Wu, J.; Li, X.; Zhang, J.; Zhao, Y.; Wang, J. Regulation of CYP450 and drug transporter mediated by gut microbiota under high-altitude hypoxia. Front. Pharmacol. 2022, 13, 977370. [Google Scholar] [CrossRef]
- Zhao, Q.; Chen, Y.; Huang, W.; Zhou, H.; Zhang, W. Drug-microbiota interactions: An emerging priority for precision medicine. Signal Transduct. Target Ther. 2023, 8, 386. [Google Scholar] [CrossRef]
- Kim, S.; Seo, S.U.; Kweon, M.N. Gut microbiota-derived metabolites tune host homeostasis fate. Semin. Immunopathol. 2024, 46, 2. [Google Scholar] [CrossRef]
- Masse, K.E.; Lu, V.B. Short-chain fatty acids, secondary bile acids and indoles: Gut microbial metabolites with effects on enteroendocrine cell function and their potential as therapies for metabolic disease. Front. Endocrinol. 2023, 14, 1169624. [Google Scholar] [CrossRef]
- Banfi, D.; Moro, E.; Bosi, A.; Bistoletti, M.; Cerantola, S.; Crema, F.; Maggi, F.; Giron, M.C.; Giaroni, C.; Baj, A. Impact of microbial metabolites on microbiota–gut–brain axis in inflammatory bowel disease. Int. J. Mol. Sci. 2021, 22, 1623. [Google Scholar] [CrossRef]
- Gasaly, N.; de Vos, P.; Hermoso, M.A. Impact of bacterial metabolites on gut barrier function and host immunity: A focus on bacterial metabolism and its relevance for intestinal inflammation. Front. Immunol. 2021, 12, 658354. [Google Scholar] [CrossRef]
- Youdim, K.A.; Dobbie, M.S.; Kuhnle, G.; Proteggente, A.R.; Abbott, N.J.; Rice-Evans, C. Interaction between flavonoids and the blood-brain barrier: In vitro studies. J. Neurochem. 2003, 85, 180–192. [Google Scholar] [CrossRef]
- Han, D.S.; Wu, W.K.; Liu, P.Y.; Yang, Y.T.; Hsu, H.C.; Kuo, C.H.; Wu, M.S.; Wang, T.G. Differences in the gut microbiome and reduced fecal butyrate in elders with low skeletal muscle mass. Clin. Nutr. 2022, 41, 1491–1500. [Google Scholar] [CrossRef] [PubMed]
- Wahlström, A.; Sayin, S.I.; Marschall, H.U.; Bäckhed, F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chiang, J.Y. Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 2014, 66, 948–983. [Google Scholar] [CrossRef] [PubMed]
- Nireeksha; Luke, A.M.; Kumari, N.S.; Hegde, M.N.; Hegde, N.N. Metabolic interplay of SCFAs in the gut and oral microbiome: A link to health and disease. Front. Oral Health 2025, 6, 1646382. [Google Scholar] [CrossRef]
- den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- Li, S.; Feng, W.; Wu, J.; Cui, H.; Wang, Y.; Liang, T.; An, J.; Chen, W.; Guo, Z.; Lei, H. A narrative review: Immunometabolic interactions of host–gut microbiota and botanical active ingredients in gastrointestinal cancers. Int. J. Mol. Sci. 2024, 25, 9096. [Google Scholar] [CrossRef]
- Salvi, P.S.; Cowles, R.A. Butyrate and the intestinal epithelium: Modulation of proliferation and inflammation in homeostasis and disease. Cells 2021, 10, 1775. [Google Scholar] [CrossRef]
- Kibbie, J.J.; Dillon, S.M.; Thompson, T.A.; Purba, C.M.; McCarter, M.D.; Wilson, C.C. Butyrate directly decreases human gut lamina propria CD4 T cell function through histone deacetylase (HDAC) inhibition and GPR43 signaling. Immunobiology 2021, 226, 152126. [Google Scholar] [CrossRef]
- Modoux, M.; Rolhion, N.; Lefevre, J.H.; Oeuvray, C.; Nádvorník, P.; Illes, P.; Emond, P.; Parc, Y.; Mani, S.; Dvorak, Z.; et al. Butyrate acts through HDAC inhibition to enhance aryl hydrocarbon receptor activation by gut microbiota-derived ligands. Gut Microbes 2022, 14, 2105637. [Google Scholar] [CrossRef]
- Sun, M.; Wu, W.; Liu, Z.; Cong, Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J. Gastroenterol. 2017, 52, 1–8. [Google Scholar] [CrossRef]
- Kayama, H.; Takeda, K. Manipulation of epithelial integrity and mucosal immunity by host and microbiota-derived metabolites. Eur. J. Immunol. 2020, 50, 921–931. [Google Scholar] [CrossRef]
- Guo, W.; Liu, J.; Sun, J.; Gong, Q.; Ma, H.; Kan, X.; Cao, Y.; Wang, J.; Fu, S. Butyrate alleviates oxidative stress by regulating NRF2 nuclear accumulation and H3K9/14 acetylation via GPR109A in bovine mammary epithelial cells and mammary glands. Free Radic. Biol. Med. 2020, 152, 728–742. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zhou, T.; He, Y.; Xie, Y.; Xu, Y.; Huang, W. The role and mechanism of butyrate in the prevention and treatment of diabetic kidney disease. Front. Microbiol. 2022, 13, 961536. [Google Scholar] [CrossRef] [PubMed]
- Smolková, K.; Mikó, E.; Kovács, T.; Leguina-Ruzzi, A.; Sipos, A.; Bai, P. Nuclear factor erythroid 2-related factor 2 in regulating cancer metabolism. Antioxid. Redox Signal. 2020, 33, 966–997. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Yuan, L.; Zhu, J. The dual role of NRF2 in colorectal cancer: Targeting NRF2 as a potential therapeutic approach. J. Inflamm. Res. 2024, 17, 5985–6004. [Google Scholar] [CrossRef]
- Frommel, T.O.; Coon, J.S.; Tsuruo, T.; Roninson, I.B. Variable effects of sodium butyrate on the expression and function of the MDR1 (P-glycoprotein) gene in colon carcinoma cell lines. Int. J. Cancer 1993, 55, 297–302. [Google Scholar] [CrossRef]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef]
- Zhou, D.; Pan, Q.; Shen, F.; Cao, H.X.; Ding, W.J.; Chen, Y.W.; Fan, J.G. Total fecal microbiota transplantation alleviates high-fat diet-induced steatohepatitis in mice via beneficial regulation of gut microbiota. Sci. Rep. 2017, 7, 1529. [Google Scholar] [CrossRef]
- Seksik, P.; Rigottier-Gois, L.; Gramet, G.; Sutren, M.; Pochart, P.; Marteau, P.; Jian, R.; Doré, J. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut 2003, 52, 237–242. [Google Scholar] [CrossRef]
- Priyamvada, S.; Anbazhagan, A.N.; Kumar, A.; Soni, V.; Alrefai, W.A.; Gill, R.K.; Dudeja, P.K.; Saksena, S. Lactobacillus acidophilus stimulates intestinal P-glycoprotein expression via a c-Fos/c-Jun-dependent mechanism in intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 310, G599–G608. [Google Scholar] [CrossRef]
- Vinolo, M.A.R.; Rodrigues, H.G.; Nachbar, R.T.; Curi, R. Regulation of inflammation by short chain fatty acids. Nutrients 2011, 3, 858–876. [Google Scholar] [CrossRef]
- Buyse, M.; Radeva, G.; Bado, A.; Farinotti, R. Intestinal inflammation induces adaptation of P-glycoprotein expression and activity. Biochem. Pharmacol. 2005, 69, 1745–1754. [Google Scholar] [CrossRef]
- Kalkan, A.E.; BinMowyna, M.N.; Raposo, A.; Ahmad, M.F.; Ahmed, F.; Otayf, A.Y.; Carrascosa, C.; Saraiva, A.; Karav, S. Beyond the gut: Unveiling butyrate’s global health impact through gut health and dysbiosis-related conditions: A narrative review. Nutrients 2025, 17, 1305. [Google Scholar] [CrossRef] [PubMed]
- Hagos, Y.; Wolff, N.A. Assessment of the role of renal organic anion transporters in drug-induced nephrotoxicity. Toxins 2010, 2, 2055–2082. [Google Scholar] [CrossRef] [PubMed]
- Magliocca, G.; Mone, P.; Di Iorio, B.R.; Heidland, A.; Marzocco, S. Short-chain fatty acids in chronic kidney disease: Focus on inflammation and oxidative stress regulation. Int. J. Mol. Sci. 2022, 23, 5354. [Google Scholar] [CrossRef] [PubMed]
- González-Bosch, C.; Boorman, E.; Zunszain, P.A.; Mann, G.E. Short-chain fatty acids as modulators of redox signaling in health and disease. Redox Biol. 2021, 47, 102165. [Google Scholar] [CrossRef]
- Shang, Q. Inulin alleviates inflammatory response and gut barrier dysfunction via modulating microbiota in lipopolysaccharide-challenged broilers. Int. J. Biol. Macromol. 2024, 282, 137208. [Google Scholar] [CrossRef]
- Liu, H.; Johnston, L.J.; Wang, F.; Ma, X. Triggers for the Nrf2/ARE signaling pathway and its nutritional regulation: Potential therapeutic applications of ulcerative colitis. Int. J. Mol. Sci. 2021, 22, 11411. [Google Scholar] [CrossRef]
- Kiriyama, Y.; Nochi, H. Physiological Role of Bile Acids Modified by the Gut Microbiome. Microorganisms 2022, 10, 68. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Harris, S.C.; Bhowmik, S.; Kang, D.J.; Hylemon, P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016, 7, 22–39, Erratum in Gut Microbes 2016, 7, 262. [Google Scholar] [CrossRef]
- Eloranta, J.J.; Kullak-Ublick, G.A. Coordinate transcriptional regulation of bile acid homeostasis and drug metabolism. Arch. Biochem. Biophys. 2005, 433, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Geick, A.; Eichelbaum, M.; Burk, O. Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J. Biol. Chem. 2001, 276, 14581–14587. [Google Scholar] [CrossRef] [PubMed]
- Swales, K.E.; Moore, R.; Truss, N.J.; Tucker, A.; Warner, T.D.; Negishi, M.; Bishop-Bailey, D. Pregnane X receptor regulates drug metabolism and transport in the vasculature and protects from oxidative stress. Cardiovasc. Res. 2012, 93, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef]
- Devlin, A.S.; Fischbach, M.A. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat. Chem. Biol. 2015, 11, 685–690. [Google Scholar] [CrossRef]
- Ding, L.; Yang, L.; Wang, Z.; Huang, W. Bile acid nuclear receptor FXR and digestive system diseases. Acta Pharm. Sin. B 2015, 5, 135–144. [Google Scholar] [CrossRef]
- Inagaki, T.; Choi, M.; Moschetta, A.; Peng, L.; Cummins, C.L.; McDonald, J.G.; Luo, G.; Jones, S.A.; Goodwin, B.; Richardson, J.A.; et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005, 2, 217–225. [Google Scholar] [CrossRef]
- Groen, A.; Romero, M.R.; Kunne, C.; Hoosdally, S.J.; Dixon, P.H.; Wooding, C.; Williamson, C.; Seppen, J.; Van den Oever, K.; Mok, K.S.; et al. Complementary functions of the flippase ATP8B1 and the floppase ABCB4 in maintaining canalicular membrane integrity. Gastroenterology 2011, 141, 1927–1937.e4. [Google Scholar] [CrossRef]
- Armstrong, L.E.; Guo, G.L. Role of FXR in liver inflammation during nonalcoholic steatohepatitis. Curr. Pharmacol. Rep. 2017, 3, 92–100. [Google Scholar] [CrossRef]
- Chiang, J.Y. Hepatocyte nuclear factor 4α regulation of bile acid and drug metabolism. Expert Opin. Drug Metab. Toxicol. 2009, 5, 137–147. [Google Scholar] [CrossRef]
- Vavassori, P.; Mencarelli, A.; Renga, B.; Distrutti, E.; Fiorucci, S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J. Immunol. 2009, 183, 6251–6261. [Google Scholar] [CrossRef]
- Gadaleta, R.M.; van Erpecum, K.J.; Oldenburg, B.; Willemsen, E.C.; Renooij, W.; Murzilli, S.; Klomp, L.W.; Siersema, P.D.; Schipper, M.E.; Danese, S.; et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 2011, 60, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, T.A.; Zalzala, M.H. FXR crosstalk with other nuclear receptors. J. Physiol. Biochem. 2025. [Google Scholar] [CrossRef] [PubMed]
- Modica, S.; Bellafante, E.; Moschetta, A. Master regulation of bile acid and xenobiotic metabolism via the FXR, PXR and CAR trio. Front. Biosci. 2009, 14, 4719–4745. [Google Scholar] [CrossRef] [PubMed]
- Fickert, P.; Fuchsbichler, A.; Marschall, H.U.; Wagner, M.; Zollner, G.; Krause, R.; Zatloukal, K.; Jaeschke, H.; Denk, H.; Trauner, M. Lithocholic acid feeding induces segmental bile duct obstruction and destructive cholangitis in mice. Am. J. Pathol. 2006, 168, 410–422. [Google Scholar] [CrossRef]
- Xie, Q.S.; Zhang, J.X.; Liu, M.; Liu, P.H.; Wang, Z.J.; Zhu, L.; Jiang, L.; Jin, M.M.; Liu, X.N.; Liu, L.; et al. Short-chain fatty acids exert opposite effects on the expression and function of P-glycoprotein and breast cancer resistance protein in rat intestine. Acta Pharmacol. Sin. 2021, 42, 470–481. [Google Scholar] [CrossRef]
- Li, T.; Liu, W.; Hui, W.; Shi, T.; Liu, H.; Feng, Y.; Gao, F. Integrated analysis of ulcerative colitis revealed an association between PHLPP2 and immune infiltration. Dis. Markers 2022, 2022, 4983471. [Google Scholar] [CrossRef]
- Bertolini, A.; Fiorotto, R.; Strazzabosco, M. Bile acids and their receptors: Modulators and therapeutic targets in liver inflammation. Semin. Immunopathol. 2022, 44, 547–564. [Google Scholar] [CrossRef]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef]
- Wlodarska, M.; Luo, C.; Kolde, R.; d’Hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic acid produced by commensal Peptostreptococcus species suppresses inflammation. Cell Host Microbe 2017, 22, 25–37.e6. [Google Scholar] [CrossRef]
- Owe-Larsson, M.; Drobek, D.; Iwaniak, P.; Kloc, R.; Urbanska, E.M.; Chwil, M. Microbiota-derived tryptophan metabolite indole-3-propionic acid—Emerging role in neuroprotection. Molecules 2025, 30, 3628. [Google Scholar] [CrossRef] [PubMed]
- Alexeev, E.E.; Lanis, J.M.; Kao, D.J.; Campbell, E.L.; Kelly, C.J.; Battista, K.D.; Gerich, M.E.; Jenkins, B.R.; Walk, S.T.; Kominsky, D.J.; et al. Microbiota-derived indole metabolites promote human and murine intestinal homeostasis through regulation of interleukin-10 receptor. Am. J. Pathol. 2018, 188, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, T.D.; Murray, I.A.; Perdew, G.H. Indole and tryptophan metabolism: Endogenous and dietary routes to Ah receptor activation. Drug Metab. Dispos. 2015, 43, 1522–1535. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Planchais, J.; Sokol, H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef]
- Ebert, B.; Seidel, A.; Lampen, A. Identification of BCRP as transporter of benzo[a]pyrene conjugates metabolically formed in Caco-2 cells and its induction by Ah-receptor agonists. Carcinogenesis 2005, 26, 1754–1763. [Google Scholar] [CrossRef]
- Kukal, S.; Guin, D.; Rawat, C.; Bora, S.; Mishra, M.K.; Sharma, P.; Paul, P.R.; Kanojia, N.; Grewal, G.K.; Kukreti, S.; et al. Multidrug efflux transporter ABCG2: Expression and regulation. Cell. Mol. Life Sci. 2021, 78, 6887–6939. [Google Scholar] [CrossRef]
- Tan, K.P.; Wang, B.; Yang, M.; Boutros, P.C.; Macaulay, J.; Xu, H.; Chuang, A.I.; Kosuge, K.; Yamamoto, M.; Takahashi, S.; et al. Aryl hydrocarbon receptor is a transcriptional activator of the human breast cancer resistance protein (BCRP/ABCG2). Mol. Pharmacol. 2010, 78, 175–185. [Google Scholar] [CrossRef]
- Zgarbová, E.; Vrzal, R. The impact of indoles activating the aryl hydrocarbon receptor on androgen receptor activity in the 22Rv1 prostate cancer cell line. Int. J. Mol. Sci. 2022, 24, 502. [Google Scholar] [CrossRef]
- Tompkins, L.M.; Li, H.; Li, L.; Lynch, C.; Xie, Y.; Nakanishi, T.; Ross, D.D.; Wang, H. A novel xenobiotic responsive element regulated by aryl hydrocarbon receptor is involved in the induction of BCRP/ABCG2 in LS174T cells. Biochem. Pharmacol. 2010, 80, 1754–1761. [Google Scholar] [CrossRef]
- Liu, J.; Tan, Y.; Cheng, H.; Zhang, D.; Feng, W.; Peng, C. Functions of gut microbiota metabolites, current status and future perspectives. Aging Dis. 2022, 13, 1106–1126. [Google Scholar] [CrossRef]
- Mackowiak, B.; Gao, B. Interplay of gut microbes and aryl hydrocarbon receptor in alcohol-associated liver disease. Cell. Mol. Gastroenterol. Hepatol. 2022, 13, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Qi, R.; Wang, J.; Liu, Z.; Wu, Z. Indole-3-propionic acid, a functional metabolite of Clostridium sporogenes, promotes muscle tissue development and reduces muscle cell inflammation. Int. J. Mol. Sci. 2021, 22, 12435. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Whitley, C.S.; Haribabu, B.; Jala, V.R. Regulation of intestinal barrier function by microbial metabolites. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 1463–1482. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tang, Y.; Guo, C.; Wang, J.; Boral, D.; Nie, D. Nuclear receptors in the multidrug resistance through the regulation of drug-metabolizing enzymes and drug transporters. Biochem. Pharmacol. 2012, 83, 1112–1126. [Google Scholar] [CrossRef]
- Reznicek, J.; Ceckova, M.; Ptackova, Z.; Martinec, O.; Tupova, L.; Cerveny, L.; Staud, F. MDR1 and BCRP transporter-mediated drug-drug interaction between rilpivirine and abacavir and effect on intestinal absorption. Antimicrob. Agents Chemother. 2017, 61, e00837-17. [Google Scholar] [CrossRef]
- Sachar, M.; Ma, X. Role of ABCG2 in liver injury associated with erythropoietic protoporphyria. Hepatology 2016, 64, 305–315. [Google Scholar] [CrossRef]
- To, K.K.; Robey, R.; Zhan, Z.; Bangiolo, L.; Bates, S.E. Upregulation of ABCG2 by romidepsin via the aryl hydrocarbon receptor pathway. Mol. Cancer Res. 2011, 9, 516–527. [Google Scholar] [CrossRef]
- Kang, H.; Chen, Z.; Wang, B.; Chen, Z. The AhR/IL-22 axis in chronic gut inflammation: Unraveling mechanisms and therapeutic prospects. Front. Immunol. 2025, 16, 1668173. [Google Scholar] [CrossRef]
- Gargaro, M.; Manni, G.; Scalisi, G.; Puccetti, P.; Fallarino, F. Tryptophan metabolites at the crossroad of immune-cell interaction via the aryl hydrocarbon receptor: Implications for tumor immunotherapy. Int. J. Mol. Sci. 2021, 22, 4644. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef]
- Krishnan, S.; Ding, Y.; Saedi, N.; Choi, M.; Sridharan, G.V.; Sherr, D.H.; Yarmush, M.L.; Alaniz, R.C.; Jayaraman, A.; Lee, K. Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep. 2018, 23, 1099–1111. [Google Scholar] [CrossRef]
- Jing, W.; Dong, S.; Xu, Y.; Liu, J.; Ren, J.; Liu, X.; Zhu, M.; Zhang, M.; Shi, H.; Li, N.; et al. Gut microbiota-derived tryptophan metabolites regulated by Wuji Wan to attenuate colitis through AhR signaling activation. Acta Pharm. Sin. B 2025, 15, 205–223. [Google Scholar] [CrossRef] [PubMed]
- García-Villalba, R.; Giménez-Bastida, J.A.; Cortés-Martín, A.; Ávila-Gálvez, M.Á.; Tomás-Barberán, F.A.; Selma, M.V.; Espín, J.C.; González-Sarrías, A. Urolithins: A comprehensive update on their metabolism, bioactivity, and associated gut microbiota. Mol. Nutr. Food Res. 2022, 66, e2101019. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Qin, M.; Zheng, Q.; Wang, K.; Han, X.; Yang, Q.; Sang, X.; Cao, G. Role of bile acid receptors in the development and function of diabetic nephropathy. Kidney Int. Rep. 2024, 9, 3116–3133. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.Y.; Chen, Z.J.; Shah, N.P.; El-Nezami, H. Modulation of intestinal epithelial defense responses by probiotic bacteria. Crit. Rev. Food Sci. Nutr. 2016, 56, 2628–2641. [Google Scholar] [CrossRef]
- Peterson, C.T.; Sharma, V.; Elmén, L.; Peterson, S.N. Immune homeostasis, dysbiosis and therapeutic modulation of the gut microbiota. Clin. Exp. Immunol. 2015, 179, 363–377. [Google Scholar] [CrossRef]
- Saksena, S.; Goyal, S.; Raheja, G.; Singh, V.; Akhtar, M.; Nazir, T.M.; Alrefai, W.A.; Gill, R.K.; Dudeja, P.K. Upregulation of P-glycoprotein by probiotics in intestinal epithelial cells and in the dextran sulfate sodium model of colitis in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G1115–G1123. [Google Scholar] [CrossRef]
- Nidhi; Iqbal, N.; Khan, N.A. Polyamines interaction with gaseous signaling molecules for resilience against drought and heat stress in plants. Plants 2025, 14, 273. [Google Scholar] [CrossRef]
- Altaany, Z.; Yang, G.; Wang, R. Crosstalk between hydrogen sulfide and nitric oxide in endothelial cells. J. Cell. Mol. Med. 2013, 17, 879–888. [Google Scholar] [CrossRef]
- Ghose, R.; Guo, T.; Haque, N. Regulation of gene expression of hepatic drug metabolizing enzymes and transporters by the Toll-like receptor 2 ligand, lipoteichoic acid. Arch. Biochem. Biophys. 2009, 481, 123–130. [Google Scholar] [CrossRef]
- Cressman, A.M.; Petrovic, V.; Piquette-Miller, M. Inflammation-mediated changes in drug transporter expression/activity: Implications for therapeutic drug response. Expert Rev. Clin. Pharmacol. 2012, 5, 69–89. [Google Scholar] [CrossRef] [PubMed]
- Wittmann, G.; Mohácsik, P.; Balkhi, M.Y.; Gereben, B.; Lechan, R.M. Endotoxin-induced inflammation down-regulates L-type amino acid transporter 1 (LAT1) expression at the blood-brain barrier of male rats and mice. Fluids Barriers CNS 2015, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, V.; Teng, S.; Piquette-Miller, M. Regulation of drug transporters during infection and inflammation. Mol. Interv. 2007, 7, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Kumar, A.; Jayawardena, D.; Priyamvada, S.; Anbazhagan, A.N.; Alrefai, W.A.; Gill, R.K.; Dudeja, P.K.; Saksena, S. Citrobacter rodentium infection inhibits colonic P-glycoprotein expression. Gene Rep. 2020, 18, 100549. [Google Scholar] [CrossRef]
- Shen, Y.; Fan, N.; Ma, S.X.; Cheng, X.; Yang, X.; Wang, G. Gut microbiota dysbiosis: Pathogenesis, diseases, prevention, and therapy. MedComm 2025, 6, e70168. [Google Scholar] [CrossRef]
- Ho, G.T.; Moodie, F.M.; Satsangi, J. Multidrug resistance 1 gene (P-glycoprotein 170): An important determinant in gastrointestinal disease? Gut 2003, 52, 759–766. [Google Scholar] [CrossRef]
- Yan, F.; Cao, H.; Cover, T.L.; Whitehead, R.; Washington, M.K.; Polk, D.B. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 2007, 132, 562–575. [Google Scholar] [CrossRef]
- Jin, P.; Duan, X.; Huang, Z.; Dong, Y.; Zhu, J.; Guo, H.; Tian, H.; Zou, C.G.; Xie, K. Nuclear receptors in health and disease: Signaling pathways, biological functions and pharmaceutical interventions. Signal Transduct. Target Ther. 2025, 10, 228. [Google Scholar] [CrossRef]
- Zeng, H.; Umar, S.; Rust, B.; Lazarova, D.; Bordonaro, M. Secondary bile acids and short chain fatty acids in the colon: A focus on colonic microbiome, cell proliferation, inflammation, and cancer. Int. J. Mol. Sci. 2019, 20, 1214. [Google Scholar] [CrossRef]
- Guo, G.L.; Lambert, G.; Negishi, M.; Ward, J.M.; Brewer, H.B., Jr.; Kliewer, S.A.; Gonzalez, F.J.; Sinal, C.J. Complementary roles of farnesoid X receptor, pregnane X receptor, and constitutive androstane receptor in protection against bile acid toxicity. J. Biol. Chem. 2003, 278, 45062–45071. [Google Scholar] [CrossRef]
- Quattrochi, L.C.; Guzelian, P.S. Cyp3A regulation: From pharmacology to nuclear receptors. Drug Metab. Dispos. 2001, 29, 615–622. [Google Scholar] [PubMed]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, M.; Robbins, D.; Chen, T. Targeting xenobiotic receptors PXR and CAR in human diseases. Drug Discov. Today 2015, 20, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Selwyn, F.P.; Cheng, S.L.; Bammler, T.K.; Prasad, B.; Vrana, M.; Klaassen, C.; Cui, J.Y. Developmental regulation of drug-processing genes in livers of germ-free mice. Toxicol. Sci. 2015, 147, 84–103. [Google Scholar] [CrossRef]
- Willson, T.M.; Kliewer, S.A. PXR, CAR and drug metabolism. Nat. Rev. Drug Discov. 2002, 1, 259–266. [Google Scholar] [CrossRef]
- Xu, C.; Li, C.Y.; Kong, A.N. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch. Pharm. Res. 2005, 28, 249–268. [Google Scholar] [CrossRef]
- Lefort, C.; Cani, P.D. The liver under the spotlight: Bile acids and oxysterols as pivotal actors controlling metabolism. Cells 2021, 10, 400. [Google Scholar] [CrossRef]
- Wei, S.; He, T.; Zhao, X.; Jing, M.; Li, H.; Chen, L.; Zheng, R.; Zhao, Y. Alterations in the gut microbiota and serum metabolomics of spontaneous cholestasis caused by loss of FXR signal in mice. Front. Pharmacol. 2023, 14, 1197847. [Google Scholar] [CrossRef]
- Ishizawa, M.; Matsunawa, M.; Adachi, R.; Uno, S.; Ikeda, K.; Masuno, H.; Shimizu, M.; Iwasaki, K.; Yamada, S.; Makishima, M. Lithocholic acid derivatives act as selective vitamin D receptor modulators without inducing hypercalcemia. J. Lipid Res. 2008, 49, 763–772. [Google Scholar] [CrossRef]
- Thummel, K.E.; Brimer, C.; Yasuda, K.; Thottassery, J.; Senn, T.; Lin, Y.; Ishizuka, H.; Kharasch, E.; Schuetz, J.; Schuetz, E. Transcriptional control of intestinal cytochrome P-4503A by 1α,25-dihydroxy vitamin D3. Mol. Pharmacol. 2001, 60, 1399–1406. [Google Scholar] [CrossRef]
- He, F.; Ru, X.; Wen, T. NRF2, a transcription factor for stress response and beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef] [PubMed]
- Maher, J.M.; Dieter, M.Z.; Aleksunes, L.M.; Slitt, A.L.; Guo, G.; Tanaka, Y.; Scheffer, G.L.; Chan, J.Y.; Manautou, J.E.; Chen, Y.; et al. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology 2007, 46, 1597–1610. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed]
- Ageeva, T.; Rizvanov, A.; Mukhamedshina, Y. NF-κB and JAK/STAT signaling pathways as crucial regulators of neuroinflammation and astrocyte modulation in spinal cord injury. Cells 2024, 13, 581. [Google Scholar] [CrossRef]
- Lawrence, T. The nuclear factor NF-κB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Ghosh, S. The NF-κB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef]
- Mariappan, N.; Elks, C.M.; Sriramula, S.; Guggilam, A.; Liu, Z.; Borkhsenious, O.; Francis, J. NF-κB-induced oxidative stress contributes to mitochondrial and cardiac dysfunction in type II diabetes. Cardiovasc. Res. 2010, 85, 473–483. [Google Scholar] [CrossRef]
- Ouahed, J.D.; Griffith, A.; Collen, L.V.; Snapper, S.B. Breaking down barriers: Epithelial contributors to monogenic IBD pathogenesis. Inflamm. Bowel Dis. 2024, 30, 1189–1206. [Google Scholar] [CrossRef]
- Song, D.; Lian, Y.; Zhang, L. The potential of activator protein 1 (AP-1) in cancer targeted therapy. Front. Immunol. 2023, 14, 1224892. [Google Scholar] [CrossRef]
- Álvarez-Carrasco, P.; Morales-Villamil, F.; Maldonado-Bernal, C. P-glycoprotein as a therapeutic target in hematological malignancies: A challenge to overcome. Int. J. Mol. Sci. 2025, 26, 4701. [Google Scholar] [CrossRef]
- Scotto, K.W.; Egan, D.A. Transcriptional regulation of MDR genes. Cytotechnology 1998, 27, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Cario, E. P-glycoprotein multidrug transporter in inflammatory bowel diseases: More questions than answers. World J. Gastroenterol. 2017, 23, 1513–1520. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, J.L.M.; Garcia, R.L. Antioxidant systems, lncRNAs, and tunneling nanotubes in cell death rescue from cigarette smoke exposure. Cells 2022, 11, 2277. [Google Scholar] [CrossRef] [PubMed]
- Hassan, F.U.; Nadeem, A.; Li, Z.; Javed, M.; Liu, Q.; Azhar, J.; Rehman, M.S.; Cui, K.; Rehman, S.U. Role of peroxisome proliferator-activated receptors (PPARs) in energy homeostasis of dairy animals: Exploiting their modulation through nutrigenomic interventions. Int. J. Mol. Sci. 2021, 22, 12463. [Google Scholar] [CrossRef]
- Ortuño Sahagún, D.; Márquez-Aguirre, A.L.; Quintero-Fabián, S.; López-Roa, R.I.; Rojas-Mayorquín, A.E. Modulation of PPAR-γ by nutraceutics as complementary treatment for obesity-related disorders and inflammatory diseases. PPAR Res. 2012, 2012, 318613. [Google Scholar] [CrossRef]
- Zhang, S.; Shen, C.; Di, H.; Wang, Y.; Guan, F. Regulatory mechanisms of phenolic acids in metabolic dysfunction-associated steatotic liver disease: A review. Antioxidants 2025, 14, 760. [Google Scholar] [CrossRef]
- Ye, X.; Zhang, T.; Han, H. PPARα: A potential therapeutic target of cholestasis. Front. Pharmacol. 2022, 13, 916866. [Google Scholar] [CrossRef]
- Pral, L.P.; Fachi, J.L.; Corrêa, R.O.; Colonna, M.; Vinolo, M.A.R. Hypoxia and HIF-1 as key regulators of gut microbiota and host interactions. Trends Immunol. 2021, 42, 604–621. [Google Scholar] [CrossRef]
- Ziello, J.E.; Jovin, I.S.; Huang, Y. Hypoxia-inducible factor (HIF)-1 regulatory pathway and its potential for therapeutic intervention in malignancy and ischemia. Yale J. Biol. Med. 2007, 80, 51–60. [Google Scholar]
- Wang, R.X.; Henen, M.A.; Lee, J.S.; Vögeli, B.; Colgan, S.P. Microbiota-derived butyrate is an endogenous HIF prolyl hydroxylase inhibitor. Gut Microbes 2021, 13, 1938380. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, T. Unveiling gut microbiota’s role: Bidirectional regulation of drug transport for improved safety. Med. Res. Rev. 2025, 45, 311–343. [Google Scholar] [CrossRef] [PubMed]
- Callaghan, R.; Luk, F.; Bebawy, M. Inhibition of the multidrug resistance P-glycoprotein: Time for a change of strategy? Drug Metab. Dispos. 2014, 42, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; Zimmermann-Kogadeeva, M.; Wegmann, R.; Goodman, A.L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019, 570, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Guthrie, B.G.H.; Alexander, M.; Noecker, C.; Ramirez, L.; Glasser, N.R.; Turnbaugh, P.J.; Balskus, E.P. Genetic manipulation of the human gut bacterium Eggerthella lenta reveals a widespread family of transcriptional regulators. Nat. Commun. 2022, 13, 7624. [Google Scholar] [CrossRef]
- Ahmed Juvale, I.I.; Abdul Hamid, A.A.; Abd Halim, K.B.; Che Has, A.T. P-glycoprotein: New insights into structure, physiological function, regulation and alterations in disease. Heliyon 2022, 8, e09777. [Google Scholar] [CrossRef]
- Seukep, A.J.; Kuete, V.; Nahar, L.; Sarker, S.D.; Guo, M. Plant-derived secondary metabolites as the main source of efflux pump inhibitors and methods for identification. J. Pharm. Anal. 2020, 10, 277–290. [Google Scholar] [CrossRef]
- Zhang, X.; Han, Y.; Huang, W.; Jin, M.; Gao, Z. The influence of the gut microbiota on the bioavailability of oral drugs. Acta Pharm. Sin. B 2021, 11, 1789–1812. [Google Scholar] [CrossRef]
- Czuba, L.C.; Hillgren, K.M.; Swaan, P.W. Post-translational modifications of transporters. Pharmacol. Ther. 2018, 192, 88–99. [Google Scholar] [CrossRef]
- Zeng, L.; Zeng, B.; Wang, H.; Li, B.; Huo, R.; Zheng, P.; Zhang, X.; Du, X.; Liu, M.; Fang, Z.; et al. Microbiota modulates behavior and protein kinase C mediated cAMP response element-binding protein signaling. Sci. Rep. 2016, 6, 29998. [Google Scholar] [CrossRef]
- Mishra, J.; Simonsen, R.; Kumar, N. Intestinal breast cancer resistance protein (BCRP) requires Janus kinase 3 activity for drug efflux and barrier functions in obesity. J. Biol. Chem. 2019, 294, 18337–18348. [Google Scholar] [CrossRef]
- Abdol Samat, H.N.; Razali, N.N.; Mahadzir, H.; Tengku Muhammad, T.S.; Ling, K.H.; Mansor, N.I.; Abidin, S.Z. The interplay of inflammation and gut-microbiota dysbiosis in Alzheimer’s disease: Mechanisms and therapeutic potential. Int. J. Mol. Sci. 2025, 26, 8905. [Google Scholar] [CrossRef]
- Vanoye, C.G.; Castro, A.F.; Pourcher, T.; Reuss, L.; Altenberg, G.A. Phosphorylation of P-glycoprotein by PKA and PKC modulates swelling-activated Cl− currents. Am. J. Physiol. 1999, 276, C370–C378. [Google Scholar] [CrossRef]
- Greer, D.A.; Ivey, S. Distinct N-glycan glycosylation of P-glycoprotein isolated from the human uterine sarcoma cell line MES-SA/Dx5. Biochim. Biophys. Acta 2007, 1770, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- Ke, X.; You, K.; Pichaud, M.; Haiser, H.J.; Graham, D.B.; Vlamakis, H.; Porter, J.A.; Xavier, R.J. Gut bacterial metabolites modulate endoplasmic reticulum stress. Genome Biol. 2021, 22, 292. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.A.; Vasi, Z.; Adhikari, R.; Gudur, A.; Ali, A.; Jiang, L.; Ferguson, R.; Liang, D.; Kuchay, S. Ubiquitin proteasome system in immune regulation and therapeutics. Curr. Opin. Pharmacol. 2022, 67, 102310. [Google Scholar] [CrossRef] [PubMed]
- Kattah, M.G.; Malynn, B.A.; Ma, A. Ubiquitin-modifying enzymes and regulation of the inflammasome. J. Mol. Biol. 2017, 429, 3471–3485. [Google Scholar] [CrossRef]
- You, M.; Xie, Z.; Zhang, N.; Zhang, Y.; Xiao, D.; Liu, S.; Zhuang, W.; Li, L.; Tao, Y. Signaling pathways in cancer metabolism: Mechanisms and therapeutic targets. Signal Transduct. Target Ther. 2023, 8, 196. [Google Scholar] [CrossRef]
- Canani, R.B.; Costanzo, M.D.; Leone, L.; Pedata, M.; Meli, R.; Calignano, A. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J. Gastroenterol. 2011, 17, 1519–1528. [Google Scholar] [CrossRef]
- Xiong, S. Gut-microbiota-driven lipid metabolism: Mechanisms and applications in swine production. Metabolites 2025, 15, 248. [Google Scholar] [CrossRef]
- Sharom, F.J. Complex interplay between the P-glycoprotein multidrug efflux pump and the membrane: Its role in modulating protein function. Front. Oncol. 2014, 4, 41. [Google Scholar] [CrossRef]
- Sakai, K.; Sakurai, T.; De Velasco, M.A.; Nagai, T.; Chikugo, T.; Ueshima, K.; Kura, Y.; Takahama, T.; Hayashi, H.; Nakagawa, K.; et al. Intestinal microbiota and gene expression reveal similarity and dissimilarity between immune-mediated colitis and ulcerative colitis. Front. Oncol. 2021, 11, 763468. [Google Scholar] [CrossRef]
- Olvera-Rosales, L.B.; Cruz-Guerrero, A.E.; Ramírez-Moreno, E.; Quintero-Lira, A.; Contreras-López, E.; Jaimez-Ordaz, J.; Castañeda-Ovando, A.; Añorve-Morga, J.; Calderón-Ramos, Z.G.; Arias-Rico, J.; et al. Impact of the gut microbiota balance on the health-disease relationship: The importance of consuming probiotics and prebiotics. Foods 2021, 10, 1261. [Google Scholar] [CrossRef]
- Lemmens, G.; Van Camp, A.; Kourula, S.; Vanuytsel, T.; Augustijns, P. Drug disposition in the lower gastrointestinal tract: Targeting and monitoring. Pharmaceutics 2021, 13, 161. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.R.; Muthukumar, T.; Dadhania, D.; Taur, Y.; Jenq, R.R.; Toussaint, N.C.; Ling, L.; Pamer, E.; Suthanthiran, M. Gut microbiota and tacrolimus dosing in kidney transplantation. PLoS ONE 2015, 10, e0122399. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Masand, A.; Wagner, M.; Kapur, S.; Dadhania, D.; Lubetzky, M.; Lee, J.R. Identification of antibiotic administration as a potentially novel factor associated with tacrolimus trough variability in kidney transplant recipients: A preliminary study. Transplant. Direct 2019, 5, e485. [Google Scholar] [CrossRef] [PubMed]
- Manes, A.; Di Renzo, T.; Dodani, L.; Reale, A.; Gautiero, C.; Di Lauro, M.; Nasti, G.; Manco, F.; Muscariello, E.; Guida, B.; et al. Pharmacomicrobiomics of classical immunosuppressant drugs: A systematic review. Biomedicines 2023, 11, 2562. [Google Scholar] [CrossRef]
- Haiser, H.J.; Seim, K.L.; Balskus, E.P.; Turnbaugh, P.J. Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics. Gut Microbes 2014, 5, 233–238. [Google Scholar] [CrossRef]
- Kvitne, K.E.; Hovd, M.; Johnson, L.K.; Wegler, C.; Karlsson, C.; Artursson, P.; Andersson, S.; Sandbu, R.; Hjelmesæth, J.; Skovlund, E.; et al. Digoxin pharmacokinetics in patients with obesity before and after a gastric bypass or a strict diet compared with normal weight individuals. Clin. Pharmacokinet. 2024, 63, 109–120. [Google Scholar] [CrossRef]
- Weiss, M.; Sermsappasuk, P.; Siegmund, W. Modeling the kinetics of digoxin absorption: Enhancement by P-glycoprotein inhibition. J. Clin. Pharmacol. 2012, 52, 381–387. [Google Scholar] [CrossRef]
- McCreight, L.J.; Stage, T.B.; Connelly, P.; Lonergan, M.; Nielsen, F.; Prehn, C.; Adamski, J.; Brøsen, K.; Pearson, E.R. Pharmacokinetics of metformin in patients with gastrointestinal intolerance. Diabetes Obes. Metab. 2018, 20, 1593–1601. [Google Scholar] [CrossRef]
- McCreight, L.J.; Bailey, C.J.; Pearson, E.R. Metformin and the gastrointestinal tract. Diabetologia 2016, 59, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Godet, M.; Meugnier, E.; Vitalis, O.; Bendridi, N.; Vieille-Marchiset, A.; Vega, N.; Benoit, B.; Pinteur, C.; Rainteau, D.; Cheillan, D.; et al. Evaluation of the effects of metformin on gut functions and microbiota and their contribution to improving glucose tolerance in diabetic mice. Mol. Metab. 2025, 102, 102263. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chen, J.; Wang, Y.; Wu, Y.; Zhang, J. Managing irinotecan-induced diarrhea: A comprehensive review of therapeutic interventions in cancer treatment. Pharmaceuticals 2025, 18, 359. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.; Boelsterli, U.A.; Redinbo, M.R. Understanding and modulating mammalian-microbial communication for improved human health. Annu. Rev. Pharmacol. Toxicol. 2014, 54, 559–580. [Google Scholar] [CrossRef]
- Hammond, W.A.; Swaika, A.; Mody, K. Pharmacologic resistance in colorectal cancer: A review. Ther. Adv. Med. Oncol. 2016, 8, 57–84. [Google Scholar] [CrossRef]
- Mego, M.; Kasperova, B.; Chovanec, J.; Danis, R.; Reckova, M.; Bystricky, B.; Konkolovsky, P.; Jurisova, S.; Porsok, S.; Vaclav, V.; et al. The beneficial effect of probiotics in the prevention of irinotecan-induced diarrhea in colorectal cancer patients with colostomy: A pooled analysis of two probiotic trials (Probio-SK-003 and Probio-SK-005) led by Slovak Cooperative Oncology Group. Front. Oncol. 2024, 14, 1438657. [Google Scholar] [CrossRef]
- Karthika, C.; Sureshkumar, R.; Zehravi, M.; Akter, R.; Ali, F.; Ramproshad, S.; Mondal, B.; Tagde, P.; Ahmed, Z.; Khan, F.S.; et al. Multidrug resistance of cancer cells and the vital role of P-glycoprotein. Life 2022, 12, 897. [Google Scholar] [CrossRef]
- Feng, W.; Liu, J.; Ao, H.; Yue, S.; Peng, C. Targeting gut microbiota for precision medicine: Focusing on the efficacy and toxicity of drugs. Theranostics 2020, 10, 11278–11301. [Google Scholar] [CrossRef]
- Xu, H.M.; Huang, H.L.; Xu, J.; He, J.; Zhao, C.; Peng, Y.; Zhao, H.L.; Huang, W.Q.; Cao, C.Y.; Zhou, Y.J.; et al. Cross-talk between butyric acid and gut microbiota in ulcerative colitis following fecal microbiota transplantation. Front. Microbiol. 2021, 12, 658292. [Google Scholar] [CrossRef]
- Karczewski, J.; Troost, F.J.; Konings, I.; Dekker, J.; Kleerebezem, M.; Brummer, R.J.; Wells, J.M. Regulation of human epithelial tight junction proteins by Lactobacillus plantarum in vivo and protective effects on the epithelial barrier. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 298, G851–G859. [Google Scholar] [CrossRef]
- López-Gómez, L.; Alcorta, A.; Abalo, R. Probiotics and probiotic-like agents against chemotherapy-induced intestinal mucositis: A narrative review. J. Pers. Med. 2023, 13, 1487. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xie, Y.; Cao, F.; Song, X. Gut microbiota-derived fatty acid and sterol metabolites: Biotransformation and immunomodulatory functions. Gut Microbes 2024, 16, 2382336. [Google Scholar] [CrossRef]
- Wang, S.; Ju, D.; Zeng, X. Mechanisms and clinical implications of human gut microbiota-drug interactions in the precision medicine era. Biomedicines 2024, 12, 194. [Google Scholar] [CrossRef] [PubMed]
- Evans, W.E.; Johnson, J.A. Pharmacogenomics: The inherited basis for interindividual differences in drug response. Annu. Rev. Genomics Hum. Genet. 2001, 2, 9–39. [Google Scholar] [CrossRef] [PubMed]
- Swanson, E.C.; Basting, C.M.; Klatt, N.R. The role of pharmacomicrobiomics in HIV prevention, treatment, and women’s health. Microbiome 2024, 12, 254. [Google Scholar] [CrossRef]
- Guo, C.; Che, X.; Briese, T.; Ranjan, A.; Allicock, O.; Yates, R.A.; Cheng, A.; March, D.; Hornig, M.; Komaroff, A.L.; et al. Deficient butyrate-producing capacity in the gut microbiome is associated with bacterial network disturbances and fatigue symptoms in ME/CFS. Cell Host Microbe 2023, 31, 288–304.e8. [Google Scholar] [CrossRef]
- Patangia, D.V.; Ryan, C.A.; Dempsey, E.; Ross, R.P.; Stanton, C. Impact of antibiotics on the human microbiome and consequences for host health. Microbiologyopen 2022, 11, e1260. [Google Scholar] [CrossRef]
- Hitch, T.C.A.; Hall, L.J.; Walsh, S.K.; Leventhal, G.E.; Slack, E.; de Wouters, T.; Walter, J.; Clavel, T. Microbiome-based interventions to modulate gut ecology and the immune system. Mucosal Immunol. 2022, 15, 1095–1113. [Google Scholar] [CrossRef]
- Zimmermann, M.; Patil, K.R.; Typas, A.; Maier, L. Towards a mechanistic understanding of reciprocal drug-microbiome interactions. Mol. Syst. Biol. 2021, 17, e10116. [Google Scholar] [CrossRef]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between drugs and the gut microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef]
- Vich Vila, A.; Collij, V.; Sanna, S.; Sinha, T.; Imhann, F.; Bourgonje, A.R.; Mujagic, Z.; Jonkers, D.M.; Masclee, A.A.; Fu, J.; et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat. Commun. 2020, 11, 362. [Google Scholar] [CrossRef]
- Li, T.; Chiang, J.Y. Rifampicin induction of CYP3A4 requires pregnane X receptor cross talk with hepatocyte nuclear factor 4α and coactivators, and suppression of small heterodimer partner gene expression. Drug Metab. Dispos. 2006, 34, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Jing, H.; Wang, J.; Zhang, S.; Chang, Q.; Li, Z.; Wu, X.; Zhang, Z. Disordered gut microbiota correlates with altered fecal bile acid metabolism and post-cholecystectomy diarrhea. Front. Microbiol. 2022, 13, 800604. [Google Scholar] [CrossRef] [PubMed]
- Härtter, S.; Koenen-Bergmann, M.; Sharma, A.; Nehmiz, G.; Lemke, U.; Timmer, W.; Reilly, P.A. Decrease in the oral bioavailability of dabigatran etexilate after co-medication with rifampicin. Br. J. Clin. Pharmacol. 2012, 74, 490–500. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.T.; Caesar, R.; Mannerås-Holm, L.; Ståhlman, M.; Olsson, L.M.; Serino, M.; Planas-Fèlix, M.; et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017, 23, 850–858. [Google Scholar] [CrossRef]
- Sayin, S.I.; Wahlström, A.; Felin, J.; Jäntti, S.; Marschall, H.U.; Bamberg, K.; Angelin, B.; Hyötyläinen, T.; Orešič, M.; Bäckhed, F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-β-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef]
- Tsunoda, S.M.; Gonzales, C.; Jarmusch, A.K.; Momper, J.D.; Ma, J.D. Contribution of the Gut Microbiome to Drug Disposition, Pharmacokinetic and Pharmacodynamic Variability. Clin. Pharmacokinet. 2021, 60, 971–984. [Google Scholar] [CrossRef]
- Jones, H.; Rowland-Yeo, K. Basic concepts in physiologically based pharmacokinetic modeling in drug discovery and development. CPT Pharmacomet. Syst. Pharmacol. 2013, 2, e63. [Google Scholar] [CrossRef]
- Rowland Yeo, K.; Venkatakrishnan, K. Physiologically-based pharmacokinetic models as enablers of precision dosing in drug development: Pivotal role of the human mass balance study. Clin. Pharmacol. Ther. 2021, 109, 51–54. [Google Scholar] [CrossRef]
- Zhuang, X.; Lu, C. PBPK modeling and simulation in drug research and development. Acta Pharm. Sin. B 2016, 6, 430–440. [Google Scholar] [CrossRef]
- Dutta, M.; Lim, J.J.; Cui, J.Y. Pregnane X receptor and the gut-liver axis: A recent update. Drug Metab. Dispos. 2022, 50, 478–491. [Google Scholar] [CrossRef] [PubMed]
- Walsh, J.; Gheorghe, C.E.; Lyte, J.M.; van de Wouw, M.; Boehme, M.; Dinan, T.G.; Cryan, J.F.; Griffin, B.T.; Clarke, G.; Hyland, N.P. Gut microbiome-mediated modulation of hepatic cytochrome P450 and P-glycoprotein: Impact of butyrate and fructo-oligosaccharide-inulin. J. Pharm. Pharmacol. 2020, 72, 1072–1081. [Google Scholar] [CrossRef] [PubMed]
- Spanogiannopoulos, P.; Bess, E.N.; Carmody, R.N.; Turnbaugh, P.J. The microbial pharmacists within us: A metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 2016, 14, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Verdegaal, A.A.; Goodman, A.L. Integrating the gut microbiome and pharmacology. Sci. Transl. Med. 2024, 16, eadg8357. [Google Scholar] [CrossRef]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef]
- Rau, S.; Gregg, A.; Yaceczko, S.; Limketkai, B. Prebiotics and probiotics for gastrointestinal disorders. Nutrients 2024, 16, 778. [Google Scholar] [CrossRef]
- Snider, N.T.; Sikora, M.J.; Sridar, C.; Feuerstein, T.J.; Rae, J.M.; Hollenberg, P.F. The endocannabinoid anandamide is a substrate for the human polymorphic cytochrome P450 2D6. J. Pharmacol. Exp. Ther. 2008, 327, 538–545. [Google Scholar] [CrossRef]
- Kalitsky-Szirtes, J.; Shayeganpour, A.; Brocks, D.R.; Piquette-Miller, M. Suppression of drug-metabolizing enzymes and efflux transporters in the intestine of endotoxin-treated rats. Drug Metab. Dispos. 2004, 32, 20–27. [Google Scholar] [CrossRef]
- Wilk, J.N.; Bilsborough, J.; Viney, J.L. The mdr1a−/− mouse model of spontaneous colitis: A relevant and appropriate animal model to study inflammatory bowel disease. Immunol. Res. 2005, 31, 151–159. [Google Scholar] [CrossRef]
- Chiang, J.Y.; Pathak, P.; Liu, H.; Donepudi, A.; Ferrell, J.; Boehme, S. Intestinal Farnesoid X Receptor and Takeda G protein-coupled receptor 5 signaling in metabolic regulation. Dig. Dis. 2017, 35, 241–245. [Google Scholar] [CrossRef]
- Wallace, B.D.; Wang, H.; Lane, K.T.; Scott, J.E.; Orans, J.; Koo, J.S.; Venkatesh, M.; Jobin, C.; Yeh, L.A.; Mani, S.; et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 2010, 330, 831–835. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).