Manipulation with Mutational Status of VHL Regulates Hypoxic Metabolism and Pro-Angiogenic Phenotypes in ccRCC Caki-1 Cells
Abstract
1. Introduction
2. Results
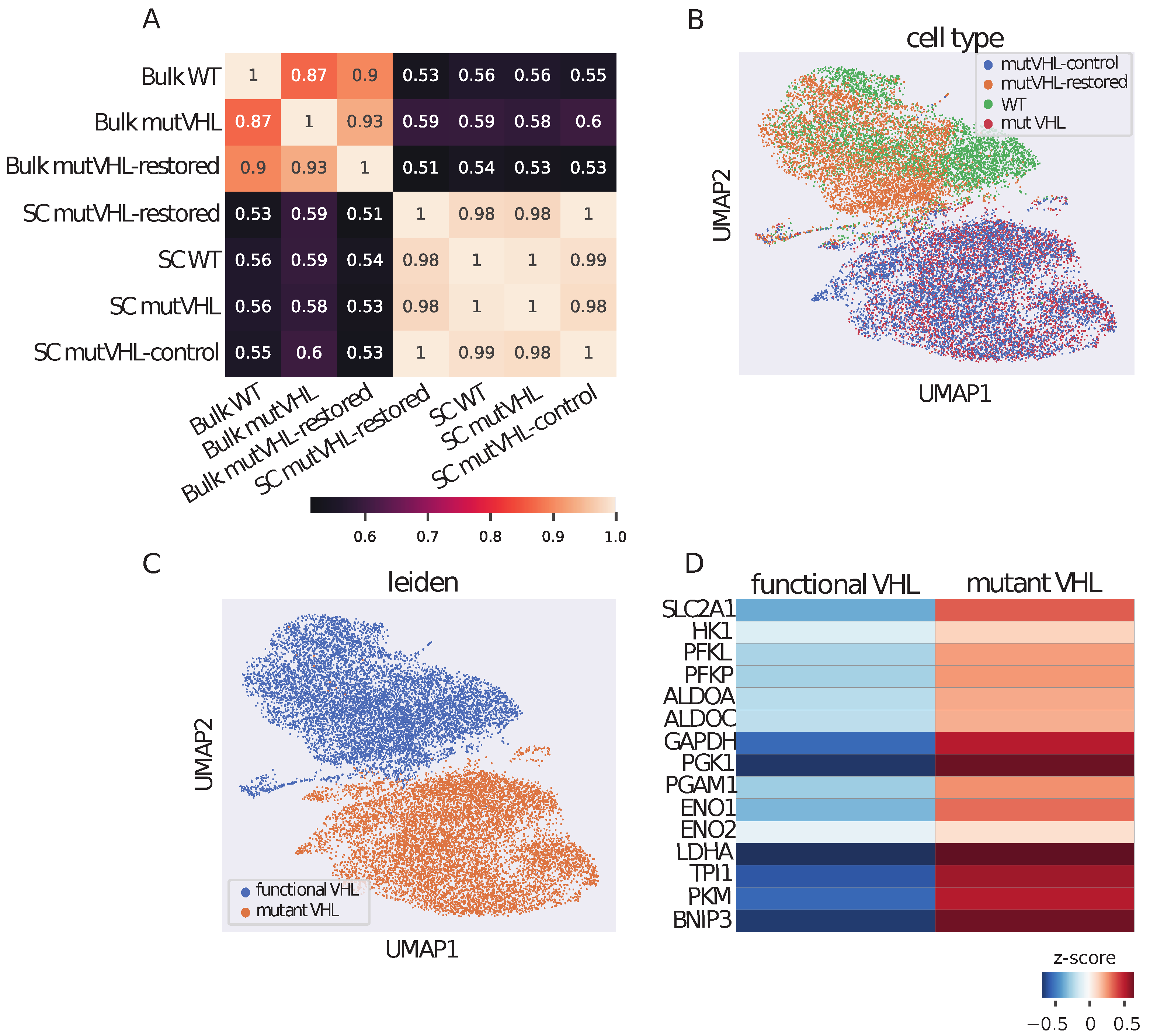
2.1. Integrated Single-Cell and Bulk Transcriptomic Analysis Reveals VHL-Dependent Gene Expression Programs in Caki-1 Cells
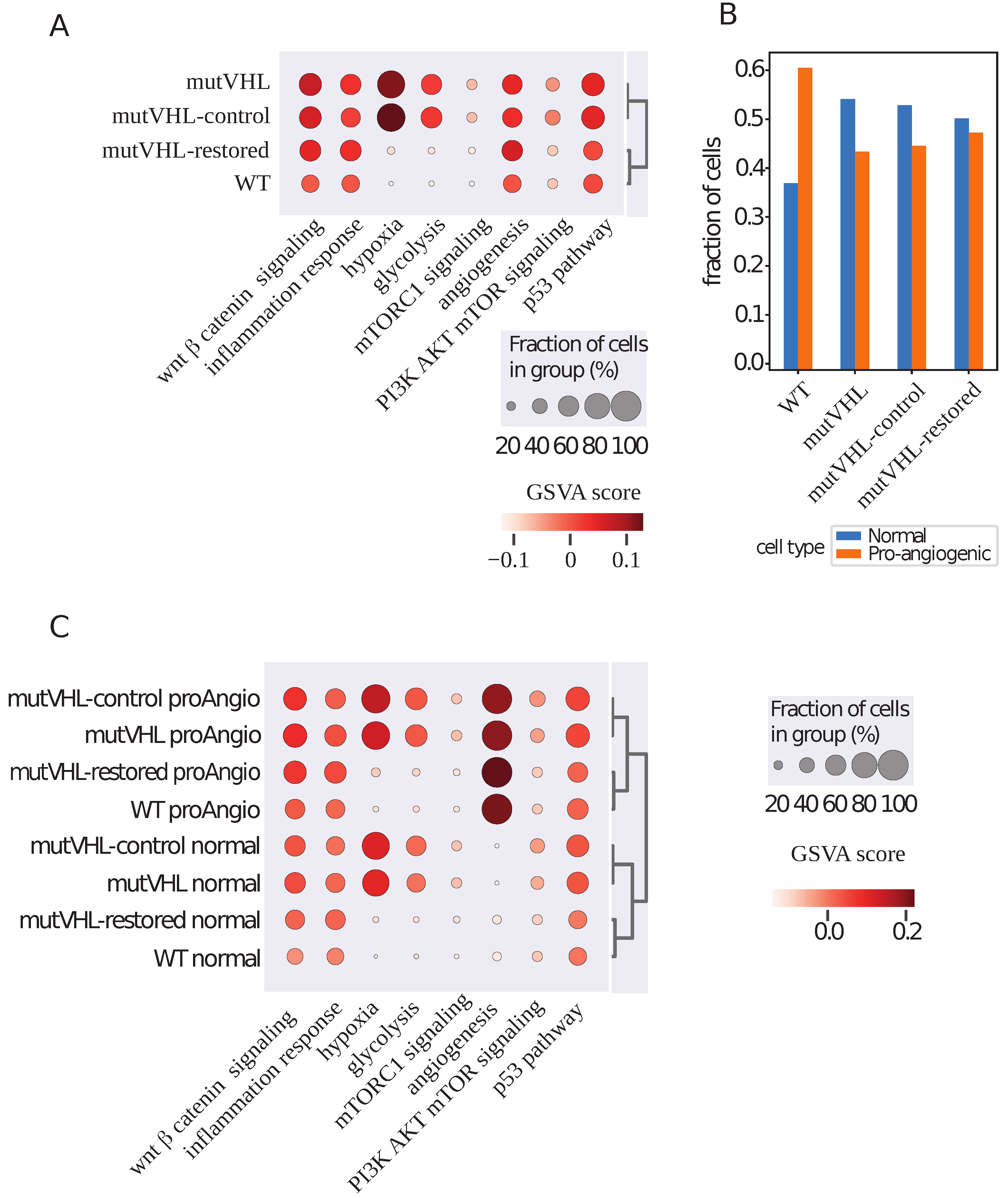
2.2. Gene Set Variational Analysis Shows Subsets of Cells with a Different Malignancy Phenotype
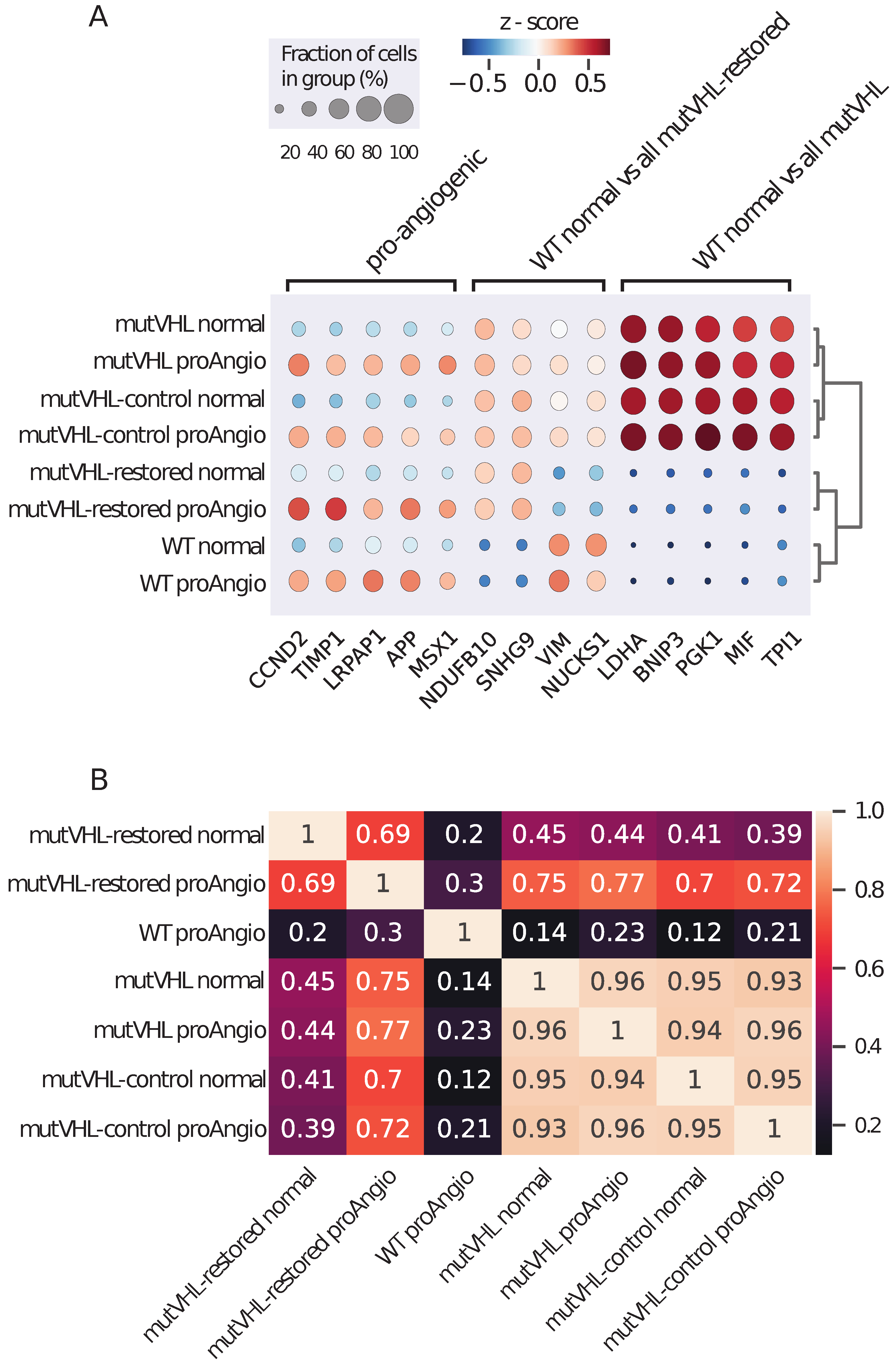
2.3. Comparison of Pro-Angiogenic and Normal Cells Gene Expressions
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Single-Cell RNA Sequencing Data Analysis
4.2.1. Single-Cell RNA Sequencing (scRNA-seq) Data Processing
4.2.2. Feature Selection and Dimensionality Reduction
4.2.3. Differential Expression and Pathway Analysis
4.2.4. Code Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef]
- Axel, E.M.; Matveev, V.B. Statistics of malignant tumors of urinary and male urogenital organs in Russia and the countries of the former USSR. Cancer Urol. 2019, 15, 15–24. [Google Scholar] [CrossRef]
- Padala, S.A.; Barsouk, A.; Thandra, K.C.; Saginala, K.; Mohammed, A.; Vakiti, A.; Rawla, P.; Barsouk, A. Epidemiology of renal cell carcinoma. World J. Oncol. 2020, 11, 79–87. [Google Scholar] [CrossRef]
- Gossage, L.; Eisen, T.; Maher, E.R. VHL, the story of a tumour suppressor gene. Nat. Rev. Cancer 2015, 15, 55–64. [Google Scholar] [CrossRef]
- Maxwell, P.H.; Wiesener, M.S.; Chang, G.W.; Clifford, S.C.; Vaux, E.C.; Cockman, M.E.; Wykoff, C.C.; Pugh, C.W.; Maher, E.R.; Ratcliffe, P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 1999, 399, 271–275. [Google Scholar] [CrossRef]
- Chen, Z.; Han, F.; Du, Y.; Shi, H.; Zhou, W. Hypoxic microenvironment in cancer: Molecular mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2023, 8, 70. [Google Scholar] [CrossRef]
- Hu, J.; Tan, P.; Ishihara, M.; Bayley, N.A.; Schokrpur, S.; Reynoso, J.G.; Zhang, Y.; Lim, R.J.; Dumitras, C.; Yang, L.; et al. Tumor heterogeneity in VHL drives metastasis in clear cell renal cell carcinoma. Signal Transduct. Target. Ther. 2023, 8, 155. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Song, J.; Zheng, S.; Wang, S. Advancements in understanding the molecular mechanisms and clinical implications of Von Hippel-Lindau syndrome: A comprehensive review. Transl. Oncol. 2025, 51, 102193. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Hu, S.; Yue, Y. Ultrasound microbubble-mediated VHL regulates the biological behavior of ovarian cancer cells. Ultrasound Med. Biol. 2021, 47, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Shen, G.; Xia, X.; Zhao, X.; Zhang, P.; Wu, H.; Guo, Q.; Qian, Z.; Wei, Y.; Liang, S. Comparisons of three polyethyleneimine-derived nanoparticles as a gene therapy delivery system for renal cell carcinoma. J. Transl. Med. 2011, 9, 46. [Google Scholar] [CrossRef]
- Artemov, A.V.; Zhenilo, S.; Kaplun, D.; Starshin, A.; Sokolov, A.; Mazur, A.M.; Szpotan, J.; Gawronski, M.; Modrzejewska, M.; Gackowski, D.; et al. An IDH-independent mechanism of DNA hypermethylation upon VHL inactivation in cancer. Epigenetics 2022, 17, 894–905. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhigalova, N.A.; Zhenilo, S.V.; Artemov, A.V.; Prokhortchouk, E.B. [CRISPR/Cas9-editing-based modeling of hypoxia in renal cancer cells]. Mol. Biol. 2017, 51, 836–840. [Google Scholar] [CrossRef]
- Artemov, A.V.; Zhigalova, N.; Zhenilo, S.; Mazur, A.M.; Prokhortchouk, E.B. VHL inactivation without hypoxia is sufficient to achieve genome hypermethylation. Sci. Rep. 2018, 8, 10667. [Google Scholar] [CrossRef] [PubMed]
- Schödel, J.; Grampp, S.; Maher, E.R.; Moch, H.; Ratcliffe, P.J.; Russo, P.; Mole, D.R. Hypoxia, hypoxia-inducible transcription factors, and renal cancer. Eur. Urol. 2016, 69, 646–657. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Wykoff, C.C.; Sotiriou, C.; Cockman, M.E.; Ratcliffe, P.J.; Maxwell, P.; Liu, E.; Harris, A.L. Gene array of VHL mutation and hypoxia shows novel hypoxia-induced genes and that cyclin D1 is a VHL target gene. Br. J. Cancer 2004, 90, 1235–1243. [Google Scholar] [CrossRef]
- Goul, C.; Peruzzo, R.; Zoncu, R. The molecular basis of nutrient sensing and signalling by mTORC1 in metabolism regulation and disease. Nat. Rev. Mol. Cell Biol. 2023, 24, 857–875. [Google Scholar] [CrossRef]
- Sullivan, K.D.; Galbraith, M.D.; Andrysik, Z.; Espinosa, J.M. Mechanisms of transcriptional regulation by p53. Cell Death Differ. 2018, 25, 133–143. [Google Scholar] [CrossRef]
- Misiewicz-Krzeminska, I.; Sarasquete, M.E.; Vicente-Dueñas, C.; Krzeminski, P.; Wiktorska, K.; Corchete, L.A.; Quwaider, D.; Rojas, E.A.; Corral, R.; Martín, A.A.; et al. Post-transcriptional modifications contribute to the upregulation of cyclin D2 in multiple myeloma. Clin. Cancer Res. 2016, 22, 207–217. [Google Scholar] [CrossRef]
- Jung, Y.S.; Liu, X.W.; Chirco, R.; Warner, R.B.; Fridman, R.; Kim, H.R.C. TIMP-1 induces an EMT-like phenotypic conversion in MDCK cells independent of its MMP-inhibitory domain. PLoS ONE 2012, 7, e38773. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.J.; Lin, S.C.; Xu, S.H.; Gao, Y.B.; Zhou, B.J.; Zhou, R.; Chen, F.M.; Li, F.R. Proteomic analysis reveals LRPAP1 as a key player in the micropapillary pattern metastasis of lung adenocarcinoma. Heliyon 2024, 10, e23913. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.N.; Jeong, M.S.; Jang, S.B. Molecular characteristics of amyloid precursor protein (APP) and its effects in cancer. Int. J. Mol. Sci. 2021, 22, 4999. [Google Scholar] [CrossRef]
- Yue, Y.; Yuan, Y.; Li, L.; Fan, J.; Li, C.; Peng, W.; Ren, G. Homeobox protein MSX1 inhibits the growth and metastasis of breast cancer cells and is frequently silenced by promoter methylation. Int. J. Mol. Med. 2018, 41, 2986–2996. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Zhou, K.; Li, J.; Jiang, S.; Li, C.; Men, H. MSX1 induces G0/G1 arrest and apoptosis by suppressing Notch signaling and is frequently methylated in cervical cancer. Onco Targets Ther. 2018, 11, 4769–4780. [Google Scholar] [CrossRef]
- Arroum, T.; Borowski, M.T.; Marx, N.; Schmelter, F.; Scholz, M.; Psathaki, O.E.; Hippler, M.; Enriquez, J.A.; Busch, K.B. Loss of respiratory complex I subunit NDUFB10 affects complex I assembly and supercomplex formation. Biol. Chem. 2023, 404, 399–415. [Google Scholar] [CrossRef]
- Ye, S.; Ni, Y. LncRNA SNHG9 promotes cell proliferation, migration, and invasion in human hepatocellular carcinoma cells by increasing GSTP1 methylation, as revealed by CRISPR-dCas9. Front. Mol. Biosci. 2021, 8, 649976. [Google Scholar] [CrossRef]
- Feng, S.G.; Bhandari, R.; Ya, L.; Zhixuan, B.; Qiuhui, P.; Jiabei, Z.; Sewi, M.; Ni, Z.; Jing, W.; Fenyong, S.; et al. SNHG9 promotes Hepatoblastoma Tumorigenesis via miR-23a-5p/Wnt3a Axis. J. Cancer 2021, 12, 6031–6049. [Google Scholar] [CrossRef]
- Li, C.; Hu, J.; Hu, X.; Zhao, C.; Mo, M.; Zu, X.; Li, Y. LncRNA SNHG9 is a prognostic biomarker and correlated with immune infiltrates in prostate cancer. Transl. Androl. Urol. 2021, 10, 215–226. [Google Scholar] [CrossRef]
- Chen, G.Y.; Zhang, Z.S.; Chen, Y.; Li, Y. Long non-coding RNA SNHG9 inhibits ovarian cancer progression by sponging microRNA-214-5p. Oncol. Lett. 2021, 21, 80. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Prim. 2017, 3, 17009. [Google Scholar] [CrossRef]
- Usman, S.; Waseem, N.H.; Nguyen, T.K.N.; Mohsin, S.; Jamal, A.; Teh, M.T.; Waseem, A. Vimentin is at the heart of epithelial mesenchymal transition (EMT) mediated metastasis. Cancers 2021, 13, 4985. [Google Scholar] [CrossRef]
- Zhao, S.; Wang, B.; Ma, Y.; Kuang, J.; Liang, J.; Yuan, Y. NUCKS1 promotes proliferation, invasion and migration of non-small cell lung cancer by upregulating CDK1 expression. Cancer Manag. Res. 2020, 12, 13311–13323. [Google Scholar] [CrossRef]
- Zheng, S.; Ji, R.; He, H.; Li, N.; Han, C.; Han, J.; Li, X.; Zhang, L.; Wang, Y.; Zhao, W. NUCKS1, a LINC00629-upregulated gene, facilitated osteosarcoma progression and metastasis by elevating asparagine synthesis. Cell Death Dis. 2023, 14, 489. [Google Scholar] [CrossRef]
- Wang, L.; Cui, Y.; Zhang, L.; Sheng, J.; Yang, Y.; Kuang, G.; Fan, Y.; Zhang, Q.; Jin, J. The silencing of CCND2 by promoter aberrant methylation in renal cell cancer and analysis of the correlation between CCND2 methylation status and clinical features. PLoS ONE 2016, 11, e0161859. [Google Scholar] [CrossRef]
- Liu, Z.; Guo, S.; Sun, H.; Bai, Y.; Song, Z.; Liu, X. Circular RNA CircHIPK3 elevates CCND2 expression and promotes cell proliferation and invasion through miR-124 in glioma. Front. Genet. 2020, 11, 1013. [Google Scholar] [CrossRef]
- Shou, Y.; Liu, Y.; Xu, J.; Liu, J.; Xu, T.; Tong, J.; Liu, L.; Hou, Y.; Liu, D.; Yang, H.; et al. TIMP1 indicates poor prognosis of renal cell carcinoma and accelerates tumorigenesis via EMT signaling pathway. Front. Genet. 2022, 13, 648134. [Google Scholar] [CrossRef]
- Gong, Y.; Scott, E.; Lu, R.; Xu, Y.; Oh, W.K.; Yu, Q. TIMP-1 promotes accumulation of cancer associated fibroblasts and cancer progression. PLoS ONE 2013, 8, e77366. [Google Scholar] [CrossRef] [PubMed]
- Nothnick, W.B.; Soloway, P.; Curry, T.E., Jr. Assessment of the role of tissue inhibitor of metalloproteinase-1 (TIMP-1) during the periovulatory period in female mice lacking a functional TIMP-1 gene. Biol. Reprod. 1997, 56, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Tarhini, A.A.; Lin, Y.; Yeku, O.; LaFramboise, W.A.; Ashraf, M.; Sander, C.; Lee, S.; Kirkwood, J.M. A four-marker signature of TNF-RII, TGF-α, TIMP-1 and CRP is prognostic of worse survival in high-risk surgically resected melanoma. J. Transl. Med. 2014, 12, 19. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Dho, S.H.; Lee, J.; Hwang, J.H.; Kim, M.; Choi, W.Y.; Lee, J.Y.; Lee, J.; Chang, W.; Lee, M.Y.; et al. Hypermethylation of PDX1, EN2, and MSX1 predicts the prognosis of colorectal cancer. Exp. Mol. Med. 2022, 54, 156–168. [Google Scholar] [CrossRef]
- Wolf, F.A.; Angerer, P.; Theis, F.J. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 2018, 19, 15. [Google Scholar] [CrossRef]
- Lause, J.; Berens, P.; Kobak, D. Analytic Pearson residuals for normalization of single-cell RNA-seq UMI data. Genome Biol. 2021, 22, 258. [Google Scholar] [CrossRef] [PubMed]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Wolf, F.A.; Hamey, F.K.; Plass, M.; Solana, J.; Dahlin, J.S.; Göttgens, B.; Rajewsky, N.; Simon, L.; Theis, F.J. PAGA: Graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019, 20, 59. [Google Scholar] [CrossRef]
- Traag, V.A.; Waltman, L.; van Eck, N.J. From Louvain to Leiden: Guaranteeing well-connected communities. Sci. Rep. 2019, 9, 5233. [Google Scholar] [CrossRef]
- Fang, Z.; Liu, X.; Peltz, G. GSEApy: A comprehensive package for performing gene set enrichment analysis in Python. Bioinformatics 2022, 39, btac757. [Google Scholar] [CrossRef] [PubMed]
| mutVHL-Restored vs. WT Pathway Names | mutVHL-Restored vs. WT Z-Values | mutVHL vs. WT Pathway Names | mutVHL vs. WT Z-Values | mutVHL-Control vs. WT Pathway Names | mutVHL-Control vs. WT Z-Values |
|---|---|---|---|---|---|
| WNT--Catenin | 14.74 | Hypoxia | 74.73 | Hypoxia | 78.52 |
| Inflammatory response | 13.59 | Glycolysis | 57.53 | Glycolysis | 60.81 |
| Angiogenesis | 12.81 | MTORC1 | 25.12 | MTORC1 | 24.63 |
| TNF via NFB | 12.31 | WNT--Catenin | 21.58 | PI3K-AKT-MTOR | 20.42 |
| Hypoxia | 12.20 | P53 | 19.0 | P53 | 19.89 |
| Cell Line | Normal Cell Counts | Pro-Angiogenic Cell Counts | Normal/Pro-Angiogenic Cell Counts Ratio |
|---|---|---|---|
| mutVHL-restored | 2086 | 2226 | 0.93 |
| WT | 2644 | 1659 | 1.59 |
| mutVHL | 1973 | 2368 | 0.83 |
| Ctr | 2327 | 2665 | 0.87 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abramov, P.; Mazur, A.; Starshin, A.; Zhenilo, S.; Prokhortchouk, E. Manipulation with Mutational Status of VHL Regulates Hypoxic Metabolism and Pro-Angiogenic Phenotypes in ccRCC Caki-1 Cells. Int. J. Mol. Sci. 2025, 26, 10629. https://doi.org/10.3390/ijms262110629
Abramov P, Mazur A, Starshin A, Zhenilo S, Prokhortchouk E. Manipulation with Mutational Status of VHL Regulates Hypoxic Metabolism and Pro-Angiogenic Phenotypes in ccRCC Caki-1 Cells. International Journal of Molecular Sciences. 2025; 26(21):10629. https://doi.org/10.3390/ijms262110629
Chicago/Turabian StyleAbramov, Pavel, Alexandr Mazur, Aleksey Starshin, Svetlana Zhenilo, and Egor Prokhortchouk. 2025. "Manipulation with Mutational Status of VHL Regulates Hypoxic Metabolism and Pro-Angiogenic Phenotypes in ccRCC Caki-1 Cells" International Journal of Molecular Sciences 26, no. 21: 10629. https://doi.org/10.3390/ijms262110629
APA StyleAbramov, P., Mazur, A., Starshin, A., Zhenilo, S., & Prokhortchouk, E. (2025). Manipulation with Mutational Status of VHL Regulates Hypoxic Metabolism and Pro-Angiogenic Phenotypes in ccRCC Caki-1 Cells. International Journal of Molecular Sciences, 26(21), 10629. https://doi.org/10.3390/ijms262110629







