Pathological and Functional Brain Amyloids: A New Concept Explaining the Differences
Abstract
1. Introduction
2. Functional Amyloids
2.1. Myelin Basic Protein
2.2. FXR1 and Orb2
2.3. Peptide and Protein Hormones in Secretory Granules
3. Pathological Amyloids
3.1. Prion Protein
3.2. Aβ Peptide
3.3. Tau
3.4. α-Synuclein
3.5. Huntingtin
3.6. TDP-43
4. Comparative Analysis of Pathological and Functional Brain Amyloids Based on Their Amino Acid Composition
5. Factors Determining the Toxicity and Functionality of Amyloids
5.1. Amyloids’ Localization
5.2. Interaction of Amyloids with Pathological or Functional Partners
5.3. The Concept of “Available Targets”
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wolfe, K.J.; Cyr, D.M. Amyloid in Neurodegenerative Diseases: Friend or Foe? Semin. Cell Dev. Biol. 2011, 22, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.M.; Koulov, A.V.; Alory-Jost, C.; Marks, M.S.; Balch, W.E.; Kelly, J.W. Functional Amyloid Formation within Mammalian Tissue. PLoS Biol. 2006, 4, e6. [Google Scholar] [CrossRef] [PubMed]
- Sønderby, T.V.; Najarzadeh, Z.; Otzen, D.E. Functional Bacterial Amyloids: Understanding Fibrillation, Regulating Biofilm Fibril Formation and Organizing Surface Assemblies. Molecules 2022, 27, 4080. [Google Scholar] [CrossRef]
- Lipke, P.N.; Klotz, S.A.; Dufrene, Y.F.; Jackson, D.N.; Garcia-Sherman, M.C. Amyloid-Like β-Aggregates as Force-Sensitive Switches in Fungal Biofilms and Infections. Microbiol. Mol. Biol. Rev. 2017, 82, e00035-17. [Google Scholar] [CrossRef] [PubMed]
- Ryzhova, T.A.; Sopova, J.V.; Zadorsky, S.P.; Siniukova, V.A.; Sergeeva, A.V.; Galkina, S.A.; Nizhnikov, A.A.; Shenfeld, A.A.; Volkov, K.V.; Galkin, A.P. Screening for Amyloid Proteins in the Yeast Proteome. Curr. Genet. 2018, 64, 469–478. [Google Scholar] [CrossRef]
- Sergeeva, A.V.; Sopova, J.V.; Belashova, T.A.; Siniukova, V.A.; Chirinskaite, A.V.; Galkin, A.P.; Zadorsky, S.P. Amyloid Properties of the Yeast Cell Wall Protein Toh1 and Its Interaction with Prion Proteins Rnq1 and Sup35. Prion 2019, 13, 21–32. [Google Scholar] [CrossRef]
- Sopova, J.V.; Koshel, E.I.; Belashova, T.A.; Zadorsky, S.P.; Sergeeva, A.V.; Siniukova, V.A.; Shenfeld, A.A.; Velizhanina, M.E.; Volkov, K.V.; Nizhnikov, A.A.; et al. RNA-Binding Protein FXR1 Is Presented in Rat Brain in Amyloid Form. Sci. Rep. 2019, 9, 18983. [Google Scholar] [CrossRef]
- Hervas, R.; Rau, M.J.; Park, Y.; Zhang, W.; Murzin, A.G.; Fitzpatrick, J.A.J.; Scheres, S.H.W.; Si, K. Cryo-EM Structure of a Neuronal Functional Amyloid Implicated in Memory Persistence in Drosophila. Science 2020, 367, 1230–1234. [Google Scholar] [CrossRef]
- Antonets, K.S.; Belousov, M.V.; Sulatskaya, A.I.; Belousova, M.E.; Kosolapova, A.O.; Sulatsky, M.I.; Andreeva, E.A.; Zykin, P.A.; Malovichko, Y.V.; Shtark, O.Y.; et al. Accumulation of Storage Proteins in Plant Seeds Is Mediated by Amyloid Formation. PLoS Biol. 2020, 18, e3000564. [Google Scholar] [CrossRef]
- Sysoev, E.I.; Shenfeld, A.A.; Belashova, T.A.; Valina, A.A.; Zadorsky, S.P.; Galkin, A.P. Amyloid Fibrils of the Myelin Basic Protein Are an Integral Component of Myelin in the Vertebrate Brain. Sci. Rep. 2025, 15, 29053. [Google Scholar] [CrossRef]
- Maji, S.K.; Perrin, M.H.; Sawaya, M.R.; Jessberger, S.; Vadodaria, K.; Rissman, R.A.; Singru, P.S.; Nilsson, K.P.R.; Simon, R.; Schubert, D.; et al. Functional Amyloids as Natural Storage of Peptide Hormones in Pituitary Secretory Granules. Science 2009, 325, 328–332. [Google Scholar] [CrossRef]
- Valina, A.A.; Siniukova, V.A.; Belashova, T.A.; Kanapin, A.A.; Samsonova, A.A.; Masharsky, A.E.; Lykholay, A.N.; Galkina, S.A.; Zadorsky, S.P.; Galkin, A.P. Amyloid Fibrils of the S36 Protein Modulate the Morphogenesis of Drosophila Melanogaster Eggshell. Int. J. Mol. Sci. 2024, 25, 12499. [Google Scholar] [CrossRef] [PubMed]
- Tetter, S.; Arseni, D.; Murzin, A.G.; Buhidma, Y.; Peak-Chew, S.Y.; Garringer, H.J.; Newell, K.L.; Vidal, R.; Apostolova, L.G.; Lashley, T.; et al. TAF15 Amyloid Filaments in Frontotemporal Lobar Degeneration. Nature 2024, 625, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Arseni, D.; Nonaka, T.; Jacobsen, M.H.; Murzin, A.G.; Cracco, L.; Peak-Chew, S.Y.; Garringer, H.J.; Kawakami, I.; Suzuki, H.; Onaya, M.; et al. Heteromeric Amyloid Filaments of ANXA11 and TDP-43 in FTLD-TDP Type C. Nature 2024, 634, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Boggs, J.M. Myelin Basic Protein: A Multifunctional Protein. Cell. Mol. Life Sci. 2006, 63, 1945–1961. [Google Scholar] [CrossRef]
- Landry, C.F.; Ellison, J.A.; Pribyl, T.M.; Campagnoni, C.; Kampf, K.; Campagnoni, A.T. Myelin Basic Protein Gene Expression in Neurons: Developmental and Regional Changes in Protein Targeting within Neuronal Nuclei, Cell Bodies, and Processes. J. Neurosci. 1996, 16, 2452–2462. [Google Scholar] [CrossRef]
- Campagnoni, A.T.; Skoff, R.P. The Pathobiology of Myelin Mutants Reveal Novel Biological Functions of the MBP and PLP Genes. Brain Pathol. 2001, 11, 74–91. [Google Scholar] [CrossRef]
- Müller, C.; Bauer, N.M.; Schäfer, I.; White, R. Making Myelin Basic Protein—From MRNA Transport to Localized Translation. Front. Cell. Neurosci. 2013, 7, 169. [Google Scholar] [CrossRef]
- Kosturko, L.D.; Maggipinto, M.J.; Korza, G.; Lee, J.W.; Carson, J.H.; Barbarese, E. Heterogeneous Nuclear Ribonucleoprotein (HnRNP) E1 Binds to HnRNP A2 and Inhibits Translation of A2 Response Element MRNAs. Mol. Biol. Cell 2006, 17, 3521–3533. [Google Scholar] [CrossRef]
- Bauer, N.M.; Moos, C.; Van Horssen, J.; Witte, M.; Van Der Valk, P.; Altenhein, B.; Luhmann, H.J.; White, R. Myelin Basic Protein Synthesis Is Regulated by Small Non-Coding RNA 715. EMBO Rep. 2012, 13, 827–834. [Google Scholar] [CrossRef]
- Kattnig, D.R.; Bund, T.; Boggs, J.M.; Harauz, G.; Hinderberger, D. Lateral Self-Assembly of 18.5-KDa Myelin Basic Protein (MBP) Charge Component-C1 on Membranes. Biochim. Biophys. Acta 2012, 1818, 2636–2647. [Google Scholar] [CrossRef]
- Aggarwal, S.; Snaidero, N.; Pähler, G.; Frey, S.; Sánchez, P.; Zweckstetter, M.; Janshoff, A.; Schneider, A.; Weil, M.T.; Schaap, I.A.T.; et al. Myelin Membrane Assembly Is Driven by a Phase Transition of Myelin Basic Proteins into a Cohesive Protein Meshwork. PLoS Biol. 2013, 11, e1001577. [Google Scholar] [CrossRef]
- Raasakka, A.; Ruskamo, S.; Kowal, J.; Barker, R.; Baumann, A.; Martel, A.; Tuusa, J.; Myllykoski, M.; Bürck, J.; Ulrich, A.S.; et al. Membrane Association Landscape of Myelin Basic Protein Portrays Formation of the Myelin Major Dense Line. Sci. Rep. 2017, 7, 4974. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, X.; Zheng, S.; Liu, X.; Jin, J.; Ren, Y.; Luo, J. Myelin Basic Protein Induces Neuron-Specific Toxicity by Directly Damaging the Neuronal Plasma Membrane. PLoS ONE 2014, 9, e108646. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Jickling, G.C.; Ander, B.P.; Stamova, B.; Liu, D.; Kao, P.F.; Zelin, M.A.; Jin, L.W.; DeCarli, C.; Sharp, F.R. Myelin basic protein associates with AβPP, Aβ1-42, and amyloid plaques in cortex of Alzheimer’s disease brain. J. Alzheimer’s Dis. 2015, 44, 1213–1229. [Google Scholar] [CrossRef]
- Majumder, M.; Johnson, R.H.; Palanisamy, V. Fragile X-Related Protein Family: A Double-Edged Sword in Neurodevelopmental Disorders and Cancer. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Tamanini, F.; Willemsen, R.; Van Unen, L.; Bontekoe, C.; Galjaard, H.; Oostra, B.A.; Hoogeveen, A.T. Differential Expression of FMR1, FXR1 and FXR2 Proteins in Human Brain and Testis. Hum. Mol. Genet. 1997, 6, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Meintjes, E.J.; Willemsen, R.; Kirkpatrick, L.L.; Niuewenhuizen, I.M.; Hoogeveen-Westerveld, M.; Verweij, M.; Reis, S.; Bardoni, B.; Hoogeveen, A.T.; Oostra, B.A.; et al. Fxr1 Knockout Mice Show a Striated Muscle Phenotype: Implications for Fxr1p Function In Vivo. Hum. Mol. Genet. 2004, 13, 1291–1302. [Google Scholar] [CrossRef]
- Cook, D.; Nuro, E.; Jones, E.V.; Altimimi, H.F.; Farmer, W.T.; Gandin, V.; Hanna, E.; Zong, R.; Barbon, A.; Nelson, D.L.; et al. FXR1P Limits Long-Term Memory, Long-Lasting Synaptic Potentiation, and de Novo GluA2 Translation. Cell Rep. 2014, 9, 1402–1416. [Google Scholar] [CrossRef]
- Garnon, J.; Lachance, C.; Di Marco, S.; Hel, Z.; Marion, D.; Ruiz, M.C.; Newkirk, M.M.; Khandjian, E.W.; Radzioch, D. Fragile X-Related Protein FXR1P Regulates Proinflammatory Cytokine Tumor Necrosis Factor Expression at the Post-Transcriptional Level. J. Biol. Chem. 2005, 280, 5750–5763. [Google Scholar] [CrossRef]
- Herman, A.B.; Vrakas, C.N.; Ray, M.; Kelemen, S.E.; Sweredoski, M.J.; Moradian, A.; Haines, D.S.; Autieri, M.V. FXR1 Is an IL-19-Responsive RNA-Binding Protein That Destabilizes Pro-Inflammatory Transcripts in Vascular Smooth Muscle Cells. Cell Rep. 2018, 24, 1176–1189. [Google Scholar] [CrossRef]
- George, J.; Li, Y.; Kadamberi, I.P.; Parashar, D.; Tsaih, S.W.; Gupta, P.; Geethadevi, A.; Chen, C.; Ghosh, C.; Sun, Y.; et al. RNA-Binding Protein FXR1 Drives CMYC Translation by Recruiting EIF4F Complex to the Translation Start Site. Cell Rep. 2021, 37, 109934. [Google Scholar] [CrossRef]
- Schaeffer, C.; Bardoni, B.; Mandel, J.L.; Ehresmann, B.; Ehresmann, C.; Moine, H. The Fragile X Mental Retardation Protein Binds Specifically to Its MRNA via a Purine Quartet Motif. EMBO J. 2001, 20, 4803–4813. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.C.; Fraser, C.E.; Mostovetsky, O.; Darnell, R.B. Discrimination of Common and Unique RNA-Binding Activities among Fragile X Mental Retardation Protein Paralogs. Hum. Mol. Genet. 2009, 18, 3164–3177. [Google Scholar] [CrossRef] [PubMed]
- Winograd, C.; Ceman, S. Fragile X Family Members Have Important and Non-Overlapping Functions. Biomol. Concepts 2011, 2, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Adinolfi, S.; Ramos, A.; Martin, S.R.; Dal Piaz, F.; Pucci, P.; Bardoni, B.; Mandel, J.L.; Pastore, A. The N-Terminus of the Fragile X Mental Retardation Protein Contains a Novel Domain Involved in Dimerization and RNA Binding. Biochemistry 2003, 42, 10437–10444. [Google Scholar] [CrossRef]
- Adams-Cioaba, M.A.; Guo, Y.; Bian, C.B.; Amaya, M.F.; Lam, R.; Wasney, G.A.; Vedadi, M.; Xu, C.; Min, J. Structural Studies of the Tandem Tudor Domains of Fragile X Mental Retardation Related Proteins FXR1 and FXR2. PLoS ONE 2010, 5, e13559. [Google Scholar] [CrossRef]
- Myrick, L.K.; Hashimoto, H.; Cheng, X.; Warren, S.T. Human FMRP Contains an Integral Tandem Agenet (Tudor) and KH Motif in the Amino Terminal Domain. Hum. Mol. Genet. 2015, 24, 1733–1740. [Google Scholar] [CrossRef]
- St. Paul, A.; Corbett, C.; Peluzzo, A.; Kelemen, S.; Okune, R.; Haines, D.S.; Preston, K.; Eguchi, S.; Autieri, M.V. FXR1 Regulates Vascular Smooth Muscle Cell Cytoskeleton, VSMC Contractility, and Blood Pressure by Multiple Mechanisms. Cell Rep. 2023, 42, 112381. [Google Scholar] [CrossRef]
- Vasudevan, S.; Steitz, J.A. AU-Rich-Element-Mediated Upregulation of Translation by FXR1 and Argonaute 2. Cell 2007, 128, 1105–1118. [Google Scholar] [CrossRef]
- Yang, J.; Chen, Z.; He, J.; Zou, B.; Si, Y.; Ma, Y.; Yu, J. Modular RNA Interactions Shape FXR1 Condensates Involved in MRNA Localization and Translation. Nat. Commun. 2025, 16, 8589. [Google Scholar] [CrossRef]
- Velizhanina, M.E.; Galkin, A.P. Amyloid Properties of the FXR1 Protein Are Conserved in Evolution of Vertebrates. Int. J. Mol. Sci. 2022, 23, 7997. [Google Scholar] [CrossRef]
- Valina, A.A.; Belashova, T.A.; Yuzman, A.K.; Zadorsky, S.P.; Sysoev, E.I.; Mitkevich, V.A.; Makarov, A.A.; Galkin, A.P. Functional Amyloid Protein FXR1 Is Recruited into Neuronal Stress Granules. Prion 2025, 19, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.R.; Li, L.; Pérez-Sánchez, C.; Saraf, A.; Florens, L.; Slaughter, B.D.; Unruh, J.R.; Si, K. Amyloidogenic Oligomerization Transforms Drosophila Orb2 from a Translation Repressor to an Activator. Cell 2015, 163, 1468–1483. [Google Scholar] [CrossRef]
- Charlesworth, A.; Meijer, H.A.; De Moor, C.H. Specificity Factors in Cytoplasmic Polyadenylation. Wiley Interdiscip. Rev. RNA 2013, 4, 437–461. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, E.; Shidlovskii, Y.V.; Gilmutdinov, R.; Schedl, P.; Zhukova, M. The Role of CPEB Family Proteins in the Nervous System Function in the Norm and Pathology. Cell Biosci. 2021, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Stepien, B.K.; Oppitz, C.; Gerlach, D.; Dag, U.; Novatchkova, M.; Krüttner, S.; Stark, A.; Keleman, K. RNA-Binding Profiles of Drosophila CPEB Proteins Orb and Orb2. Proc. Natl. Acad. Sci. USA 2016, 113, E7030–E7038. [Google Scholar] [CrossRef]
- White-Grindley, E.; Li, L.; Mohammad Khan, R.; Ren, F.; Saraf, A.; Florens, L.; Si, K. Contribution of Orb2A Stability in Regulated Amyloid-Like Oligomerization of Drosophila Orb2. PLoS Biol. 2014, 12, e1001786. [Google Scholar] [CrossRef]
- Dannies, P.S. Concentrating Hormones into Secretory Granules: Layers of Control. Mol. Cell Endocrinol. 2001, 177, 87–93. [Google Scholar] [CrossRef]
- Kim, T.; Gondré-Lewis, M.C.; Arnaoutova, I.; Loh, Y.P. Dense-Core Secretory Granule Biogenesis. Physiology 2006, 21, 124–133. [Google Scholar] [CrossRef]
- Tesar, J.T.; Koenig, H.; Hughes, C. Hormone Storage Granules in the Beef Anterior Pituitary. I. Isolation, Ultrastructure, and Some Biochemical Properties. J. Cell Biol. 1969, 40, 225–235. [Google Scholar] [CrossRef]
- Perdue, J.F.; McShan, W.H. Association of Adrenocorticotropic Hormone Activity with Small Secretory Granules from Rat Anterior Pituitary Glands. Endocrinology 1966, 78, 406–408. [Google Scholar] [CrossRef] [PubMed]
- Tasso, F.; Picard, D.; Dreifuss, J.J. Ultrastructural Identification of Granules Containing Oxytocin and Vasopressin. Nature 1976, 260, 621–622. [Google Scholar] [CrossRef] [PubMed]
- Weber, E.; Martin, R.; Voigt, K.H. Corticotropin/β-Endorphin Precursor: Concomitant Storage of Its Fragments in the Secretory Granules of Anterior Pituitary Corticotropin/Endorphin Cells. Life Sci. 1979, 25, 1111–1118. [Google Scholar] [CrossRef] [PubMed]
- Tooze, S.A. Biogenesis of Secretory Granules in the Trans-Golgi Network of Neuroendocrine and Endocrine Cells. Biochim. Biophys. Acta Mol. Cell Res. 1998, 1404, 231–244. [Google Scholar] [CrossRef]
- Farquhar, M.G.; Reid, J.A.J.; Daniell, L.W. Intracellular Transport and Packaging of Prolactin: A Quantitative Electron Microscope Autoradiographic Study of Mammotrophs Dissociated from Rat Pituitaries. Endocrinology 1978, 102, 296–311. [Google Scholar] [CrossRef]
- Gumbiner, B.; Kelly, R.B. Secretory Granules of an Anterior Pituitary Cell Line, AtT-20, Contain Only Mature Forms of Corticotropin and β-Lipotropin. Proc. Natl. Acad. Sci. USA 1981, 78, 318–322. [Google Scholar] [CrossRef]
- Dannies, P.S. Prolactin and Growth Hormone Aggregates in Secretory Granules: The Need to Understand the Structure of the Aggregate. Endocr. Rev. 2012, 33, 254–270. [Google Scholar] [CrossRef]
- Chanat, E.; Huttner, W.B. Milieu-Induced, Selective Aggregation of Regulated Secretory Proteins in the Trans-Golgi Network. J. Cell Biol. 1991, 115, 1505–1519. [Google Scholar] [CrossRef]
- Birk, J.; Friberg, M.A.; Prescianotto-Baschong, C.; Spiess, M.; Rutishauser, J. Dominant Pro-Vasopressin Mutants That Cause Diabetes Insipidus Form Disulfide-Linked Fibrillar Aggregates in the Endoplasmic Reticulum. J. Cell Sci. 2009, 221, 3994–4002. [Google Scholar] [CrossRef]
- Bendheim, P.E.; Brown, H.R.; Rudelli, R.D.; Scala, L.J.; Goller, N.L.; Wen, G.Y.; Kascsak, R.J.; Cashman, N.R.; Bolton, D.C. Nearly Ubiquitous Tissue Distribution of the Scrapie Agent Precursor Protein. Neurology 1992, 42, 149–156. [Google Scholar] [CrossRef]
- McDonald, A.J.; Leon, D.R.; Markham, K.A.; Wu, B.; Heckendorf, C.F.; Schilling, K.; Showalter, H.D.; Andrews, P.C.; McComb, M.E.; Pushie, M.J.; et al. Altered Domain Structure of the Prion Protein Caused by Cu2+ Binding and Functionally Relevant Mutations: Analysis by Cross-Linking, MS/MS, and NMR. Structure 2019, 27, 907–922. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.R.; Mu, T.C.; Gao, Z.X.; Wang, J.; Sy, M.S.; Li, C.Y. Prion Protein Is Required for Tumor Necrosis Factor α (TNFα)-Triggered Nuclear Factor Κb (NF-ΚB) Signaling and Cytokine Production. J. Biol. Chem. 2017, 292, 18747–18759. [Google Scholar] [CrossRef]
- Brown, D.R.; Clive, C.; Haswell, S.J. Antioxidant Activity Related to Copper Binding of Native Prion Protein. J. Neurochem. 2001, 76, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Mercer, R.C.C.; Harris, D.A. Mechanisms of Prion-Induced Toxicity. Cell Tissue Res. 2023, 392, 81–96. [Google Scholar] [CrossRef] [PubMed]
- Legname, G. Elucidating the Function of the Prion Protein. PLoS Pathog. 2017, 13, e1006458. [Google Scholar] [CrossRef]
- Prusiner, S.B.; Scott, M.R.; DeArmond, S.J.; Cohen, F.E. Prion Protein Biology. Cell 1998, 93, 337–348. [Google Scholar] [CrossRef]
- Sakudo, A.; Ano, Y.; Onodera, T.; Nitta, K.; Shintani, H.; Ikuta, K.; Tanaka, Y. Fundamentals of Prions and Their Inactivation (Review). Int. J. Mol. Med. 2011, 27, 483–489. [Google Scholar] [CrossRef]
- Kujala, P.; Raymond, C.R.; Romeijn, M.; Godsave, S.F.; van Kasteren, S.I.; Wille, H.; Prusiner, S.B.; Mabbott, N.A.; Peters, P.J. Prion Uptake in the Gut: Identification of the First Uptake and Replication Sites. PLoS Pathog. 2011, 7, e1002449. [Google Scholar] [CrossRef]
- Prinz, M.; Helkenwalder, M.; Junt, T.; Schwarz, P.; Glatzel, M.; Heppner, F.L.; Fu, Y.X.; Lipp, M.; Aguzzi, A. Positioning of Follicular Dendritic Cells within the Spleen Controls Prion Neuroinvasion. Nature 2003, 425, 957–962. [Google Scholar] [CrossRef]
- Aguzzi, A.; Weissmann, C. Spongiform encephalopathies: A suspicious Signature. Nature 1996, 383, 666–667. [Google Scholar] [CrossRef]
- Ferguson, N.M.; Donnelly, C.A.; Woolhouse, M.E.J.; Anderson, R.M. The Epidemiology of BSE in Cattle Herds in Great Britain. II. Model Construction and Analysis of Transmission Dynamics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1997, 352, 803–838. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.A.; Vorberg, I.; Priola, S.A. Species Barriers in Prion Diseases–Brief Review. Arch. Virol. Suppl. 2005, 19, 187–202. [Google Scholar] [CrossRef]
- Johnson, R.T. Prion Diseases. Lancet Neurol. 2005, 4, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Simoneau, S.; Rezaei, H.; Salès, N.; Kaiser-Schulz, G.; Lefebvre-Roque, M.; Vidal, C.; Fournier, J.G.; Comte, J.; Wopfner, F.; Grosclaude, J.; et al. In Vitro and in Vivo Neurotoxicity of Prion Protein Oligomers. PLoS Pathog. 2007, 3, e125. [Google Scholar] [CrossRef]
- Resenberger, U.K.; Harmeier, A.; Woerner, A.C.; Goodman, J.L.; Müller, V.; Krishnan, R.; Vabulas, R.M.; Kretzschmar, H.A.; Lindquist, S.; Hartl, F.U.; et al. The Cellular Prion Protein Mediates Neurotoxic Signalling of Î 2-Sheet-Rich Conformers Independent of Prion Replication. EMBO J. 2011, 30, 2057–2070. [Google Scholar] [CrossRef]
- Goold, R.; Rabbanian, S.; Sutton, L.; Andre, R.; Arora, P.; Moonga, J.; Clarke, A.R.; Schiavo, G.; Jat, P.; Collinge, J.; et al. Rapid Cell-Surface Prion Protein Conversion Revealed Using a Novel Cell System. Nat. Commun. 2011, 2, 281. [Google Scholar] [CrossRef]
- Fang, C.; Wu, B.; Le, N.T.T.; Imberdis, T.; Mercer, R.C.C.; Harris, D.A. Prions Activate a P38 MAPK Synaptotoxic Signaling Pathway. PLoS Pathog. 2018, 14, e1007283. [Google Scholar] [CrossRef]
- Moreno, J.A.; Radford, H.; Peretti, D.; Steinert, J.R.; Verity, N.; Martin, M.G.; Halliday, M.; Morgan, J.; Dinsdale, D.; Ortori, C.A.; et al. Sustained Translational Repression by EIF2α-P Mediates Prion Neurodegeneration. Nature 2012, 485, 507–511, Erratum in: Nature 2014, 511, 370. [Google Scholar] [CrossRef]
- Moreno, J.A.; Halliday, M.; Molloy, C.; Radford, H.; Verity, N.; Axten, J.M.; Ortori, C.A.; Willis, A.E.; Fischer, P.M.; Barrett, D.A.; et al. Oral Treatment Targeting the Unfolded Protein Response Prevents Neurodegeneration and Clinical Disease in Prion-Infected Mice. Sci. Transl. Med. 2013, 5, 206ra138. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, H.J.; Jang, B.; Kim, H.J.; Mostafa, M.N.; Park, S.J.; Kim, Y.S.; Choi, E.K. Impairment of Neuronal Mitochondrial Quality Control in Prion-Induced Neurodegeneration. Cells 2022, 11, 2744. [Google Scholar] [CrossRef]
- Carroll, J.A.; Striebel, J.F.; Rangel, A.; Woods, T.; Phillips, K.; Peterson, K.E.; Race, B.; Chesebro, B. Prion Strain Differences in Accumulation of PrPSc on Neurons and Glia Are Associated with Similar Expression Profiles of Neuroinflammatory Genes: Comparison of Three Prion Strains. PLoS Pathog. 2016, 12, e1005551. [Google Scholar] [CrossRef]
- Makarava, N.; Chang, J.C.Y.; Molesworth, K.; Baskakov, I.V. Region-Specific Glial Homeostatic Signature in Prion Diseases Is Replaced by a Uniform Neuroinflammation Signature, Common for Brain Regions and Prion Strains with Different Cell Tropism. Neurobiol. Dis. 2020, 137, 104783. [Google Scholar] [CrossRef] [PubMed]
- Makarava, N.; Mychko, O.; Chang, J.C.Y.; Molesworth, K.; Baskakov, I.V. The Degree of Astrocyte Activation Is Predictive of the Incubation Time to Prion Disease. Acta Neuropathol. Commun. 2021, 9, 87. [Google Scholar] [CrossRef] [PubMed]
- Makarava, N.; Kushwaha, R.; Baskakov, I.V. Reactive Astrocytes in Prion Diseases: Friend or Foe? PLoS Pathog. 2024, 20, e1012286. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Thomassen, J.Q.; Bellenguez, C.; Grenier-Boley, B.; De Rojas, I.; Castillo, A.; Parveen, K.; Küçükali, F.; Nicolas, A.; Peters, O.; et al. Genetic Associations between Modifiable Risk Factors and Alzheimer Disease. JAMA Netw. Open 2023, 6, e2313734. [Google Scholar] [CrossRef]
- Armstrong, R.A. Risk Factors for Alzheimer’s Disease. Folia Neuropathol. 2019, 57, 87–105. [Google Scholar] [CrossRef]
- Chen, G.F.; Xu, T.H.; Yan, Y.; Zhou, Y.R.; Jiang, Y.; Melcher, K.; Xu, H.E. Amyloid Beta: Structure, Biology and Structure-Based Therapeutic Development. Acta Pharmacol. Sin. 2017, 38, 1205–1235. [Google Scholar] [CrossRef]
- Palop, J.J.; Mucke, L. Amyloid-Β-Induced Neuronal Dysfunction in Alzheimer’s Disease: From Synapses toward Neural Networks. Nat. Neurosci. 2010, 13, 812–818. [Google Scholar] [CrossRef]
- Petrushanko, I.Y.; Tverskoi, A.M.; Barykin, E.P.; Petrovskaya, A.V.; Strelkova, M.A.; Leonova, O.G.; Anashkina, A.A.; Tolstova, A.P.; Adzhubei, A.A.; Bogdanova, A.Y.; et al. Na,K-ATPase Acts as a Beta-Amyloid Receptor Triggering Src Kinase Activation. Cells 2022, 11, 2753. [Google Scholar] [CrossRef]
- Kamenetz, F.; Tomita, T.; Hsieh, H.; Seabrook, G.; Borchelt, D.; Iwatsubo, T.; Sisodia, S.; Malinow, R. APP Processing and Synaptic Function. Neuron 2003, 37, 925–937. [Google Scholar] [CrossRef]
- Kumar, D.K.V.; Choi, H.S.; Washicosky, K.J.; Eimer, W.A.; Tucker, S.; Ghofrani, J.; Lefkowitz, A.; McColl, G.; Goldstein, L.E.; Tanzi, R.E.; et al. Amyloid-β Peptide Protects against Microbial Infection in Mouse and Worm Models of Alzheimer’s Disease. Sci. Transl. Med. 2016, 8, 340ra72. [Google Scholar] [CrossRef]
- Pike, C.J.; Burdick, D.; Walencewicz, A.J.; Glabe, C.G.; Cotman, C.W. Neurodegeneration Induced by β-Amyloid Peptides in Vitro: The Role of Peptide Assembly State. J. Neurosci. 1993, 13, 1676–1687. [Google Scholar] [CrossRef]
- Ezkurdia, A.; Ramírez, M.J.; Solas, M. Metabolic Syndrome as a Risk Factor for Alzheimer’s Disease: A Focus on Insulin Resistance. Int. J. Mol. Sci. 2023, 24, 4354. [Google Scholar] [CrossRef]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a Risk Factor for Neurodegenerative Disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef]
- Yoon, S.S.; Jo, S.A. Mechanisms of Amyloid-β Peptide Clearance: Potential Therapeutic Targets for Alzheimer’s Disease. Biomol. Ther. 2012, 20, 245–255. [Google Scholar] [CrossRef]
- Nedergaard, M.; Goldman, S.A. Glymphatic Failure as a Final Common Pathway to Dementia. Science 2020, 370, 50–56. [Google Scholar] [CrossRef]
- Sweeney, M.D.; Zhao, Z.; Montagne, A.; Nelson, A.R.; Zlokovic, B.V. Blood-Brain Barrier: From Physiology to Disease and Back. Physiol. Rev. 2019, 99, 21–78. [Google Scholar] [CrossRef] [PubMed]
- Sikanyika, N.L.; Parkington, H.C.; Smith, A.I.; Kuruppu, S. Powering Amyloid Beta Degrading Enzymes: A Possible Therapy for Alzheimer’s Disease. Neurochem. Res. 2019, 44, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Laurén, J.; Gimbel, D.A.; Nygaard, H.B.; Gilbert, J.W.; Strittmatter, S.M. Cellular Prion Protein Mediates Impairment of Synaptic Plasticity by Amyloid-Β Oligomers. Nature 2009, 457, 1128–1132. [Google Scholar] [CrossRef] [PubMed]
- Stoner, A.; Fu, L.; Nicholson, L.S.; Zheng, C.; Toyonaga, T.; Spurrier, J.; Laird, W.; Cai, Z.; Strittmatter, S.M. Neuronal Transcriptome, Tau and Synapse Loss in Alzheimer’s Knock-in Mice Require Prion Protein. Alzheimer’s Res. Ther. 2023, 15, 201. [Google Scholar] [CrossRef] [PubMed]
- Um, J.W.; Stephen, M.S. Amyloid-β Induced Signaling by Cellular Prion Protein and Fyn Kinase in Alzheimer Disease. Prion 2013, 7, 37–41. [Google Scholar] [CrossRef]
- Taniguchi, K.; Yamamoto, F.; Amano, A.; Tamaoka, A.; Sanjo, N.; Yokota, T.; Kametani, F.; Araki, W. Amyloid-β Oligomers Interact with NMDA Receptors Containing GluN2B Subunits and Metabotropic Glutamate Receptor 1 in Primary Cortical Neurons: Relevance to the Synapse Pathology of Alzheimer’s Disease. Neurosci. Res. 2022, 180, 90–98. [Google Scholar] [CrossRef]
- Bode, D.C.; Baker, M.D.; Viles, J.H. Ion Channel Formation by Amyloid-Β42 Oligomers but Not Amyloid-Β40 in Cellular Membranes. J. Biol. Chem. 2017, 292, 1404–1413. [Google Scholar] [CrossRef]
- Lustbader, J.W.; Cirilli, M.; Lin, C.; Xu, H.W.; Takuma, K.; Wang, N.; Caspersen, C.; Chen, X.; Pollak, S.; Chaney, M.; et al. ABAD Directly Links Aβ to Mitochondrial Toxicity in Alzheimer’s Disease. Science 2004, 304, 448–452. [Google Scholar] [CrossRef]
- Hansson Petersen, C.A.; Alikhani, N.; Behbahani, H.; Wiehager, B.; Pavlov, P.F.; Alafuzoff, I.; Leinonen, V.; Ito, A.; Winblad, B.; Glaser, E.; et al. The Amyloid β-Peptide Is Imported into Mitochondria via the TOM Import Machinery and Localized to Mitochondrial Cristae. Proc. Natl. Acad. Sci. USA 2008, 105, 13145–13150. [Google Scholar] [CrossRef]
- Meng, X.; Song, Q.; Liu, Z.; Liu, X.; Wang, Y.; Liu, J. Neurotoxic β-Amyloid Oligomers Cause Mitochondrial Dysfunction—The Trigger for PANoptosis in Neurons. Front. Aging Neurosci. 2024, 16, 1400544. [Google Scholar] [CrossRef]
- Deng, Q.; Wu, C.; Parker, E.; Liu, T.C.Y.; Duan, R.; Yang, L. Microglia and Astrocytes in Alzheimer’s Disease: Significance and Summary of Recent Advances. Aging Dis. 2024, 15, 1537–1564. [Google Scholar] [CrossRef]
- Weingarten, M.D.; Lockwood, A.H.; Hwo, S.Y.; Kirschner, M.W. A Protein Factor Essential for Microtubule Assembly. Proc. Natl. Acad. Sci. USA 1975, 72, 1858–1862. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.V.W.; Stoothoff, W.H. Tau Phosphorylation in Neuronal Cell Function and Dysfunction. J. Cell Sci. 2004, 117, 5721–5729. [Google Scholar] [CrossRef] [PubMed]
- Trinczek, B.; Ebneth, A.; Mandelkow, E.M.; Mandelkow, E. Tau Regulates the Attachment/Detachment but Not the Speed of Motors in Microtubule-Dependent Transport of Single Vesicles and Organelles. J. Cell Sci. 1999, 112, 2355–2367. [Google Scholar] [CrossRef]
- Aranda-Abreu, G.E.; Rojas-Durán, F.; Hernández-Aguilar, M.E.; Herrera-Covarrubias, D.; García-Hernández, L.I.; Toledo-Cárdenas, M.R.; Chi-Castañeda, D. The Role of Tau in Neuronal Function and Neurodegeneration. Neurol. Int. 2025, 17, 75. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, A.W.P.; Falcon, B.; He, S.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Crowther, R.A.; Ghetti, B.; Goedert, M.; Scheres, S.H.W. Cryo-EM Structures of Tau Filaments from Alzheimer’s Disease. Nature 2017, 547, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, A.; Hoerndli, F.; Baechi, T.; Nitsch, R.M.; Götz, J. β-Amyloid Induces Paired Helical Filament-like Tau Filaments in Tissue Culture. J. Biol. Chem. 2003, 278, 40162–40168. [Google Scholar] [CrossRef] [PubMed]
- Ising, C.; Venegas, C.; Zhang, S.; Scheiblich, H.; Schmidt, S.V.; Vieira-Saecker, A.; Schwartz, S.; Albasset, S.; McManus, R.M.; Tejera, D.; et al. NLRP3 Inflammasome Activation Drives Tau Pathology. Nature 2019, 575, 669–673. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, K.M.; Yang, L.; Dong, Q.; Yu, J.T. Tauopathies: New Perspectives and Challenges. Mol. Neurodegener. 2022, 17, 28. [Google Scholar] [CrossRef]
- Rodríguez-Martín, T.; Cuchillo-Ibáñez, I.; Noble, W.; Nyenya, F.; Anderton, B.H.; Hanger, D.P. Tau Phosphorylation Affects Its Axonal Transport and Degradation. Neurobiol. Aging 2013, 34, 2146–2157. [Google Scholar] [CrossRef]
- Min, S.W.; Chen, X.; Tracy, T.E.; Li, Y.; Zhou, Y.; Wang, C.; Shirakawa, K.; Minami, S.S.; Defensor, E.; Mok, S.A.; et al. Critical Role of Acetylation in Tau-Mediated Neurodegeneration and Cognitive Deficits. Nat. Med. 2015, 21, 1154–1162. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, Y.; Ma, L.; Wei, Y.; Li, H. Possible Mechanisms of Tau Spread and Toxicity in Alzheimer’s Disease. Front. Cell Dev. Biol. 2021, 9, 707268. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Y.; Gao, D.; Ye, J.; Wang, X.; Fang, L.; Wu, D.; Pi, G.; Lu, C.; Zhou, X.W.; et al. Accumulation of Human Full-Length Tau Induces Degradation of Nicotinic Acetylcholine Receptor A4 via Activating Calpain-2. Sci. Rep. 2016, 6, 27283. [Google Scholar] [CrossRef]
- Parra Bravo, C.; Naguib, S.A.; Gan, L. Cellular and Pathological Functions of Tau. Nat. Rev. Mol. Cell Biol. 2024, 25, 845–864. [Google Scholar] [CrossRef] [PubMed]
- Berger, Z.; Roder, H.; Hanna, A.; Carlson, A.; Rangachari, V.; Yue, M.; Wszolek, Z.; Ashe, K.; Knight, J.; Dickson, D.; et al. Accumulation of Pathological Tau Species and Memory Loss in a Conditional Model of Tauopathy. J. Neurosci. 2007, 27, 3650–3662. [Google Scholar] [CrossRef] [PubMed]
- Lasagna-Reeves, C.A.; Castillo-Carranza, D.L.; Sengupta, U.; Clos, A.L.; Jackson, G.R.; Kayed, R. Tau Oligomers Impair Memory and Induce Synaptic and Mitochondrial Dysfunction in Wild-Type Mice. Mol. Neurodegener. 2011, 6, 39. [Google Scholar] [CrossRef] [PubMed]
- Manczak, M.; Reddy, P.H. Abnormal Interaction between the Mitochondrial Fission Protein Drp1 and Hyperphosphorylated Tau in Alzheimer’s Disease Neurons: Implications for Mitochondrial Dysfunction and Neuronal Damage. Hum. Mol. Genet. 2012, 21, 2538–2547. [Google Scholar] [CrossRef]
- Manczak, M.; Reddy, P.H. Abnormal Interaction of VDAC1 with Amyloid Beta and Phosphorylated Tau Causes Mitochondrial Dysfunction in Alzheimer’s Disease. Hum. Mol. Genet. 2012, 21, 5131–5146. [Google Scholar] [CrossRef]
- Eckert, A.; Nisbet, R.; Grimm, A.; Götz, J. March Separate, Strike Together—Role of Phosphorylated TAU in Mitochondrial Dysfunction in Alzheimer’s Disease. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 1258–1266. [Google Scholar] [CrossRef]
- Hoover, B.R.; Reed, M.N.; Su, J.; Penrod, R.D.; Kotilinek, L.A.; Grant, M.K.; Pitstick, R.; Carlson, G.A.; Lanier, L.M.; Yuan, L.L.; et al. Tau Mislocalization to Dendritic Spines Mediates Synaptic Dysfunction Independently of Neurodegeneration. Neuron 2010, 68, 1067–1081. [Google Scholar] [CrossRef]
- Younas, N.; Saleem, T.; Younas, A.; Zerr, I. Nuclear Face of Tau: An inside Player in Neurodegeneration. Acta Neuropathol. Commun. 2023, 11, 196. [Google Scholar] [CrossRef]
- Bolós, M.; Llorens-Martín, M.; Jurado-Arjona, J.; Hernández, F.; Rábano, A.; Avila, J. Direct Evidence of Internalization of Tau by Microglia in Vitro and in Vivo. J. Alzheimer’s Dis. 2016, 50, 77–87. [Google Scholar] [CrossRef]
- Perea, J.R.; López, E.; Diéz-Ballesteros, J.C.; Ávila, J.; Hernández, F.; Bolós, M. Extracellular Monomeric Tau Is Internalized by Astrocytes. Front. Neurosci. 2019, 13, 442. [Google Scholar] [CrossRef]
- Piacentini, R.; Li Puma, D.D.; Mainardi, M.; Lazzarino, G.; Tavazzi, B.; Arancio, O.; Grassi, C. Reduced Gliotransmitter Release from Astrocytes Mediates Tau-Induced Synaptic Dysfunction in Cultured Hippocampal Neurons. Glia 2017, 65, 1302–1316. [Google Scholar] [CrossRef]
- Corbett, G.T.; Wang, Z.; Hong, W.; Colom-Cadena, M.; Rose, J.; Liao, M.; Asfaw, A.; Hall, T.C.; Ding, L.; DeSousa, A.; et al. PrP Is a Central Player in Toxicity Mediated by Soluble Aggregates of Neurodegeneration-Causing Proteins. Acta Neuropathol. 2020, 139, 503–526. [Google Scholar] [CrossRef]
- Wysocka, A.; Palasz, E.; Steczkowska, M.; Niewiadomska, G. Dangerous Liaisons: Tau Interaction with Muscarinic Receptors. Curr. Alzheimer Res. 2020, 17, 224–237. [Google Scholar] [CrossRef]
- Ruan, Z.; Pathak, D.; Venkatesan Kalavai, S.; Yoshii-Kitahara, A.; Muraoka, S.; Bhatt, N.; Takamatsu-Yukawa, K.; Hu, J.; Wang, Y.; Hersh, S.; et al. Alzheimer’s Disease Brain-Derived Extracellular Vesicles Spread Tau Pathology in Interneurons. Brain 2021, 144, 288–309, Erratum in: Brain 2021, 144, e42. [Google Scholar] [CrossRef]
- Bigi, A.; Cascella, R.; Cecchi, C. α-Synuclein Oligomers and Fibrils: Partners in Crime in Synucleinopathies. Neural Regen. Res. 2023, 18, 2332–2342. [Google Scholar] [CrossRef]
- Guerrero-Ferreira, R.; Taylor, N.M.I.; Mona, D.; Ringler, P.; Lauer, M.E.; Riek, R.; Britschgi, M.; Stahlberg, H. Cryo-EM Structure of Alpha-Synuclein Fibrils. eLife 2018, 7, e36402. [Google Scholar] [CrossRef]
- Brieger, K.; Schiavone, S.; Miller, F.J.; Krause, K.H. Reactive Oxygen Species: From Health to Disease. Swiss Med. Wkly. 2012, 142, w13659. [Google Scholar] [CrossRef]
- Zhang, P.; Tian, B. Metabolic Syndrome: An Important Risk Factor for Parkinson’s Disease. Oxid. Med. Cell. Longev. 2014, 2014, 729194. [Google Scholar] [CrossRef]
- Brás, I.C.; Xylaki, M.; Outeiro, T.F. Mechanisms of Alpha-Synuclein Toxicity: An Update and Outlook. Prog. Brain Res. 2020, 252, 91–129. [Google Scholar] [CrossRef]
- Bousset, L.; Pieri, L.; Ruiz-Arlandis, G.; Gath, J.; Jensen, P.H.; Habenstein, B.; Madiona, K.; Olieric, V.; Böckmann, A.; Meier, B.H.; et al. Structural and Functional Characterization of Two Alpha-Synuclein Strains. Nat. Commun. 2013, 4, 2575. [Google Scholar] [CrossRef]
- Choi, B.K.; Choi, M.G.; Kim, J.Y.; Yang, Y.; Lai, Y.; Kweon, D.H.; Lee, N.K.; Shin, Y.K. Large α-Synuclein Oligomers Inhibit Neuronal SNARE-Mediated Vesicle Docking. Proc. Natl. Acad. Sci. USA 2013, 110, 4087–4092. [Google Scholar] [CrossRef]
- Wang, L.; Das, U.; Scott, D.A.; Tang, Y.; McLean, P.J.; Roy, S. α-Synuclein Multimers Cluster Synaptic Vesicles and Attenuate Recycling. Curr. Biol. 2014, 24, 2319–2326. [Google Scholar] [CrossRef]
- Perez, R.G.; Waymire, J.C.; Lin, E.; Liu, J.J.; Guo, F.; Zigmond, M.J. A Role for α-Synuclein in the Regulation of Dopamine Biosynthesis. J. Neurosci. 2002, 22, 3090–3099. [Google Scholar] [CrossRef]
- Wersinger, C.; Sidhu, A. Attenuation of Dopamine Transporter Activity by α-Synuclein. Neurosci. Lett. 2003, 340, 189–192. [Google Scholar] [CrossRef]
- Bro̷chner, B.V.; Zhang, X.; Nielsen, J.; Kjems, J.; Otzen, D.E.; Malle, M.G. Single-Vesicle Tracking of α-Synuclein Oligomers Reveals Pore Formation by a Three-Stage Model. ACS Nano 2025, 19, 32108–32122. [Google Scholar] [CrossRef]
- Pozo Devoto, V.M.; Falzone, T.L. Mitochondrial Dynamics in Parkinson’s Disease: A Role for α-Synuclein? Dis. Model. Mech. 2017, 10, 1075–1087. [Google Scholar] [CrossRef]
- Klivenyi, P.; Siwek, D.; Gardian, G.; Yang, L.; Starkov, A.; Cleren, C.; Ferrante, R.J.; Kowall, N.W.; Abeliovich, A.; Beal, M.F. Mice Lacking Alpha-Synuclein Are Resistant to Mitochondrial Toxins. Neurobiol. Dis. 2006, 21, 541–548. [Google Scholar] [CrossRef]
- Koch, J.C.; Bitow, F.; Haack, J.; Hedouville, Z.; Zhang, J.N.; Tönges, L.; Michel, U.; Oliveira, L.M.A.; Jovin, T.M.; Liman, J.; et al. Alpha-Synuclein Affects Neurite Morphology, Autophagy, Vesicle Transport and Axonal Degeneration in CNS Neurons. Cell Death Dis. 2015, 6, e1811. [Google Scholar] [CrossRef]
- Saramowicz, K.; Siwecka, N.; Galita, G.; Kucharska-Lusina, A.; Rozpędek-Kamińska, W.; Majsterek, I. Alpha-Synuclein Contribution to Neuronal and Glial Damage in Parkinson’s Disease. Int. J. Mol. Sci. 2024, 25, 360. [Google Scholar] [CrossRef]
- Guo, M.; Wang, J.; Zhao, Y.; Feng, Y.; Han, S.; Dong, Q.; Cui, M.; Tieu, K. Microglial Exosomes Facilitate A-Synuclein Transmission in Parkinson’s Disease. Brain 2020, 143, 1476–1497. [Google Scholar] [CrossRef]
- Boza-Serrano, A.; Reyes, J.F.; Rey, N.L.; Leffler, H.; Bousset, L.; Nilsson, U.; Brundin, P.; Venero, J.L.; Burguillos, M.A.; Deierborg, T. The Role of Galectin-3 in α-Synuclein-Induced Microglial Activation. Acta Neuropathol. Commun. 2014, 2, 156. [Google Scholar] [CrossRef]
- George, S.; Rey, N.L.; Tyson, T.; Esquibel, C.; Meyerdirk, L.; Schulz, E.; Pierce, S.; Burmeister, A.R.; Madaj, Z.; Steiner, J.A.; et al. Microglia Affect α-Synuclein Cell-to-Cell Transfer in a Mouse Model of Parkinson’s Disease. Mol. Neurodegener. 2019, 14, 34. [Google Scholar] [CrossRef]
- Chavarría, C.; Rodríguez-Bottero, S.; Quijano, C.; Cassina, P.; Souza, J.M. Impact of Monomeric, Oligomeric and Fibrillar Alpha-Synuclein on Astrocyte Reactivity and Toxicity to Neurons. Biochem. J. 2018, 475, 3153–3169. [Google Scholar] [CrossRef]
- Gil, J.M.; Rego, A.C. Mechanisms of Neurodegeneration in Huntington’s Disease. Eur. J. Neurosci. 2008, 27, 2803–2820. [Google Scholar] [CrossRef]
- DiFiglia, M.; Sapp, E.; Chase, K.O.; Davies, S.W.; Bates, G.P.; Vonsattel, J.P.; Aronin, N. Aggregation of Huntingtin in Neuronal Intranuclear Inclusions and Dystrophic Neurites in Brain. Science 1997, 277, 1990–1993. [Google Scholar] [CrossRef]
- Sapp, E.; Penney, J.; Young, A.; Aronin, N.; Vonsattel, J.P.; DiFiglia, M. Axonal Transport of N-Terminal Huntingtin Suggests Early Pathology of Corticostriatal Projections in Huntington Disease. J. Neuropathol. Exp. Neurol. 1999, 58, 165–173. [Google Scholar] [CrossRef]
- MacDonald, M.E.; Ambrose, C.M.; Duyao, M.P.; Myers, R.H.; Lin, C.; Srinidhi, L.; Barnes, G.; Taylor, S.A.; James, M.; Groot, N.; et al. A Novel Gene Containing a Trinucleotide Repeat That Is Expanded and Unstable on Huntington’s Disease Chromosomes. Cell 1993, 72, 971–983. [Google Scholar] [CrossRef]
- Ranen, N.G.; Stine, O.C.; Abbott, M.H.; Sherr, M.; Codori, A.M.; Franz, M.L.; Chao, N.I.; Chung, A.S.; Pleasant, N.; Callahan, C.; et al. Anticipation and Instability of IT-15 (CAG)N Repeats in Parent-Offspring Pairs with Huntington Disease. Am. J. Hum. Genet. 1995, 57, 593–602. [Google Scholar]
- Kaltenbach, L.S.; Romero, E.; Becklin, R.R.; Chettier, R.; Bell, R.; Phansalkar, A.; Strand, A.; Torcassi, C.; Savage, J.; Hurlburt, A.; et al. Huntingtin Interacting Proteins Are Genetic Modifiers of Neurodegeneration. PLoS Genet. 2007, 3, e82. [Google Scholar] [CrossRef]
- Zuccato, C.; Tartari, M.; Crotti, A.; Goffredo, D.; Valenza, M.; Conti, L.; Cataudella, T.; Leavitt, B.R.; Hayden, M.R.; Timmusk, T.; et al. Huntingtin Interacts with REST/NRSF to Modulate the Transcription of NRSE-Controlled Neuronal Genes. Nat. Genet. 2003, 35, 76–83. [Google Scholar] [CrossRef]
- Marcora, E.; Gowan, K.; Lee, J.E. Stimulation of NeuroD Activity by Huntingtin and Huntingtin-Associated Proteins HAP1 and MLK2. Proc. Natl. Acad. Sci. USA 2003, 100, 9578–9583. [Google Scholar] [CrossRef]
- Rigamonti, D.; Bauer, J.H.; De-Fraja, C.; Conti, L.; Sipione, S.; Sciorati, C.; Clementi, E.; Hackam, A.; Hayden, M.R.; Li, Y.; et al. Wild-Type Huntingtin Protects from Apoptosis Upstream of Caspase-3. J. Neurosci. 2000, 20, 3705–3713. [Google Scholar] [CrossRef]
- Gervais, F.G.; Singaraja, R.; Xanthoudakis, S.; Gutekunst, C.A.; Leavitt, B.R.; Metzler, M.; Hackam, A.S.; Tam, J.; Vaillancourt, J.P.; Houtzager, V.; et al. Recruitment and Activation of Caspase-8 by the Huntingtin-Interacting Protein Hip-1 and a Novel Partner Hippi. Nat. Cell Biol. 2002, 4, 95–105. [Google Scholar] [CrossRef]
- Joel, D. Open Interconnected Model of Basal Ganglia-Thalamocortical Circuitry and Its Relevance to the Clinical Syndrome of Huntington’s Disease. Mov. Disord. 2001, 16, 407–423. [Google Scholar] [CrossRef]
- Cowan, C.M.; Fan, M.M.Y.; Fan, J.; Shehadeh, J.; Zhang, L.Y.J.; Graham, R.K.; Hayden, M.R.; Raymond, L.A. Polyglutamine-Modulated Striatal Calpain Activity in YAC Transgenic Huntington Disease Mouse Model: Impact on NMDA Receptor Function and Toxicity. J. Neurosci. 2008, 28, 12725–12735. [Google Scholar] [CrossRef]
- Miller, J.P.; Holcomb, J.; Al-Ramahi, I.; de Haro, M.; Gafni, J.; Zhang, N.; Kim, E.; Sanhueza, M.; Torcassi, C.; Kwak, S.; et al. Matrix Metalloproteinases Are Modifiers of Huntingtin Proteolysis and Toxicity in Huntington’s Disease. Neuron 2010, 67, 199–212. [Google Scholar] [CrossRef]
- Martin, D.D.O.; Schmidt, M.E.; Nguyen, Y.T.; Lazic, N.; Hayden, M.R. Identification of a Novel Caspase Cleavage Site in Huntingtin That Regulates Mutant Huntingtin Clearance. FASEB J. 2019, 33, 3190–3197. [Google Scholar] [CrossRef]
- Wellington, C.L.; Ellerby, L.M.; Gutekunst, C.A.; Rogers, D.; Warby, S.; Graham, R.K.; Loubser, O.; Van Raamsdonk, J.; Singaraja, R.; Yang, Y.Z.; et al. Caspase Cleavage of Mutant Huntingtin Precedes Neurodegeneration in Huntington’s Disease. J. Neurosci. 2002, 22, 7862–7872. [Google Scholar] [CrossRef]
- Goldberg, Y.P.; Nicholson, D.W.; Rasper, D.M.; Kalchman, M.A.; Koide, H.B.; Graham, R.K.; Bromm, M.; Kazemi-Esfarjani, P.; Thornberry, N.A.; Vaillancourt, J.P.; et al. Cleavage of Huntingtin by Apopain, a Proapoptotic Cysteine Protease, Is Modulated by the Polyglutamine Tract. Nat. Genet. 1996, 13, 442–449. [Google Scholar] [CrossRef]
- Bano, D.; Zanetti, F.; Mende, Y.; Nicotera, P. Neurodegenerative Processes in Huntington’s Disease. Cell Death Dis. 2011, 2, e228. [Google Scholar] [CrossRef]
- Huang, C.C.; Faber, P.W.; Persichetti, F.; Mittal, V.; Vonsattel, J.P.; MacDonald, M.E.; Gusella, J.F. Amyloid Formation by Mutant Huntingtin: Threshold, Progressivity and Recruitment of Normal Polyglutamine Proteins. Somat. Cell Mol. Genet. 1998, 24, 217–233. [Google Scholar] [CrossRef]
- Scherzinger, E.; Lurz, R.; Turmaine, M.; Mangiarini, L.; Hollenbach, B.; Hasenbank, R.; Bates, G.P.; Davies, S.W.; Lehrach, H.; Wanker, E.E. Huntingtin-Encoded Polyglutamine Expansions Form Amyloid-like Protein Aggregates In Vitro and In Vivo. Cell 1997, 90, 549–558. [Google Scholar] [CrossRef]
- Xiong, K.; Punihaole, D.; Asher, S.A. UV Resonance Raman Spectroscopy Monitors Polyglutamine Backbone and Side Chain Hydrogen Bonding and Fibrillization. Biochemistry 2012, 51, 5822–5830. [Google Scholar] [CrossRef]
- Buchanan, L.E.; Carr, J.K.; Fluitt, A.M.; Hoganson, A.J.; Moran, S.D.; De Pablo, J.J.; Skinner, J.L.; Zanni, M.T. Structural Motif of Polyglutamine Amyloid Fibrils Discerned with Mixed-Isotope Infrared Spectroscopy. Proc. Natl. Acad. Sci. USA 2014, 111, 5796–5801. [Google Scholar] [CrossRef]
- Bagherpoor Helabad, M.; Matlahov, I.; Kumar, R.; Daldrop, J.O.; Jain, G.; Weingarth, M.; van der Wel, P.C.A.; Miettinen, M.S. Integrative Determination of Atomic Structure of Mutant Huntingtin Exon 1 Fibrils Implicated in Huntington Disease. Nat. Commun. 2024, 15, 10793. [Google Scholar] [CrossRef]
- Kuemmerle, S.; Gutekunst, C.A.; Klein, A.M.; Li, X.J.; Li, S.H.; Beal, M.F.; Hersch, S.M.; Ferrante, R.J. Huntingtin Aggregates May Not Predict Neuronal Death in Huntington’s Disease. Ann. Neurol. 1999, 46, 842–849. [Google Scholar] [CrossRef]
- Saudou, F.; Finkbeiner, S.; Devys, D.; Greenberg, M.E. Huntingtin Acts in the Nucleus to Induce Apoptosis but Death Does Not Correlate with the Formation of Intranuclear Inclusions. Cell 1998, 95, 55–66. [Google Scholar] [CrossRef]
- Arrasate, M.; Mitra, S.; Schweitzer, E.S.; Segal, M.R.; Finkbeiner, S. Inclusion Body Formation Reduces Levels of Mutant Huntingtin and the Risk of Neuronal Death. Nature 2004, 431, 805–810. [Google Scholar] [CrossRef]
- Gauthier, L.R.; Charrin, B.C.; Borrell-Pagès, M.; Dompierre, J.P.; Rangone, H.; Cordelières, F.P.; De Mey, J.; MacDonald, M.E.; Leßmann, V.; Humbert, S.; et al. Huntingtin Controls Neurotrophic Support and Survival of Neurons by Enhancing BDNF Vesicular Transport along Microtubules. Cell 2004, 118, 127–138. [Google Scholar] [CrossRef]
- Morton, A.J.; Edwardson, J.M. Progressive Depletion of Complexin II in a Transgenic Mouse Model of Huntington’s Disease. J. Neurochem. 2001, 76, 166–172. [Google Scholar] [CrossRef]
- Modregger, J.; DiProspero, N.A.; Charles, V.; Tagle, D.A.; Plomann, M. PACSIN 1 Interacts with Huntingtin and Is Absent from Synaptic Varicosities in Presymptomatic Huntington’s Disease Brains. Hum. Mol. Genet. 2002, 11, 2547–2558. [Google Scholar] [CrossRef]
- Jurcau, A. Molecular Pathophysiological Mechanisms in Huntington’s Disease. Biomedicines 2022, 10, 1432. [Google Scholar] [CrossRef]
- Maglione, V.; Cannella, M.; Gradini, R.; Cislaghi, G.; Squitieri, F. Huntingtin Fragmentation and Increased Caspase 3, 8 and 9 Activities in Lymphoblasts with Heterozygous and Homozygous Huntington’s Disease Mutation. Mech. Ageing Dev. 2006, 127, 213–216. [Google Scholar] [CrossRef]
- Schapira, A.H.V.; Olanow, C.W.; Greenamyre, J.T.; Bezard, E. Slowing of Neurodegeneration in Parkinson’s Disease and Huntington’s Disease: Future Therapeutic Perspectives. Lancet 2014, 384, 545–555. [Google Scholar] [CrossRef]
- Zarei, S.; Carr, K.; Reiley, L.; Diaz, K.; Guerra, O.; Altamirano, P.F.; Pagani, W.; Lodin, D.; Orozco, G.; Chinea, A. A Comprehensive Review of Amyotrophic Lateral Sclerosis. Surg. Neurol. Int. 2015, 6, 171. [Google Scholar] [CrossRef]
- Rabinovici, G.D.; Miller, B.L. Frontotemporal Lobar Degeneration: Epidemiology, Pathophysiology, Diagnosis and Management. CNS Drugs 2010, 24, 375–398. [Google Scholar] [CrossRef]
- Lillo, P.; Hodges, J.R. Frontotemporal Dementia and Motor Neurone Disease: Overlapping Clinic-Pathological Disorders. J. Clin. Neurosci. 2009, 16, 1131–1135. [Google Scholar] [CrossRef]
- Van Langenhove, T.; Van Der Zee, J.; Van Broeckhoven, C. The Molecular Basis of the Frontotemporal Lobar Degeneration-Amyotrophic Lateral Sclerosis Spectrum. Ann. Med. 2012, 44, 817–828. [Google Scholar] [CrossRef]
- Andersen, P.M.; Al-Chalabi, A. Clinical Genetics of Amyotrophic Lateral Sclerosis: What Do We Really Know? Nat. Rev. Neurol. 2011, 7, 603–615. [Google Scholar] [CrossRef]
- Logroscino, G.; Piccininni, M.; Graff, C.; Hardiman, O.; Ludolph, A.C.; Moreno, F.; Otto, M.; Remes, A.M.; Rowe, J.B.; Seelaar, H.; et al. Incidence of Syndromes Associated With Frontotemporal Lobar Degeneration in 9 European Countries. JAMA Neurol. 2023, 80, 279–286. [Google Scholar] [CrossRef]
- Tzeplaeff, L.; Jürs, A.V.; Wohnrade, C.; Demleitner, A.F. Unraveling the Heterogeneity of ALS—A Call to Redefine Patient Stratification for Better Outcomes in Clinical Trials. Cells 2024, 13, 452. [Google Scholar] [CrossRef]
- Arseni, D.; Hasegawa, M.; Murzin, A.G.; Kametani, F.; Arai, M.; Yoshida, M.; Ryskeldi-Falcon, B. Structure of Pathological TDP-43 Filaments from ALS with FTLD. Nature 2022, 601, 139–143. [Google Scholar] [CrossRef]
- Alami, N.H.; Smith, R.B.; Carrasco, M.A.; Williams, L.A.; Winborn, C.S.; Han, S.S.W.; Kiskinis, E.; Winborn, B.; Freibaum, B.D.; Kanagaraj, A.; et al. Axonal Transport of TDP-43 MRNA Granules Is Impaired by ALS-Causing Mutations. Neuron 2014, 81, 536–543. [Google Scholar] [CrossRef]
- Higashi, S.; Kabuta, T.; Nagai, Y.; Tsuchiya, Y.; Akiyama, H.; Wada, K. TDP-43 Associates with Stalled Ribosomes and Contributes to Cell Survival during Cellular Stress. J. Neurochem. 2013, 126, 288–300. [Google Scholar] [CrossRef]
- Kawahara, Y.; Mieda-Sato, A. TDP-43 Promotes MicroRNA Biogenesis as a Component of the Drosha and Dicer Complexes. Proc. Natl. Acad. Sci. USA 2012, 109, 3347–3352. [Google Scholar] [CrossRef]
- Fukushima, M.; Hosoda, N.; Chifu, K.; Hoshino, S. ichi TDP-43 Accelerates Deadenylation of Target MRNAs by Recruiting Caf1 Deadenylase. FEBS Lett. 2019, 593, 277–287. [Google Scholar] [CrossRef]
- Sephton, C.F.; Cenik, C.; Kucukural, A.; Dammer, E.B.; Cenik, B.; Han, Y.H.; Dewey, C.M.; Roth, F.P.; Herz, J.; Peng, J.; et al. Identification of Neuronal RNA Targets of TDP-43-Containing Ribonucleoprotein Complexes. J. Biol. Chem. 2011, 286, 1204–1215. [Google Scholar] [CrossRef]
- Ayala, Y.M.; De Conti, L.; Avendaño-Vázquez, S.E.; Dhir, A.; Romano, M.; D’Ambrogio, A.; Tollervey, J.; Ule, J.; Baralle, M.; Buratti, E.; et al. TDP-43 Regulates Its MRNA Levels through a Negative Feedback Loop. EMBO J. 2011, 30, 277–288. [Google Scholar] [CrossRef]
- Gu, J.; Wu, F.; Xu, W.; Shi, J.; Hu, W.; Jin, N.; Qian, W.; Wang, X.; Iqbal, K.; Gong, C.X.; et al. TDP-43 Suppresses Tau Expression via Promoting Its MRNA Instability. Nucleic Acids Res. 2017, 45, 6177–6193. [Google Scholar] [CrossRef]
- Fang, Y.S.; Tsai, K.J.; Chang, Y.J.; Kao, P.; Woods, R.; Kuo, P.H.; Wu, C.C.; Liao, J.Y.; Chou, S.C.; Lin, V.; et al. Full-Length TDP-43 Forms Toxic Amyloid Oligomers That Are Present in Frontotemporal Lobar Dementia-TDP Patients. Nat. Commun. 2014, 5, 4824. [Google Scholar] [CrossRef]
- Johnson, B.S.; Snead, D.; Lee, J.J.; McCaffery, J.M.; Shorter, J.; Gitler, A.D. TDP-43 Is Intrinsically Aggregation-Prone, and Amyotrophic Lateral Sclerosis-Linked Mutations Accelerate Aggregation and Increase Toxicity. J. Biol. Chem. 2009, 284, 20329–20339. [Google Scholar] [CrossRef]
- Wood, A.; Gurfinkel, Y.; Polain, N.; Lamont, W.; Lyn Rea, S. Molecular Mechanisms Underlying TDP-43 Pathology in Cellular and Animal Models of ALS and FTLD. Int. J. Mol. Sci. 2021, 22, 4705. [Google Scholar] [CrossRef]
- Davis, S.A.; Itaman, S.; Khalid-Janney, C.M.; Sherard, J.A.; Dowell, J.A.; Cairns, N.J.; Gitcho, M.A. TDP-43 interacts with mitochondrial proteins critical for mitophagy and mitochondrial dynamics. Neurosci. Lett. 2018, 678, 8–15. [Google Scholar] [CrossRef]
- Ulamec, S.M.; Radford, S.E. Spot the Difference: Function versus Toxicity in Amyloid Fibrils. Trends Biochem. Sci. 2020, 45, 635–636. [Google Scholar] [CrossRef]
- Seuring, C.; Verasdonck, J.; Gath, J.; Ghosh, D.; Nespovitaya, N.; Wälti, M.A.; Maji, S.K.; Cadalbert, R.; Güntert, P.; Meier, B.H.; et al. The Three-Dimensional Structure of Human β-Endorphin Amyloid Fibrils. Nat. Struct. Mol. Biol. 2020, 27, 1178–1184. [Google Scholar] [CrossRef]
- Sun, C.; Zhou, K.; DePaola, P.; Shin, W.S.; Hillyer, T.; Sawaya, M.R.; Zhu, R.; Peng, C.; Zhou, Z.H.; Jiang, L. Cryo-EM Structure of Amyloid Fibril Formed by α-Synuclein Hereditary A53E Mutation Reveals a Distinct Protofilament Interface. J. Biol. Chem. 2023, 299, 104566. [Google Scholar] [CrossRef]
- Manka, S.W.; Zhang, W.; Wenborn, A.; Betts, J.; Joiner, S.; Saibil, H.R.; Collinge, J.; Wadsworth, J.D.F. 2.7 Å Cryo-EM Structure of Ex Vivo RML Prion Fibrils. Nat. Commun. 2022, 13, 4004. [Google Scholar] [CrossRef]
- Arseni, D.; Chen, R.; Murzin, A.G.; Peak-Chew, S.Y.; Garringer, H.J.; Newell, K.L.; Kametani, F.; Robinson, A.C.; Vidal, R.; Ghetti, B.; et al. TDP-43 Forms Amyloid Filaments with a Distinct Fold in Type A FTLD-TDP. Nature 2023, 620, 898–903. [Google Scholar] [CrossRef]
- Yang, Y.; Arseni, D.; Zhang, W.; Huang, M.; Lövestam, S.; Schweighauser, M.; Kotecha, A.; Murzin, A.G.; Peak-Chew, S.Y.; MacDonald, J.; et al. Cryo-EM Structures of Amyloid-b 42 Filaments from Human Brains. Science 2022, 375, 167–172. [Google Scholar] [CrossRef]
- Joshi, D.C.; Chavan, M.B.; Gurow, K.; Gupta, M.; Dhaliwal, J.S.; Ming, L.C. The Role of Mitochondrial Dysfunction in Huntington’s Disease: Implications for Therapeutic Targeting. Biomed. Pharmacother. 2025, 183, 117827. [Google Scholar] [CrossRef]
- Lurette, O.; Martín-Jiménez, R.; Khan, M.; Sheta, R.; Jean, S.; Schofield, M.; Teixeira, M.; Rodriguez-Aller, R.; Perron, I.; Oueslati, A.; et al. Aggregation of Alpha-Synuclein Disrupts Mitochondrial Metabolism and Induce Mitophagy via Cardiolipin Externalization. Cell Death Dis. 2023, 14, 729. [Google Scholar] [CrossRef]
- Needs, H.I.; Wilkinson, K.A.; Henley, J.M.; Collinson, I. Aggregation-Prone Tau Impairs Mitochondrial Import, Which Affects Organelle Morphology and Neuronal Complexity. J. Cell Sci. 2023, 136, jcs260993. [Google Scholar] [CrossRef] [PubMed]
- Chong, Z.Z.; Souayah, N. Pathogenic TDP-43 in Amyotrophic Lateral Sclerosis. Drug Discov. Today 2025, 30, 104351. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.A.; Fang, N.; Zhang, W.; Ji, S. The Multifaceted Role of Fragile X-Related Protein 1 (FXR1) in Cellular Processes: An Updated Review on Cancer and Clinical Applications. Cell Death Dis. 2024, 15, 72. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Fansler, M.M.; Janjoš, U.; Ule, J.; Mayr, C. The FXR1 Network Acts as a Signaling Scaffold for Actomyosin Remodeling. Cell 2024, 187, 5048–5063.e25. [Google Scholar] [CrossRef]
- Nuvolone, M.; Merlini, G. Systemic amyloidosis: Novel therapies and role of biomarkers. Nephrol. Dial. Transplant. 2017, 32, 770–780. [Google Scholar] [CrossRef][Green Version]
- Galkin, A.P.; Sysoev, E.I. Stress Response Is the Main Trigger of Sporadic Amyloidoses. Int. J. Mol. Sci. 2021, 22, 4092. [Google Scholar] [CrossRef]


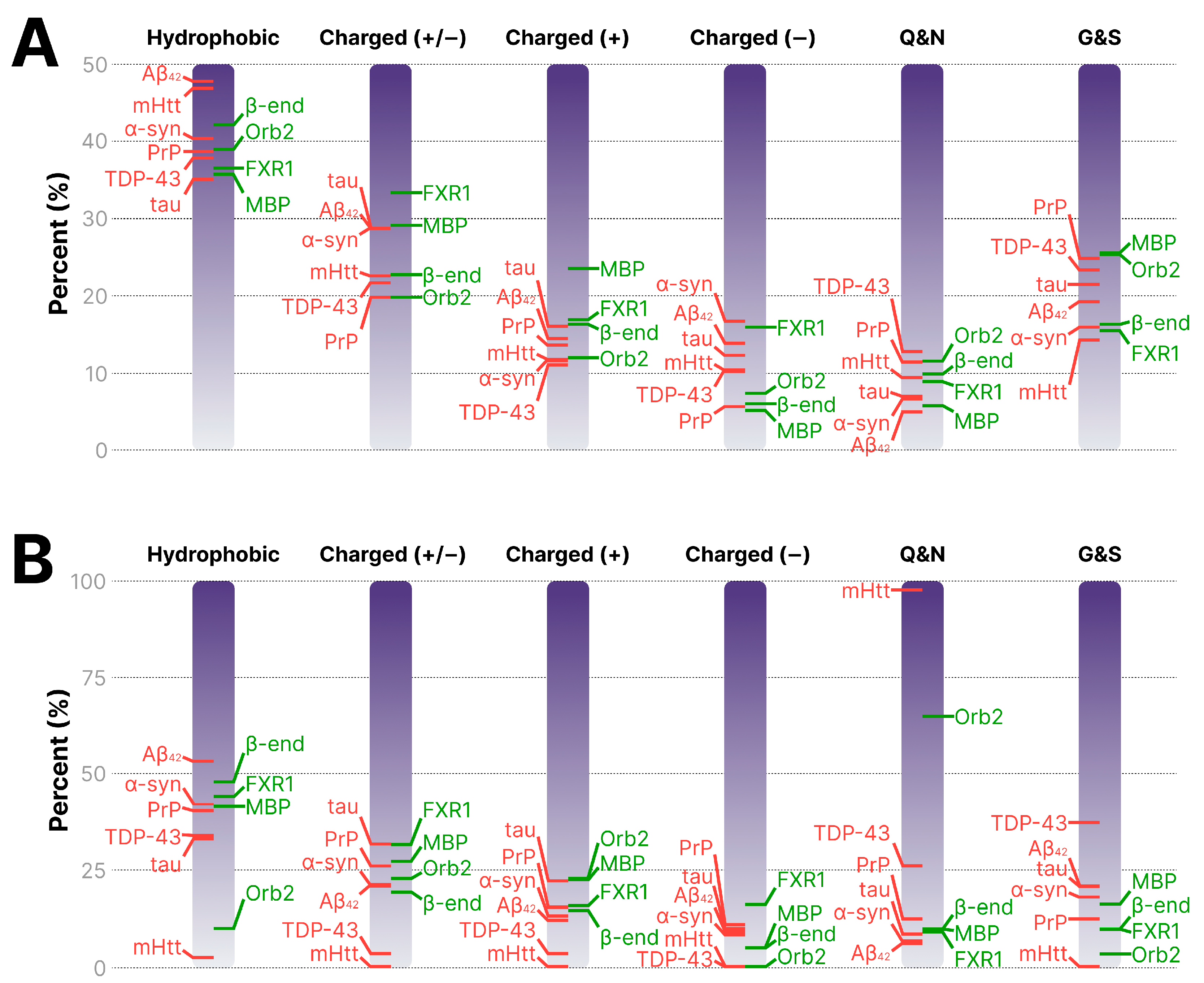
| Protein | Uniprot ID | Hydrophobic, % | Charged (+/−), % | Charged (+), % | Charged (−), % | Q/N, % | G/S, % |
|---|---|---|---|---|---|---|---|
| Functional amyloids | |||||||
| Orb2 | Q9VSR3 | 38.77 | 19.62 | 11.8 | 7.82 | 11.36 | 25.15 |
| FXR1 | P51114 | 36.38 | 33.17 | 16.75 | 16.42 | 8.69 | 15.3 |
| MBP | P02686-3 | 35.54 | 28.94 | 23.35 | 5.59 | 5.58 | 25.38 |
| β-endorphin | PRO_0000024975 | 41.94 | 22.58 | 16.13 | 6.45 | 9.68 | 16.13 |
| Pathological amyloids | |||||||
| PrP | P04156 | 38.53 | 19.48 | 13.42 | 6.06 | 11.25 | 24.67 |
| mHtt (Q44) | P42858 | 46.71 | 22.4 | 11.6 | 10.8 | 9.25 | 14.06 |
| TDP-43 | Q13148 | 37.67 | 21.49 | 10.87 | 10.62 | 12.56 | 23.19 |
| tau | P10636-8 | 34.91 | 28.57 | 15.87 | 12.7 | 6.8 | 21.31 |
| α-syn | P37840 | 42.15 | 28.57 | 11.42 | 17.15 | 6.43 | 15.72 |
| Aβ42 | PRO_0000000095 | 47.61 | 28.56 | 14.28 | 14.28 | 4.76 | 19.05 |
| Amyloidogenic Core * | Core, % of Full Protein | Hydrophobic, % | Charged (+/−), % | Charged (+), % | Charged (−), % | Q/N, % | G/S, % |
|---|---|---|---|---|---|---|---|
| Orb2 [8] | 4.4 | 9.68 | 22.58 | 22.58 | 0 | 64.52 | 3.23 |
| FXR1 [7] | 61.0 | 43.79 | 31.39 | 15.56 | 15.83 | 8.97 | 9.5 |
| MBP [10] | 32.0 | 41.27 | 26.98 | 22.22 | 4.76 | 9.52 | 15.87 |
| β-endorphin [205] | 67.7 | 47.6 | 19.05 | 14.29 | 4.76 | 9.52 | 9.52 |
| PrP [207] | 57.1 | 40.16 | 25.76 | 15.15 | 10.61 | 12.12 | 12.12 |
| mHtt (Q44) [175] | 1.4 | 2.22 | 0 | 0 | 0 | 97.8 | 0 |
| TDP-43 [208] | 21.5 | 33.7 | 3.37 | 3.37 | 0 | 25.85 | 37.08 |
| tau [113] | 16.6 | 32.88 | 31.51 | 21.92 | 9.59 | 8.22 | 20.55 |
| α-syn [206] | 44.3 | 41.93 | 20.96 | 12.9 | 8.06 | 6.45 | 17.75 |
| Aβ42 [209] | 81.0 | 52.93 | 20.58 | 11.76 | 8.82 | 5.88 | 20.59 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galkin, A.P.; Mitkevich, V.A.; Makarov, A.A.; Valina, A.A.; Sysoev, E.I. Pathological and Functional Brain Amyloids: A New Concept Explaining the Differences. Int. J. Mol. Sci. 2025, 26, 10459. https://doi.org/10.3390/ijms262110459
Galkin AP, Mitkevich VA, Makarov AA, Valina AA, Sysoev EI. Pathological and Functional Brain Amyloids: A New Concept Explaining the Differences. International Journal of Molecular Sciences. 2025; 26(21):10459. https://doi.org/10.3390/ijms262110459
Chicago/Turabian StyleGalkin, Alexey P., Vladimir A. Mitkevich, Alexander A. Makarov, Anna A. Valina, and Evgeniy I. Sysoev. 2025. "Pathological and Functional Brain Amyloids: A New Concept Explaining the Differences" International Journal of Molecular Sciences 26, no. 21: 10459. https://doi.org/10.3390/ijms262110459
APA StyleGalkin, A. P., Mitkevich, V. A., Makarov, A. A., Valina, A. A., & Sysoev, E. I. (2025). Pathological and Functional Brain Amyloids: A New Concept Explaining the Differences. International Journal of Molecular Sciences, 26(21), 10459. https://doi.org/10.3390/ijms262110459






