Population Genetics of Pharmacogenetic Variants in a Greek Psychiatric Cohort of over 3000 Individuals
Abstract
1. Introduction
2. Results
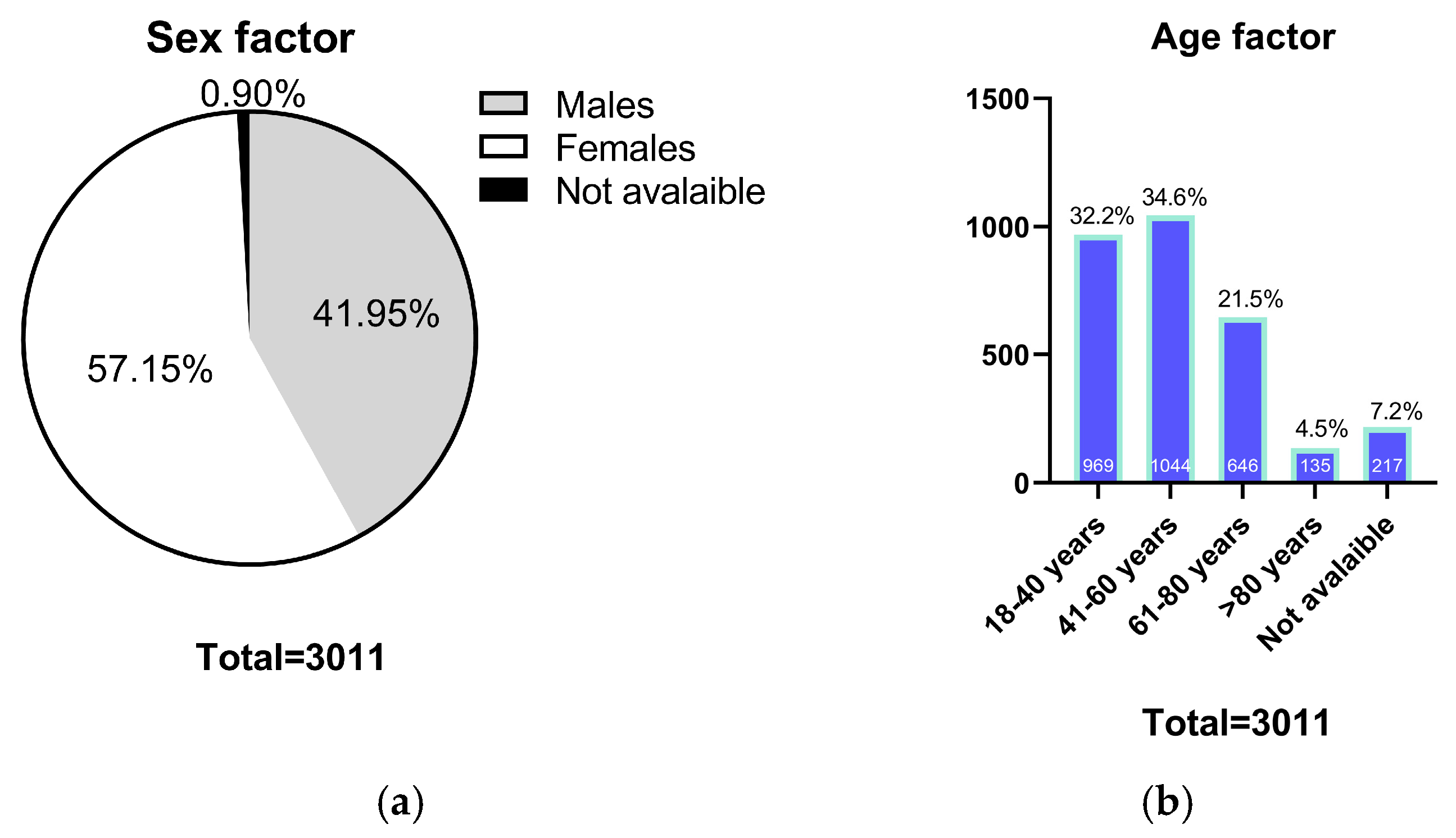
2.1. Population Characteristics
2.2. Pharmacogenetic Variant Frequencies Distributions Between Greek and Other Continental Populations
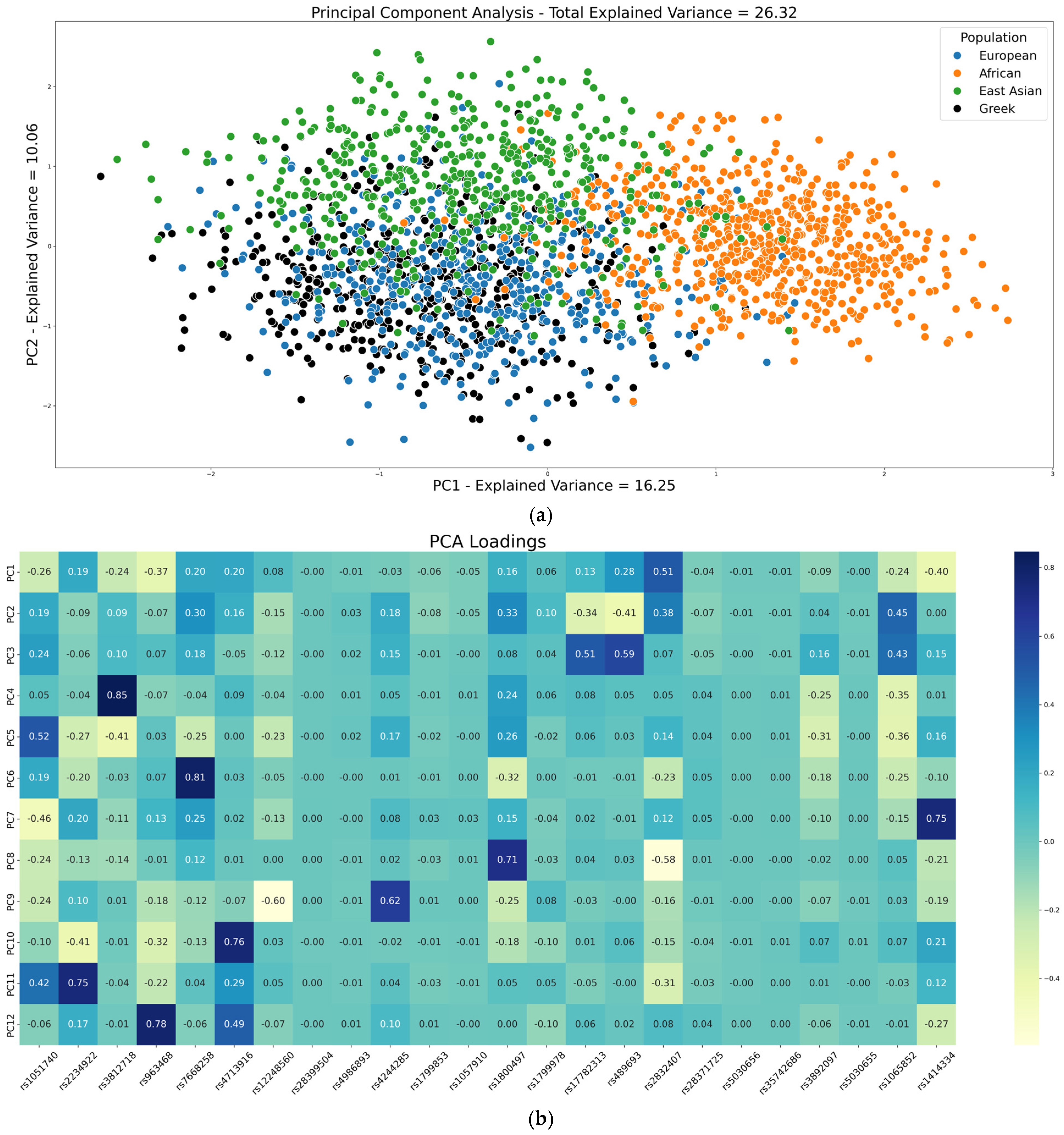
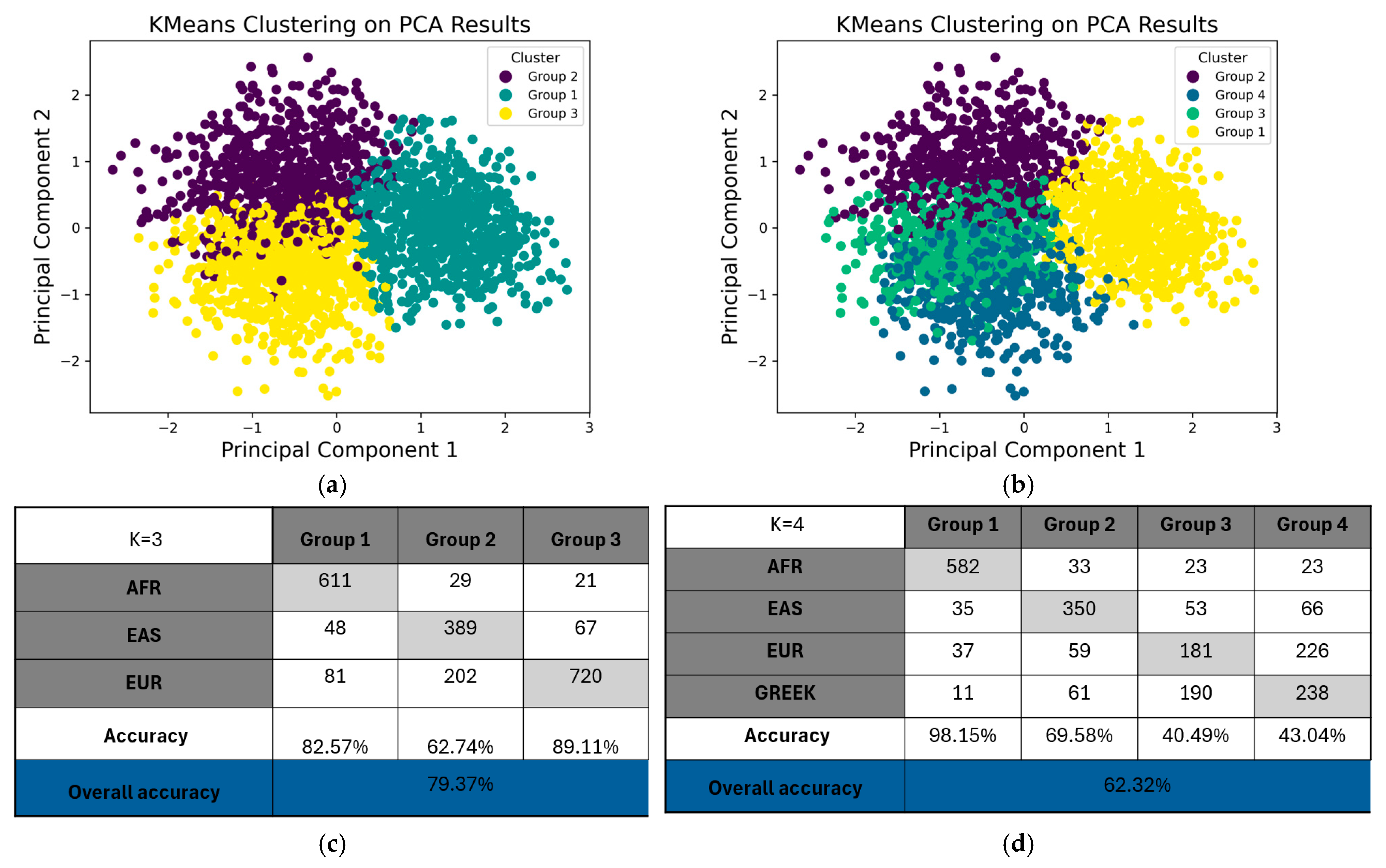
2.3. Population Structure Analysis Through PCA and K-Means Clustering of PGx Variants
2.4. Assessment of Population Divergence Between Greeks and Global Populations Based on the FST and STRUCTURE
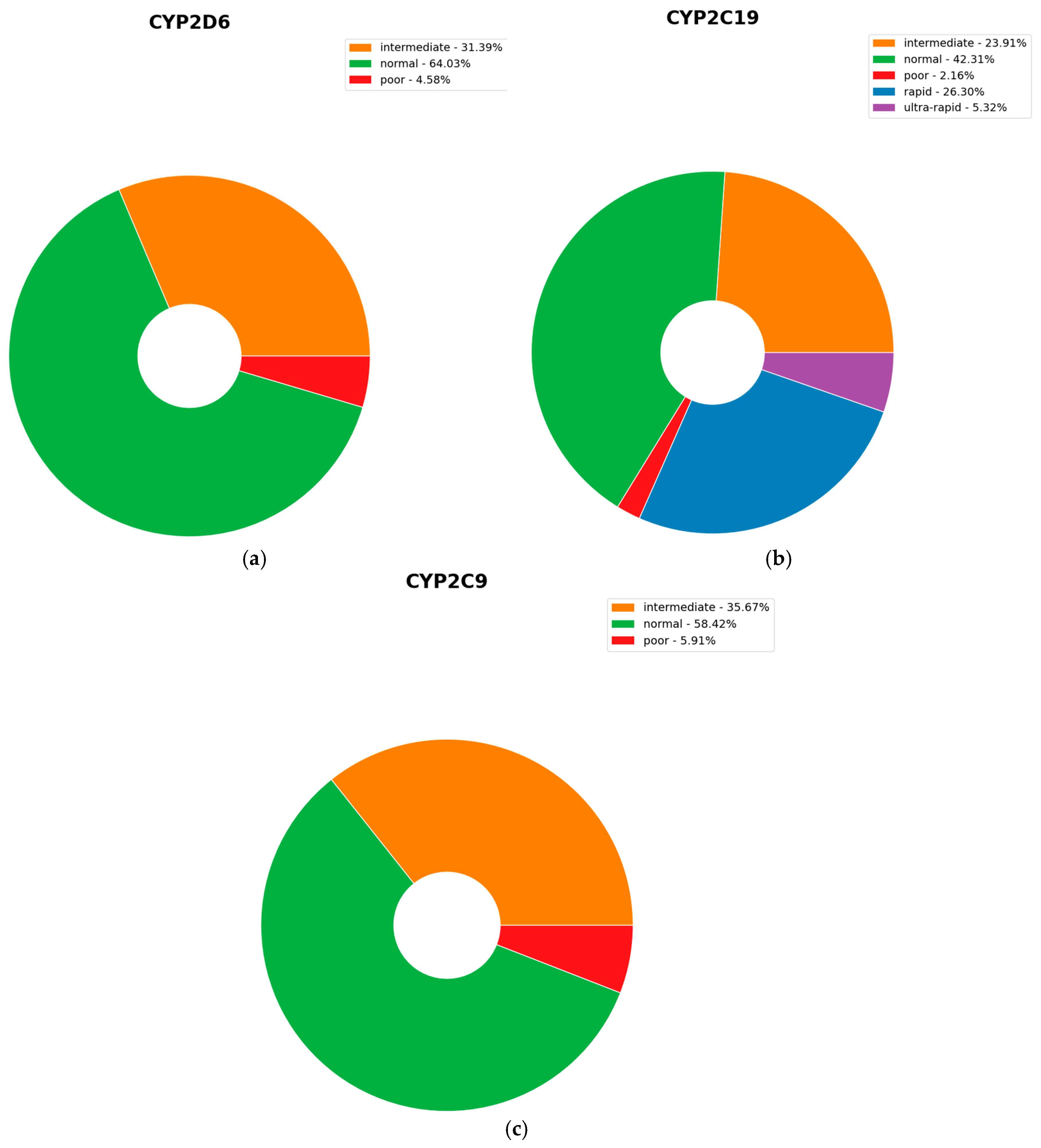
2.5. Clinically Actionable Variants
3. Discussion
4. Materials and Methods
4.1. Samples and Data
4.2. Genotyping
4.3. Comparative Analysis of Variant Frequencies Across Different Populations
4.3.1. Statistical and Bioinformatic Analysis
4.3.2. Population Genetics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PGx | Pharmacogenetics |
| MDD | Major Depressive Disorder |
| BD | Bipolar Disorder |
| CPIC | Clinical Pharmacogenetics Implementation Consortium |
| DPWG | Dutch Pharmacogenetics Working Group |
| AMP | Association for Molecular Pathology |
| SNPs | Single-Nucleotide Polymorphisms |
| PBS | Phosphate-Buffered Saline |
| NTC | No-Template Control |
| VCF | Variant Call Format |
| CNS | Central Nervous System |
| PCA | Principal Component Analysis |
| EUR | European |
| AFR | African |
| EAS | East Asian |
| ARI | Adjusted Rand Index |
| NMI | Normalized Mutual Information |
| FST | Fixation Index |
| IGA | Individual Genetic Ancestry |
References
- James, S.L.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, Regional, and National Incidence, Prevalence, and Years Lived with Disability for 354 Diseases and Injuries for 195 Countries and Territories, 1990–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar] [CrossRef] [PubMed]
- WHO Mental Disorders. Available online: https://www.who.int/news-room/fact-sheets/detail/mental-disorders (accessed on 13 June 2025).
- Rush, A.J.; Trivedi, M.H.; Wisniewski, S.R.; Nierenberg, A.A.; Stewart, J.W.; Warden, D.; Niederehe, G.; Thase, M.E.; Lavori, P.W.; Lebowitz, B.D.; et al. Acute and Longer-Term Outcomes in Depressed Outpatients Requiring One or Several Treatment Steps: A STAR*D Report. Am. J. Psychiatry 2006, 163, 1905–1917. [Google Scholar] [CrossRef]
- Geddes, J.R.; Miklowitz, D.J. Treatment of Bipolar Disorder. Lancet 2013, 381, 1672–1682. [Google Scholar] [CrossRef]
- Howes, O.D.; McCutcheon, R.; Agid, O.; De Bartolomeis, A.; Van Beveren, N.J.M.; Birnbaum, M.L.; Bloomfield, M.A.P.; Bressan, R.A.; Buchanan, R.W.; Carpenter, W.T.; et al. Treatment-ResistantSchizophrenia: TreatmentResponse and Resistance in Psychosis (TRRIP) Working Group Consensus Guidelines on Diagnosis and Terminology. Am. J. Psychiatry 2017, 174, 216–229. [Google Scholar] [CrossRef]
- Caudle, K.E.; Dunnenberger, H.M.; Freimuth, R.R.; Peterson, J.F.; Burlison, J.D.; Whirl-Carrillo, M.; Scott, S.A.; Rehm, H.L.; Williams, M.S.; Klein, T.E.; et al. Standardizing Terms for Clinical Pharmacogenetic Test Results: Consensus Terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet. Med. 2016, 19, 215. [Google Scholar] [CrossRef]
- Qahwaji, R.; Ashankyty, I.; Sannan, N.S.; Hazzazi, M.S.; Basabrain, A.A.; Mobashir, M. Pharmacogenomics: A Genetic Approach to Drug Development and Therapy. Pharmaceuticals 2024, 17, 940. [Google Scholar] [CrossRef]
- Truong, T.T.T.; Lago, J.; Neil, J.; Wilkes, F.A.; Barnes, R.; Hopwood, M.; Singh, A.B. Psychotropic Pharmacogenetics in Adult Populations: From Basic Science to Clinical Trials and Health Economics—An Evidence-Based Overview for Decision Makers. Aust. N. Z. J. Psychiatry 2025. [Google Scholar] [CrossRef]
- Dalton, R.; Lee, S.; Claw, K.G.; Prasad, B.; Phillips, B.R.; Shen, D.D.; Wong, L.H.; Fade, M.; McDonald, M.G.; Dunham, M.J.; et al. Interrogation of CYP2D6 Structural Variant Alleles Improves the Correlation Between CYP2D6 Genotype and CYP2D6-Mediated Metabolic Activity. Clin. Transl. Sci. 2020, 13, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Petrović, J.; Pešić, V.; Lauschke, V.M. Frequencies of Clinically Important CYP2C19 and CYP2D6 Alleles Are Graded across Europe. Eur. J. Hum. Genet. 2020, 28, 88–94. [Google Scholar] [CrossRef]
- van Schaik, R.H.N.; Müller, D.J.; Serretti, A.; Ingelman-Sundberg, M. Pharmacogenetics in Psychiatry: An Update on Clinical Usability. Front. Pharmacol. 2020, 11, 575540. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.A. The Genetic Testing Reference Materials Coordination Program: Over 10 Years of Support for Pharmacogenomic Testing. J. Mol. Diagn. 2023, 25, 630–633. [Google Scholar] [CrossRef]
- Pratt, V.M.; Cavallari, L.H.; Del Tredici, A.L.; Gaedigk, A.; Hachad, H.; Ji, Y.; Kalman, L.V.; Ly, R.C.; Moyer, A.M.; Scott, S.A.; et al. Recommendations for Clinical CYP2D6 Genotyping Allele Selection: A Joint Consensus Recommendation of the Association for Molecular Pathology, College of American Pathologists, Dutch Pharmacogenetics Working Group of the Royal Dutch Pharmacists Association, and the European Society for Pharmacogenomics and Personalized Therapy. J. Mol. Diagn. 2021, 23, 1047–1064. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, J.M.J.L.; Nijenhuis, M.; Soree, B.; Guchelaar, H.-J.; Swen, J.J.; van Schaik, R.H.N.; van der Weide, J.; Rongen, G.A.P.J.M.; Buunk, A.-M.; de Boer-Veger, N.J.; et al. Dutch Pharmacogenetics Working Group (DPWG) Guideline for the Gene-Drug Interaction between CYP2C19 and CYP2D6 and SSRIs. Eur. J. Hum. Genet. 2022, 30, 1114–1120. [Google Scholar] [CrossRef]
- Bousman, C.A.; Stevenson, J.M.; Ramsey, L.B.; Sangkuhl, K.; Hicks, J.K.; Strawn, J.R.; Singh, A.B.; Ruaño, G.; Mueller, D.J.; Tsermpini, E.E.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6, CYP2C19, CYP2B6, SLC6A4, and HTR2A Genotypes and Serotonin Reuptake Inhibitor Antidepressants. Clin. Pharmacol. Ther. 2023, 114, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Park, Y.; Camara, M.D.; Lauschke, V.M. Opportunities and Challenges of Population Pharmacogenomics. Ann. Hum. Genet. 2025, 89, 384–397. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Uno, T.; Yamazaki, H.; Tateishi, T. Limited Frequency of the CYP2C19*17 Allele and Its Minor Role in a Japanese Population. Br. J. Clin. Pharmacol. 2007, 65, 437. [Google Scholar] [CrossRef]
- Sim, S.C.; Risinger, C.; Dahl, M.L.; Aklillu, E.; Christensen, M.; Bertilsson, L.; Ingelman-Sundberg, M. A Common Novel CYP2C19 Gene Variant Causes Ultrarapid Drug Metabolism Relevant for the Drug Response to Proton Pump Inhibitors and Antidepressants. Clin. Pharmacol. Ther. 2006, 79, 103–113. [Google Scholar] [CrossRef]
- Ragia, G.; Pallikarou, M.; Manolopoulou, Y.; Vorvolakos, T.; Manolopoulos, V.G. Genetic Diversity of Cytochrome P450 in Patients Receiving Psychiatric Care in Greece: A Step towards Clinical Implementation. Pharmacogenomics 2024, 25, 217. [Google Scholar] [CrossRef]
- Koopmans, A.B.; Braakman, M.H.; Vinkers, D.J.; Hoek, H.W.; van Harten, P.N. Meta-Analysis of Probability Estimates of Worldwide Variation of CYP2D6 and CYP2C19. Transl. Psychiatry 2021, 11, 141. [Google Scholar] [CrossRef]
- Jukić, M.M.; Haslemo, T.; Molden, E.; Ingelman-Sundberg, M. Impact of CYP2C19 Genotype on Escitalopram Exposure and Therapeutic Failure: A Retrospective Study Based on 2,087 Patients. Am. J. Psychiatry 2018, 175, 463–470. [Google Scholar] [CrossRef]
- Ramamoorthy, A.; Kim, H.H.; Shah-Williams, E.; Zhang, L. Racial and Ethnic Differences in Drug Disposition and Response: Review of New Molecular Entities Approved Between 2014 and 2019. J. Clin. Pharmacol. 2022, 62, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Petrovski, S.; Goldstein, D.B. Unequal Representation of Genetic Variation across Ancestry Groups Creates Healthcare Inequality in the Application of Precision Medicine. Genome Biol. 2016, 17, 157. [Google Scholar] [CrossRef]
- Zhou, Y.; Lauschke, V.M. Population Pharmacogenomics: An Update on Ethnogeographic Differences and Opportunities for Precision Public Health. Hum. Genet. 2021, 141, 1113. [Google Scholar] [CrossRef] [PubMed]
- Stojanović Marković, A.; Zajc Petranović, M.; Škarić-Jurić, T.; Celinšćak, Ž.; Šetinc, M.; Tomas, Ž.; Peričić Salihović, M. Relevance of CYP2D6 Gene Variants in Population Genetic Differentiation. Pharmaceutics 2022, 14, 2481. [Google Scholar] [CrossRef]
- Lo, C.; Nguyen, S.; Yang, C.; Witt, L.; Wen, A.; Liao, T.V.; Nguyen, J.; Lin, B.; Altman, R.B.; Palaniappan, L. Pharmacogenomics in Asian Subpopulations and Impacts on Commonly Prescribed Medications. Clin. Transl. Sci. 2020, 13, 861. [Google Scholar] [CrossRef]
- Bothos, E.; Ntoumou, E.; Kelaidoni, K.; Roukas, D.; Drakoulis, N.; Papasavva, M.; Karakostis, F.A.; Moulos, P.; Karakostis, K. Clinical Pharmacogenomics in Action: Design, Assessment and Implementation of a Novel Pharmacogenetic Panel Supporting Drug Selection for Diseases of the Central Nervous System (CNS). J. Transl. Med. 2021, 19, 151. [Google Scholar] [CrossRef]
- Nagar, S.D.; Conley, A.B.; Jordan, I.K. Population Structure and Pharmacogenomic Risk Stratification in the United States. BMC Biol. 2020, 18, 140. [Google Scholar] [CrossRef]
- Hernandez, W.; Danahey, K.; Pei, X.; Yeo, K.T.J.; Leung, E.; Volchenboum, S.L.; Ratain, M.J.; Meltzer, D.O.; Stranger, B.E.; Perera, M.A.; et al. Pharmacogenomic Genotypes Define Genetic Ancestry in Patients and Enable Population-Specific Genomic Implementation. Pharmacogenomics J. 2020, 20, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Stathias, V.; Sotiris, G.R.; Karagiannidis, I.; Bourikas, G.; Martinis, G.; Papazoglou, D.; Tavridou, A.; Papanas, N.; Maltezos, E.; Theodoridis, M.; et al. Exploring Genomic Structure Differences and Similarities between the Greek and European HapMap Populations: Implications for Association Studies. Ann. Hum. Genet. 2012, 76, 472–483. [Google Scholar] [CrossRef]
- Relling, M.V.; Klein, T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 2011, 89, 464. [Google Scholar] [CrossRef]
- Zhou, Y.; Ingelman-Sundberg, M.; Lauschke, V.M. Worldwide Distribution of Cytochrome P450 Alleles: A Meta-Analysis of Population-Scale Sequencing Projects. Clin. Pharmacol. Ther. 2017, 102, 688–700. [Google Scholar] [CrossRef]
- Hicks, J.K.; Sangkuhl, K.; Swen, J.J.; Ellingrod, V.L.; Müller, D.J.; Shimoda, K.; Bishop, J.R.; Kharasch, E.D.; Skaar, T.C.; Gaedigk, A.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2D6 and CYP2C19 Genotypes and Dosing of Tricyclic Antidepressants: 2016 Update. Clin. Pharmacol. Ther. 2017, 102, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Han, J.M.; Lee, J.; Oh, J.Y.; Kim, J.S.; Gwak, H.S.; Choi, K.H. Comparison of Pharmacogenomic Information for Drug Approvals Provided by the National Regulatory Agencies in Korea, Europe, Japan, and the United States. Front. Pharmacol. 2023, 14, 1205624. [Google Scholar] [CrossRef] [PubMed]
- Fairley, S.; Lowy-Gallego, E.; Perry, E.; Flicek, P. The International Genome Sample Resource (IGSR) Collection of Open Human Genomic Variation Resources. Nucleic Acids Res. 2020, 48, D941–D947. [Google Scholar] [CrossRef]
- Hicks, J.K.; Bishop, J.R.; Sangkuhl, K.; Muller, D.J.; Ji, Y.; Leckband, S.G.; Leeder, J.S.; Graham, R.L.; Chiulli, D.L.; LLerena, A.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin. Pharmacol. Ther. 2015, 98, 127–134. [Google Scholar] [CrossRef]
- Meulman, J.J.; van der Kooij, A.J.; Babinec, A. New Features of Categorical Principal Components Analysis for Complicated Data Sets, Including Data Mining; Springer: Berlin/Heidelberg, Germany, 2002; pp. 207–217. [Google Scholar] [CrossRef]
- Linting, M.; Meulman, J.J.; Groenen, P.J.F.; van der Kooij, A.J. Nonlinear Principal Components Analysis: Introduction and Application. Psychol. Methods 2007, 12, 336–358. [Google Scholar] [CrossRef]
- Song, Y.; Westerhuis, J.A.; Aben, N.; Michaut, M.; Wessels, L.F.A.; Smilde, A.K. Principal Component Analysis of Binary Genomics Data. Brief. Bioinform. 2019, 20, 317–329. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of Population Structure Using Multilocus Genotype Data: Dominant Markers and Null Alleles. Mol. Ecol. Notes 2007, 7, 574. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of Population Structure Using Multilocus Genotype Data: Linked Loci and Correlated Allele Frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef] [PubMed]
- Krishna, K.; Murty, M.N. Genetic K-Means Algorithm. IEEE Trans. Syst. Man Cybern. Part B (Cybern.) 1999, 29, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Likas, A.; Vlassis, N.; Verbeek, J.J. The Global K-Means Clustering Algorithm. Pattern Recognit. 2003, 36, 451–461. [Google Scholar] [CrossRef]

| Gene Variants | Allele_1 | Frequencies (Greeks) | Allele_2 | Frequencies (Greeks) | Greeks-EUR | Greeks-AFR | Greeks-EAS |
|---|---|---|---|---|---|---|---|
| rs1799853 | C | 85.6% | T | 14.4% | >0.99 | <0.01 | <0.01 |
| rs28371725 | C | 87.7% | T | 12.3% | 0.2 | <0.01 | <0.01 |
| rs2832407 | A | 33.6% | C | 66.4% | 0.66 | <0.01 | <0.01 |
| rs1414334 | C | 13.3% | G | 86.7% | >0.99 | <0.01 | <0.01 |
| rs1065852 | A | 20.0% | G | 80.0% | >0.99 | <0.01 | <0.01 |
| rs35742686 | T | 98.4% | - | 1.6% | >0.99 | <0.01 | <0.01 |
| rs28399504 | A | 99.5% | G | 0.5% | >0.99 * | 0.08 | 0.22 * |
| rs4244285 | A | 13.6% | G | 86.4% | >0.99 | <0.01 | <0.01 |
| rs5030656 | CTTCT | 99.1% | CT | 0.9% | <0.01 | 0.02 | 0.02 |
| rs1799978 | C | 7.8% | T | 92.2% | 0.84 | <0.01 | <0.01 |
| rs4713916 | A | 28.4% | G | 71.6% | 0.66 | <0.01 | <0.01 |
| rs5030655 | A | 99.3% | - | 0.7% | <0.01 | 0.06 | 0.04 |
| rs3892097 | C | 83.6% | T | 16.4% | >0.99 | <0.01 | <0.01 |
| rs4986893 | G | 99.9% | A | 0.1% | >0.99 * | 0.21 * | <0.01 |
| rs2234922 | A | 79.3% | G | 20.7% | 0.04 | <0.01 | <0.01 |
| rs1051740 | C | 29.3% | T | 70.7% | >0.99 | <0.01 | <0.01 |
| rs489693 | A | 30.7% | C | 69.3% | >0.99 | <0.01 | <0.01 |
| rs12248560 | C | 79.0% | T | 21.0% | >0.99 | 0.09 | <0.01 |
| rs7668258 | C | 52.4% | T | 47.6% | >0.99 | <0.01 | <0.01 |
| rs3812718 | C | 48.5% | T | 51.5% | >0.99 | <0.01 | <0.01 |
| rs1057910 | A | 90.6% | C | 9.4% | 0.66 | <0.01 | <0.01 |
| rs963468 | A | 37.5% | G | 62.5% | >0.99 | <0.01 | 0.27 |
| rs1800497 | A | 16.6% | G | 83.4% | >0.99 | <0.01 | <0.01 |
| rs17782313 | C | 24.4% | T | 75.6% | >0.99 | 0.06 | <0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ntoumou, E.; Papailia, S.; Vrachnos, D.M.; Fotis, T.; Salata, E.; Kapellou, A.; Vittas, S. Population Genetics of Pharmacogenetic Variants in a Greek Psychiatric Cohort of over 3000 Individuals. Int. J. Mol. Sci. 2025, 26, 9896. https://doi.org/10.3390/ijms26209896
Ntoumou E, Papailia S, Vrachnos DM, Fotis T, Salata E, Kapellou A, Vittas S. Population Genetics of Pharmacogenetic Variants in a Greek Psychiatric Cohort of over 3000 Individuals. International Journal of Molecular Sciences. 2025; 26(20):9896. https://doi.org/10.3390/ijms26209896
Chicago/Turabian StyleNtoumou, Eleni, Sevastiani Papailia, Dimitrios Miltiadis Vrachnos, Thanasis Fotis, Effie Salata, Angeliki Kapellou, and Spiros Vittas. 2025. "Population Genetics of Pharmacogenetic Variants in a Greek Psychiatric Cohort of over 3000 Individuals" International Journal of Molecular Sciences 26, no. 20: 9896. https://doi.org/10.3390/ijms26209896
APA StyleNtoumou, E., Papailia, S., Vrachnos, D. M., Fotis, T., Salata, E., Kapellou, A., & Vittas, S. (2025). Population Genetics of Pharmacogenetic Variants in a Greek Psychiatric Cohort of over 3000 Individuals. International Journal of Molecular Sciences, 26(20), 9896. https://doi.org/10.3390/ijms26209896







