Abstract
Bee pollen is a primary and secondary metabolite-rich natural product collected by pollinators such as honeybees. Polyphenols, particularly flavonoids, are well known for their potent antioxidant activities. Numerous phytochemical and biological studies have focused on Quercus mongolica, a member of the Fagaceae family. However, research focusing specifically on pollen is limited. Moreover, bee pollen chemical composition varies significantly depending on its geographical origin and cultivation conditions. In this study, the flavonoid glycosides of Q. mongolica pollen were profiled using LC–MS/MS-based molecular networking, which revealed that the largest molecular cluster corresponded to flavonoid glycosides. A total of 69 flavonoid glycosides, primarily comprising 2 kaempferol derivatives, 14 quercetin derivatives, and 46 isorhamnetin derivatives, were annotated based on MS/MS fragmentation patterns, spectral library matches in GNPS (Global Natural Products Social Molecular Networking), and comparison with previously reported data. Two primary compounds, isorhamnetin 3-O-β-D-xylopyranosyl (1→6)-β-D-glucopyranoside and isorhamnetin-3-O-neohesperidoside, were identified by comparison with reference standards. This study offers foundational insights into the flavonoid diversity of Q. mongolica pollen, contributing to a broad understanding of its secondary metabolite profile.
1. Introduction
Floral pollen is a male gametophyte produced in the anthers of flowers and mainly transferred by pollinators, such as honeybees, during pollination. Honeybees collect floral pollen and mix it with their secretions and nectar, forming bee pollen [1,2,3,4,5]. Bee secretions contain various enzymes such as amylase, invertase, and glucosidase, which assist in binding and compacting the pollen [4]. During this transformation, bee pollen may exhibit morphological and chemical characteristics distinct from the original floral pollen [6]. Bee pollen is used as a raw material for pharmaceuticals and cosmetics, and mainly consumed as a health supplement [1,7,8].
Previous studies have identified essential oils, phenolic compounds, polyamines, and their metabolites in bee pollen [7,9,10]. Among these, polyphenols, particularly flavonoids, contribute significantly to the potent antioxidant activity of bee pollen [1,8]. Flavonoids have attracted considerable attention for their diverse bioactivities, including antioxidant, anti-inflammatory, antibacterial, and anticancer effects [11,12]. Also, our previous research has demonstrated that flavonoids derived from Quercus mongolica pollen exhibit significant antioxidant and tyrosinase-inhibitory activities, underscoring their potential application in health-promoting products [13]. Given this context, understanding the flavonoid composition of bee pollen can aid in the identification of novel bioactive compounds beneficial for pharmaceutical and nutraceutical applications.
Flavonoids are recognized as the major chemical constituents of plant pollen, predominantly occurring as flavonoid glycosides [7,8,14,15]. These flavonoid glycosides exhibit remarkable structural complexity and diversity, attributed to differences in glycosylation sites, linkage types, and sugar moieties. Glycosylation improves the chemical stability of flavonoids and increases the aqueous solubility of the aglycone backbone, thereby improving their bioavailability [15,16,17,18,19]. In this study, we primarily focused on bee pollen-derived flavonoid glycosides.
Quercus mongolica, a deciduous hardwood tree in the Fagaceae family, occurs broadly throughout Republic of Korea and accounts for approximately 3.7% of the country’s forested area [1,20,21,22,23]. In a previous study, 18 flavonoids were isolated and identified from Q. mongolica pollen, 11 of which were flavonoid glycosides. Quercetin, kaempferol, and isorhamnetin were predominantly present as their glycosides in Q. mongolica pollen, and these compounds exhibited potent antioxidant activity [13]. Numerous reports have focused on flavonoids and their glycosides in bee pollen. However, various flavonoid glycosides that protect pollen from external environmental factors have not been identified yet. The discovery of these compounds is significant, but research on flavonoid glycosides in bee pollen remains underdeveloped.
Molecular networking using the Global Natural Products Social Molecular Networking (GNPS) platform has become a powerful approach for untargeted metabolomics in natural product studies. This technique clusters compounds based on similar MS/MS fragmentation patterns, enabling visualization of their molecular relationships as networks, and facilitating efficient metabolite annotation through spectral libraries [24,25,26].
Research on the flavonoid chemical profiling of Q. mongolica pollen is limited. Moreover, the chemical composition of bee pollen can vary depending on geographic location and cultivation practices [7,8,27,28]. Therefore, this study aimed to conduct a preliminary chemical profiling of Q. mongolica pollen using molecular networking. This approach provides valuable insights into the flavonoid glycoside diversity and potential bioactivity, offering a valuable foundation for subsequent research and applications.
2. Results
2.1. Flavonoid Glycoside Profiling of Q. mongolica Pollen Using Molecular Networking Based on UPLC–QTOF–MS/MS Analysis
GNPS based on UPLC–QTOF–MS/MS. The MS/MS data were acquired in negative ion mode, which provides high selectivity and sensitivity for the LC–MS analysis of flavonoids in plant resources [29]. Figure 1 shows the base peak ion chromatogram (BPI) of Q. mongolica pollen. According to LC–MS/MS analysis, peaks eluting between 3.5 and 6.0 min were identified as flavonoid glycosides, whereas those between 6.0 and 7.8 min were attributed to polyamine compounds. Three major flavonoid glycosides were shown on the base peak ion chromatogram (BPI) of Q. mongolica pollen, in which three primary flavonoid glycosides were prominently observed (A1–A3).
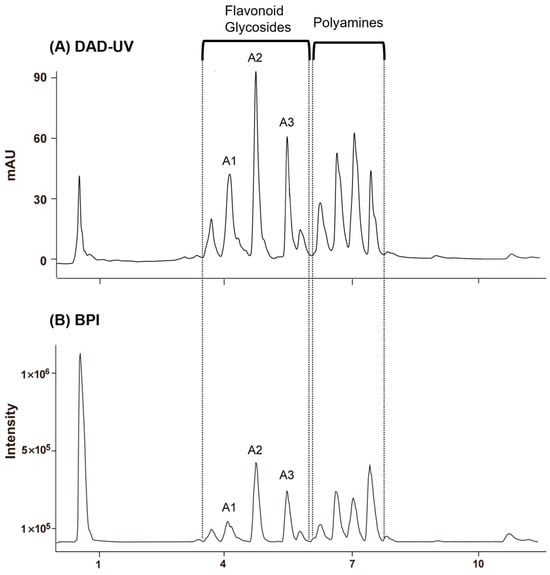
Figure 1.
(A) DAD–UV chromatograms of Quercus mongolica pollen at 254 nm. (B) Base peak ion chromatogram (BPI) with reference compounds A1–A3. Peaks observed at 3.48–5.96 min were identified as flavonoid glycosides, while those at 6.01–7.68 min corresponded to polyamines.
In this study, Q. mongolica pollen metabolites were profiled using molecular networking, a technique that facilitates the discovery of untargeted metabolites based on LC–MS/MS data. While the BPI data contained a vast amount of information that is difficult to interpret manually, molecular networking provided an intuitive visualization, enabling the identification of target compounds and their related derivative compounds. In the case of flavonoid glycosides, the MS/MS fragmentation patterns revealed distinct signals corresponding to the aglycone part (flavonoid) and the sugar moiety. By analyzing these patterns, it was confirmed that the structures could be categorized into aglycone backbones and sugar components (monosaccharides and disaccharides). Furthermore, the relative intensity of ionized peaks was used to predict the substitution positions of sugar residues, thereby assisting in the structural identification of the compounds. To annotate the flavonoid glycosides, molecular networking analysis was performed based on the UPLC–QTOF–MS/MS data using the GNPS platform. Two major clusters were observed in the MS/MS spectral network of Q. mongolica pollen (Figure 2). Based on the spectral library matches in the GNPS, Cluster A was identified as a flavonoid glycoside, whereas Cluster B was classified as a polyamine. In total, 69 flavonoid glycosides were grouped into Cluster A, with a predominance of flavonoid O-glycosides. Among the annotated compounds, kaempferol, quercetin, and isorhamnetin were identified as major aglycone backbones. Figure 3 shows that two primary compounds, isorhamnetin 3-O-β-D-xylopyranosyl (1→6)-O-β-D-glucopyranoside and isorhamnetin 3-O-neohesperidoside, were confirmed using 1H and 13C NMR and LC–MS spectroscopic data (Table S1 and Figures S2–S13). The chemical composition of bee pollen varies depending on geographic origin, collection period, and cultivation conditions, factors which are also known to influence its biological activities. Therefore, to utilize the natural resource of bee pollen, standardization studies are required. Two primary compounds, isorhamnetin 3-O-β-D-xylopyranosyl (1→6)-O-β-D-glucopyranoside and isorhamnetin 3-O-neohesperidoside, can be used as standard compounds for future standardization research. These compounds were selected because they are among the major constituents of Q. mongolica pollen, and their chromatographic signals were well separated from other metabolite peaks, making them suitable for accurate qualitative and quantitative analysis.
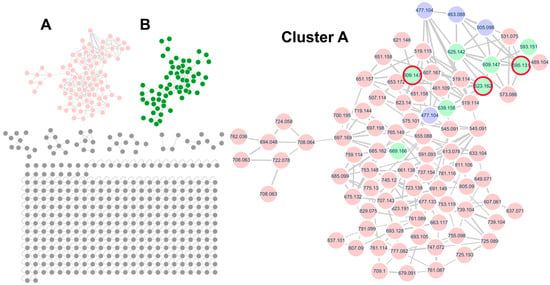
Figure 2.
The MS/MS spectral molecular network of Quercus mongolica pollen in negative ion mode and the molecular family of flavonoid glycosides showing the molecular families of Cluster (A) (flavonoid glycosides, pink), Cluster (B) (polyamines, green). In Cluster (A), purple nodes, green nodes, and red circles represent flavonoid monoglycosides matched with the GNPS spectral library, flavonoid diglycosides, and reference compounds identified in Q. mongolica pollen, respectively.
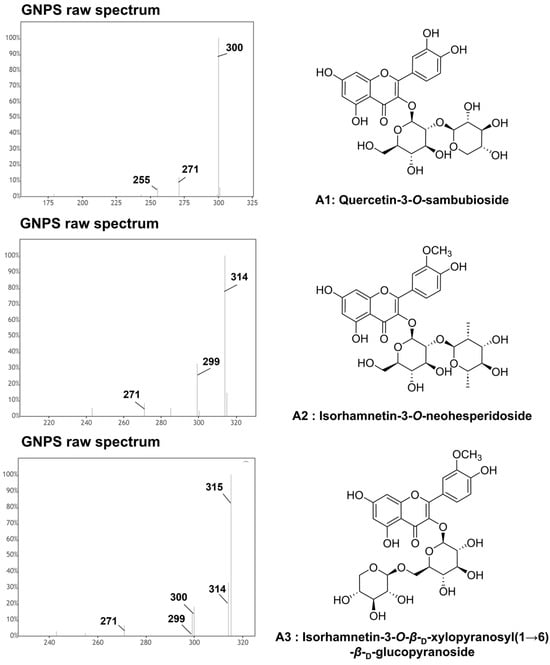
Figure 3.
Raw MS/MS spectra of primary compounds (A1, A2, and A3) from Quercus mongolica pollen obtained from GNPS. Quercetin 3-O-sambubioside (A1, compound 6; RT = 4.17 min; [M−H]− m/z 595.132), isorhamnetin 3-O-neohesperidoside (A2, compound 30; RT = 4.75 min; [M−H]− m/z 623.165), isorhamnetin 3-O-β-D-xylopyranosyl(1→6)-β-D-glucopyranoside (A3, compound 28; RT = 5.47 min; [M−H]− m/z 609.165).
Research on Flavonoid glycosides exhibits characteristic fragmentation patterns involving both aglycone and sugar moieties. The sugar moieties were primarily hexose (−162 Da), pentose (−132 Da), and deoxyhexose (−146 Da). In some cases, a neutral loss of hexose along with a water molecule (−180 Da) was observed. Additionally, some glycosides were acetylated, exhibiting fragment losses corresponding to an acetyl group (−42 Da), acetic acid (−60 Da), or acetylated hexose (−204 Da) [24,30]. All compounds were tentatively annotated based on spectral matching with the GNPS library, comparison with reference compounds, and results of previous studies [30,31].
To determine the glycosylation sites of flavonoid glycosides, we compared the relative intensities of deprotonated and radical aglycone ions. In flavonoid O-glycosides, these fragment ions are typically generated during glycoside fragmentation under negative ion mode [31]. However, the abundance ratio between these ions can vary depending on the substitution pattern on the flavonoid backbone, glycosylation site, and sugar moiety type. Glycosylation at the 3-OH position tends to increase the abundance of radical aglycone ions, whereas glycosylation at the 7-OH position increases the abundance of deprotonated aglycone ions [31,32,33]. Based on these characteristic fragmentation patterns, the glycosylation positions of the annotated flavonoid glycosides were tentatively identified.
2.2. Identification of Kaempferol Derivatives
The annotated compounds are listed in Table 1. Compounds 1 and 2 were identified as kaempferol glucosides based on their characteristic product ions at m/z 285, 255, and 227 [29,32].
Compound 1 (RT 6.93 min) exhibited a deprotonated molecular ion ([M−H]−) at m/z 489.104. In the MS/MS spectrum, a fragment ion at m/z 285 indicated the neutral loss of acetylhexoside moiety (−204 Da) [34]. The appearance of a fragment ion at m/z 429 suggested the loss of an acetyl group (−60 Da). The glycosylation site was tentatively assigned to the 3-OH position based on the higher intensity of [M−H−204]•− than that of [M−H−204]−. Consequently, compound 1 was putatively characterized as kaempferol 3-O-acetylglucoside [24].
Compound 2 (RT 4.69 min) presented a [M−H]− at m/z 593.151 and generated a fragment ion at m/z 285, indicating the loss of deoxyhexose and pentose moieties (−308 Da). The 3-OH position was proposed as the glycosylation site, inferred from the relatively high intensity of the [M−H−308]•− ion. Accordingly, compound 2 was putatively annotated as kaempferol 3-O-rutinoside based on GNPS library matches and comparison with the results of previous studies [24,35].

Table 1.
Putative identification of flavonoid glycosides in the flavonoid glycoside cluster of Quercus mongolica pollen extracts analyzed by UPLC–QTOF–MS in negative ion mode.
Table 1.
Putative identification of flavonoid glycosides in the flavonoid glycoside cluster of Quercus mongolica pollen extracts analyzed by UPLC–QTOF–MS in negative ion mode.
| No. | RT (min) | [M−H]− (m/z) | Molecular Formula (Error in ppm) | Tentatively Identification | MS2 (m/z) | Ref |
|---|---|---|---|---|---|---|
| Kaempferol derivatives | ||||||
| 1 | 6.92 | 489.104 | C23H22O12 (−0.1) | Kaempferol-3-O-acetylglucoside | 429, 309, 285, 284, 255, 227 | [24] |
| 2 | 4.69 | 593.151 | C27H30O15 (−0.2) | Kaempferol-3-O-rutinoside a | 285, 284, 255, 227 | [24,35] |
| Quercetin derivatives | ||||||
| 3 | 4.95 | 463.088 | C21H20O12 (0.0) | Quercetin-3-O-glucoside a | 301, 300, 271, 255, 243 | [34,35] |
| 4 | 6.03 | 505.098 | C23H22O13 (−0.3) | Quercetin-3-O-acetylglucoside a | 301, 300, 271, 255, 243 | [24] |
| 5 | 6.27 | 573.086 | C33H18O10 (4.9) | Quercetin derivatives | 505, 301, 300, 271, 255 | |
| 6 | 4.17 | 595.132 | C26H28O16 (1.2) | Quercetin-3-O-sambubioside a,b | 463, 445, 301, 300, 271, 255 | [36] |
| 7 | 4.13 | 609.147 | C27H30O16 (0.8) | Quercetin-3-O-rutinoside a | 463, 445, 301, 300, 271, 255 | [37] |
| 8 | 3.70 | 625.142 | C27H30O17 (1.1) | Quercetin-3-O-sophoroside a | 463, 445, 301, 300, 271, 255 | [36,37] |
| 9 | 4.17 | 663.117 | C29H28O18 (−4.1) | Quercetin derivatives | 595, 323, 301, 300 | |
| 10 | 4.19 | 679.091 | C32H24O17 (−5.1) | Quercetin derivatives | 611, 595, 301, 300, 271 | |
| 11 | 3.72 | 693.128 | C37H26O14 (4.2) | Quercetin derivatives | 647, 625, 323, 301, 300 | |
| 12 | 3.72 | 709.100 | C33H26O18 (−6.3) | Quercetin derivatives | 625, 399, 384, 301, 300 | |
| 13 | 4.27 | 725.193 | C29H26O22 (5.9) | Quercetin derivatives | 679, 633, 384, 301, 300, 284 | |
| 14 | 4.15 | 747.072 | C24H28O27 (−3.4) | Quercetin derivatives | 701, 655, 595, 523, 301, 300 | |
| 15 | 3.74 | 761.114 | C33H30O21 (−9.3) | Quercetin derivatives | 715, 647, 625, 323, 301, 300 | |
| 16 | 3.72 | 777.082 | C32H26O23 (3.6) | Quercetin derivatives | 731, 685, 625, 523, 301, 300 | |
| Isorhamnetin derivatives | ||||||
| 17 | 6.46 | 461.109 | C22H22O11 (−0.3) | Isorhamnetin-3-O-rhamnoside | 315, 314, 300, 299, 285, 271, 257, 243 | [24] |
| 18 | 5.79 | 477.104 | C22H22O12 (1.2) | Isorhamnetin-3-O-glucoside isomer a | 315, 314, 299, 285, 271, 257, 243 | [38] |
| 19 | 5.47 | 477.104 | C22H22O12 (1.4) | Isorhamnetin-3-O-glucoside isomer a | 315, 314, 300, 299, 271, 255 | [38] |
| 20 | 6.53 | 519.114 | C24H24O13 (−0.2) | Isorhamnetin-3-O-acetylglucoside isomer | 315, 314, 299, 285, 271, 257, 243 | |
| 21 | 6.79 | 519.114 | C24H24O13 (0) | Isorhamnetin-3-O-acetylglucoside isomer | 315, 314, 299, 285, 271, 257, 243 | |
| 22 | 7.11 | 519.114 | C24H24O13 (0.7) | Isorhamnetin-3-O-acetylglucoside isomer | 459, 315, 314, 300, 299, 285, 271 | |
| 23 | 5.45 | 545.091 | C25H22O14 (−3.3) | Isorhamnetin derivatives | 477, 315, 314, 300, 299, 271 | |
| 24 | 5.77 | 545.091 | C32H18O9 (5.3) | Isorhamnetin derivatives | 477, 315, 314, 299, 285, 271, | |
| 25 | 5.77 | 591.093 | C33H20O11 (−0.3) | Isorhamnetin derivatives | 477, 383, 315, 314, 285, 271 | |
| 26 | 6.58 | 607.167 | C28H32O15 (0.8) | Isorhamnetin derivatives | 315, 314, 299, 285, 271 | |
| 27 | 5.79 | 607.061 | C25H20O18 (5.8) | Isorhamnetin derivatives | 399, 383, 371, 315, 314, 299 | |
| 28 | 5.47 | 609.148 | C27H30O16 (1.5) | Isorhamnetin-3-O-β-D-xylopyranosyl(1→6)-β-D-glucopyranoside a,b | 315, 314, 300, 299, 271 | |
| 29 | 7.26 | 623.140 | C31H28O14 (0.1) | Isorhamnetin derivatives | 477, 315, 314, 300, 299, 271 | |
| 30 | 4.75 | 623.162 | C28H32O16 (1.2) | Isorhamnetin-3-O-neohesperidoside a,b | 315, 314, 300, 299, 285, 271 | [24,38] |
| 31 | 7.09 | 633.104 | C35H22O12 (0.4) | Isorhamnetin derivatives | 519, 383, 315, 314, 300, 299, 112 | |
| 32 | 4.08 | 639.158 | C28H32O17 (−0.6) | Isorhamnetin-3-O-sophoroside a | 459, 315, 314, 300, 299, 271 | [39] |
| 33 | 7.02 | 651.157 | C29H32O17 (0.5) | Isorhamnetin derivatives | 591, 315, 314, 299, 285, 243 | |
| 34 | 6.60 | 651.158 | C29H32O17 (0.5) | Isorhamnetin derivatives | 519, 383, 315, 314, 300, 299 | |
| 35 | 7.13 | 655.088 | C30H24O17 (−8.4) | Isorhamnetin derivatives | 519, 459, 315, 314, 300, 299 | |
| 36 | 4.73 | 669.166 | C29H34O18 (−1.1) | Isorhamnetin derivatives | 623, 315, 314, 299 | |
| 37 | 4.12 | 675.132 | C34H28O15 (−5.2) | Isorhamnetin derivatives | 639, 321, 315, 314, 300, 299 | |
| 38 | 5.47 | 677.133 | C30H30O18 (−5.2) | Isorhamnetin derivatives | 609, 315, 314, 300, 299 | |
| 39 | 4.15 | 685.099 | C29H34O19 (−1.0) | Isorhamnetin derivatives | 639, 315, 314, 300, 299 | |
| 40 | 4.08 | 685.162 | C29H34O19 (−1.2) | Isorhamnetin derivatives | 639, 315, 314, 300, 299 | |
| 41 | 4.73 | 691.149 | C31H32O18 (−4.2) | Isorhamnetin derivatives | 623, 337, 315, 314 | |
| 42 | 5.47 | 693.105 | C33H26O17 (−6.5) | Isorhamnetin derivatives | 647, 609, 413, 315, 314 | |
| 43 | 7.07 | 697.169 | C23H26O17 (−0.5) | Isorhamnetin derivatives | 651, 605, 591, 315, 314, 300, 299 | |
| 44 | 7.15 | 697.198 | C31H38O18 (−3.2) | Isorhamnetin derivatives | 651, 605, 591, 315, 314, 299 | |
| 45 | 4.10 | 707.143 | C38H28O17 (−3.2) | Isorhamnetin derivatives | 639, 609, 315, 314, 300, 299 | |
| 46 | 7.00 | 719.144 | C32H32O19 (−3.6) | Isorhamnetin derivatives | 651, 591, 315, 314 | |
| 47 | 5.47 | 723.138 | C38H28O15 (3.9) | Isorhamnetin derivatives | 631, 609, 315, 314 | |
| 48 | 4.73 | 737.154 | C39H30O15 (3.8) | Isorhamnetin derivatives | 691, 645, 623, 315, 314 | |
| 49 | 4.92 | 739.104 | C23H32O27 (−3.5) | Isorhamnetin derivatives | 693, 609, 399, 399, 315, 314 | |
| 50 | 5.47 | 739.104 | C23H32O27 (−2.5) | Isorhamnetin derivatives | 693, 647, 609, 479, 413, 315 | |
| 51 | 5.47 | 745.120 | C33H30O20 (−8.1) | Isorhamnetin derivatives | 653, 609, 315, 314 | |
| 52 | 4.8 | 753.119 | C24H34O27 (−2.9) | Isorhamnetin derivatives | 707, 661, 623, 399, 315, 314 | |
| 53 | 4.1 | 753.148 | C39H30O16 (2.6) | Isorhamnetin derivatives | 707, 639, 315, 314, 300, 299 | |
| 54 | 5.49 | 761.089 | C25H30O27 (−2.1) | Isorhamnetin derivatives | 669, 663, 609, 435, 315, 314 | |
| 55 | 7.05 | 765.149 | C33H34O21 (−4.2) | Isorhamnetin derivatives | 701, 673, 651, 315, 314, 112 | |
| 56 | 4.08 | 775.130 | C34H32O21 (−8.4) | Isorhamnetin derivatives | 639, 609, 315, 314, 300, 299 | |
| 57 | 7.00 | 781.116 | C25H34O28 (−1.0) | Isorhamnetin derivatives | 689, 651, 629, 413, 315, 314 | |
| 58 | 4.12 | 791.099 | C26H32O28 (−2.4) | Isorhamnetin derivatives | 699, 639, 609, 315, 314, 301, 300, 299 | |
| 59 | 5.49 | 807.090 | C33H28O24 (−0.3) | Isorhamnetin derivatives | 715, 669, 609, 435, 315, 314 | |
| 60 | 4.75 | 811.106 | C29H32O27 (−0.1) | Isorhamnetin derivatives | 765, 623, 456, 315, 314 | |
| 61 | 4.34 | 823.191 | C36H40O22 (−3.7) | Isorhamnetin derivatives | 755, 315, 314 | |
| 62 | 4.08 | 837.101 | C34H30O25 (0.9) | Isorhamnetin derivatives | 723, 699, 639, 399, 315, 314 | |
| Quercetagetin-dimethyl derivatives | ||||||
| 63 | 5.64 | 507.114 | C23H24O13 (0.2) | Quercetagetin-dimethyl 3-O-hexoside | 345, 344, 330, 329, 314, 301, 286 | [40] |
| 64 | 5.65 | 637.071 | C26H22O19 (5.1) | Quercetagetin-dimethyl derivatives | 429, 413, 386, 345, 329, 300 | |
| 65 | 4.50 | 653.172 | C29H34O17 (−0.1) | Quercetagetin-dimethyl derivatives | 345, 344, 330, 329, 301 | |
| Others | ||||||
| 66 | 5.62 | 575.101 | C26H24O15 (−4.8) | unknown | 507, 492, 344, 329, 301 | |
| 67 | 3.34 | 621.146 | C28H30O16 (−0.2) | unknown | 327, 326, 312, 311, 284, 283 | |
| 68 | 4.15 | 725.089 | C29H26O22 (5.9) | unknown | 679, 633, 595, 399, 384, 301 | |
| 69 | 3.74 | 755.098 | C30H28O23 (4.7) | unknown | 709, 663, 625, 399, 384, 301 | |
a Compounds were tentatively identified based on GNPS spectral library matches; b Identified by comparison with reference standards. The most abundant fragment ions are shown in bold.
2.3. Identification of Quercetin Derivatives
Characteristic fragment ions of quercetin were observed at m/z 301, 271, and 255 [29,32] Compound 3 (RT 4.95 min) exhibited a [M−H]− at m/z 463.088 and produced a fragment ion at m/z 301, indicating the loss of a hexose unit (−162 Da). The predominance of the [M−H−162]•− ion suggests glycosylation of 3-OH. Consequently, compound 3 was putatively characterized as quercetin 3-O-glucoside, in agreement with the GNPS library matches and results of previous reports [34,35]. Compound 4 (RT 6.03 min) showed a [M−H]− at m/z 505.098 and generated a fragment ion at m/z 301, indicating the loss of an acetylhexoside moiety (−204 Da). The 3-OH position was also proposed as a glycosylation site based on the relatively high intensity of the [M−H−204]•− ion. Consequently, compound 4 was putatively characterized as quercetin 3-O-acetylglucoside, supported by the GNPS library matches and results of previous studies [24]. Compound 6 (RT 4.17 min), identified as primary compound A1, exhibited a [M−H]− at m/z 595.131. As shown in the MS/MS spectrum in Figure S1, a fragment ion at m/z 301 indicated the loss of hexose and pentose moieties (−294 Da), and an additional fragment at m/z 463 corresponded to the loss of a pentose unit (−132 Da). The predominance of the [M−H−294]•− ion suggested glycosylation at the 3-OH position. Consequently, compound 6 was putatively annotated as quercetin 3-O-sambubioside [36]. Compound 7 (RT 4.13 min) showed a [M−H]− at m/z 609.147 and produced a fragment ion at m/z 301, consistent with the loss of deoxyhexose and pentose moieties (−308 Da). Additional fragment ions at m/z 463 and 445 were observed, indicating the loss of a deoxyhexose moiety (−146 Da) and a hexose moiety (−162 Da), respectively. The 3-OH position was tentatively suggested to be the glycosylation site, supported by the predominance of [M−H−308]•−. Consequently, compound 7 was putatively annotated as quercetin 3-O-rutinoside based on GNPS library matches and comparison with the results of previous studies [38]. Compound 8 (RT 3.70 min) exhibited a [M−H]− at m/z 625.142 and generated a fragment ion at m/z 301, indicating the loss of dihexose moieties (−324 Da). A fragment ion at m/z 465, indicating the loss of a single hexose (−162 Da). The glycosylation site was tentatively determined to be at the 3-OH position, based on the higher intensity of the [M−H−324]•− ion. Consequently, compound 8 was putatively characterized as quercetin 3-O-sophoroside, supported by GNPS library matches and results of previous reports [36,37]. Compounds 9–16 were provided as quercetin glycoside derivatives, and it is anticipated that these compounds can be identified through isolation and purification based on the preliminary information presented.
2.4. Identification of Isorhamnetin Derivatives
The characteristic fragment ions of isorhamnetin were observed at m/z 300, 271, 255, and 227 [32] Compound 17 (RT 6.46 min) exhibited a [M−H]− at m/z 461.109 and generated a fragment ion at m/z 315, indicating the loss of deoxyhexose (−146 Da). The glycosylation site was tentatively determined to be at the 3-OH position based on the higher intensity of the [M−H−146]•− ion. Consequently, compound 17 was putatively characterized as isorhamnetin 3-O-rhamnoside based on comparison of the obtained MS/MS spectra with those reported in previous studies [24]. Compounds 18 (RT 5.79 min) and 19 (RT 5.47 min) exhibited identical [M−H]− ions at m/z 477.104 and generated a fragment ion at m/z 315, indicating the loss of hexose (−162 Da). The glycosylation site was tentatively determined to be at the 3-OH position, based on the higher intensity of the [M−H−162]•− ion. Accordingly, compounds 18 and 19 were putatively characterized as isorhamnetin 3-O-glucoside isomer supported by the GNPS library matches and previous reports [38]. Compounds 20 (RT 6.53 min), 21 (RT 6.79 min), and 22 (RT 7.11 min) displayed a [M−H]− at m/z 519.114 and generated a fragment ion at m/z 315, indicating the loss of acetyl hexose [M−H−204]−. A fragment ion at m/z 459 corresponded to the loss of an acetyl group (−60 Da). The glycosylation site was tentatively determined to be at the 3-OH position, based on the higher intensity of the [M−H−204]•− ion. Accordingly, compounds 20, 21, and 22 were putatively characterized as isorhamnetin 3-O-acetyl glucoside isomers. Compound 28 (RT 5.47 min), identified as primary compound A3, presented a [M−H]− at m/z 609.148, and produced a fragment ion at m/z 315, indicating the loss of hexose and pentose moieties (−294 Da). As previously described, the tentative assignment of glycosylation positions can be inferred from a relative intensity comparison between the radical aglycone ions and regular deprotonated aglycone ions. However, these intensities may vary depending on the flavonoid backbone, glycosylation position, and the type of sugar moiety. For compound 28, although the intensity of [M−H−294]− was higher than that of [M−H−294]•−, it was ultimately confirmed as a 3-O-glycoside by NMR spectroscopy. This compound was confirmed by comparing its 1D (1H and 13C) and 2D (COSY, HSQC, HMBC) NMR spectrum with that of a reference standard (Table S1 and Figures S2–S7). Accordingly, compound 28 was identified as isorhamnetin 3-O-β-D-xylopyranosyl(1→6)-β-D-glucopyranoside. Compound 30 (RT 4.75 min), identified as the primary compound A2, exhibited a [M−H]− at m/z 623.162 and produced a fragment ion at m/z 315, indicating the loss of deoxyhexose and hexose moieties (−308 Da). Based on GNPS spectral library matching, this compound was initially annotated as isorhamnetin 3-O-robinoside. However, a comparison of the 1D (1H and 13C) and 2D (COSY, HSQC, HMBC) NMR spectra with those of a reference compound confirmed that the compound was isorhamnetin 3-O-neohesperidoside (Table S1 and Figures S8–S13). These findings suggest that although GNPS spectral library matching is a useful tool for putative annotation, definitive structural elucidation requires complementary analytical techniques with both NMR and LC–MS/MS. Accordingly, 30 was identified as isorhamnetin 3-O-neohesperidoside [24,38]. Compound 32 (RT 4.08 min) presented a [M−H]− at m/z 639.158 and generated a fragment ion at m/z 315, indicating the loss of dihexose moieties (−324 Da). A fragment ion at m/z 477, indicating the loss of a single hexose (−162 Da). The glycosylation site was tentatively determined to be at the 3-OH position, based on the higher intensity of the [M−H−324]•− ion. Consequently, compound 32 was putatively characterized as isorhamnetin 3-O-sophoroside, as supported by the GNPS library matches and previous reports [39]. Compounds 23–27, 29, 31, and 33–62 were provided as isorhamnetin glycoside derivatives, and it is anticipated that these compounds can be identified through isolation and purification based on the preliminary information presented.
2.5. Identification of Additional Flavonoid Glycosides
Quercetagetin-dimethyl is a relatively uncommon aglycone characterized by fragment ions at m/z 345, 330, and 315. Compounds 63, 64, and 65 were putatively characterized as quercetagetin-methyl derivatives, including m/z 345, 330, and 315 [40]. Compound 63 (RT 5.64 min) showed a [M−H]− at m/z 507.114 and produced a fragment ion at m/z 345, indicating the loss of hexose (−162 Da). The glycosylation site was tentatively determined to be at the 3-OH position, based on the higher intensity of the [M−H−162]•− ion. Consequently, compound 63 was putatively characterized as quercetagetin-dimethyl 3-O-hexoside by comparing its MS/MS spectrum with those reported in previous studies [40]. Compounds 64 and 65 were putatively characterized as quercetagetin-methyl derivatives based on their mass fragmentation patterns.
3. Discussion
Research on Q. mongolica pollen has been limited to data, particularly regarding its chemical composition. In this study, we performed untargeted chemical profiling using LC–MS/MS-based molecular networking to explore the metabolite diversity of Q. mongolica pollen, with a focus on flavonoid derivatives.
Flavonoids are well-known for their diverse biological activities. A previous study reported that 18 flavonoid metabolites isolated from Q. mongolica pollen exhibited strong antioxidant and tyrosinase inhibitory activities, supporting the functional potential of this material. In the present work, we tentatively annotated 69 flavonoid glycosides using GNPS-based molecular networking and spectral library matching. Based on their characteristic fragmentation patterns, kaempferol, quercetin, and isorhamnetin were identified as major aglycone backbones. Q. mongolica pollen was found to be particularly rich in isorhamnetin derivatives. The structural diversity of flavonoids observed highlights the importance of comprehensive metabolite profiling. Moreover, the presence of numerous previously unreported flavonoid glycosides suggests their potential as promising sources of novel bioactive compounds.
We anticipate that this study will lay the groundwork for future investigations into the bioactive metabolites of Q. mongolica pollen. Given that the chemical composition of Q. mongolica pollen can vary considerably depending on environmental factors and collection timing, the results of this study provide a crucial foundation for standardization studies. Standardization is essential to ensure consistent quality, reproducibility, and bioactivity in future applications, especially for pharmaceutical or functional food development. Thus, future research should aim to conduct standardization studies.
4. Materials and Methods
4.1. Reagents and Materials
HPLC-grade MeOH, H2O, and formic acid were obtained from Daejung Chemicals (Siheung, Republic of Korea). Two reference compounds, isorhamnetin 3-O-β-D-xylopyranosyl(1→6)-β-D-glucopyranoside and isorhamnetin 3-O-neohesperidoside, were isolated and structurally elucidated by 1D and 2D NMR spectroscopy (Figures S3–S7 and S9–S13).
4.2. Plant Materials and Extract Preparation
Quercus mongolica pollen collected from Yangyang, Gangwon-do, Republic of Korea, was purchased online (https://e-honey.net/goods/goods_view.php?goodsNo=127, accessed on 11 July 2023). The collected pollen was ground into a fine powder and 100 mg powder was extracted with 4 mL 80% MeOH at room temperature for 48 h. Following extraction, the solution was filtered and then concentrated under reduced pressure at 40 °C. An aliquot (5 mg) of the dried extract was dissolved in 5 mL of methanol and filtered through a 0.20 µm hydrophilic PTFE membrane filter (Advantec, Tokyo, Japan) prior to LC–MS/MS analysis.
4.3. UPLC–QTOF–MS/MS Analysis
The LC–MS/MS analysis was performed using an Agilent 1290 Infinity II UPLC system (Agilent Technologies, Santa Clara, CA, USA) coupled with a SCIEX ZenoTOF 7600 mass spectrometer (SCIEX, Framingham, MA, USA). Chromatographic separation was conducted on a Phenomenex Kinetex XB-C18 column (1.7 μm, 50 × 2.1 mm; Phenomenex, Torrance, CA, USA). The mobile phase comprised H2O (solvent A) and MeOH (solvent B) containing 0.1% formic acid. The procedure for gradient elution was as follows: 23–53% B (0–6 min), isocratic mode at 53% B (6–10 min), 53–100% (10–12 min), followed by washing with 100% B for 5 min and reconditioning with 23% B for 3 min. The flow rate was set to 0.3 mL/min, and the injection volume was 3 μL. UV detection was performed at 254 nm. MS data were collected in negative ionization mode using a TOF/MS–IDA–TOF MS/MS acquisition. The TOF/MS and TOF MS/MS ranges were set to 100−2000 and 80−2000 Da, respectively. The ion source parameters were as follows: curtain gas 35 psi; CAD gas 7; ion source gas 50 psi; source temperature 500 °C; spray voltage −4500 V. In the CID mode, 45 eV collision energy was applied. The chemical formula of each compound was predicted using the Formula Finder function in SCIEX OS software.
4.4. Data Processing
The raw LC–MS/MS data were acquired using SCIEX OS 3.3.1 software and subsequently converted to mzML files using the ProteoWizard MSconvert software version 3.0.24164. Data processing was carried out in MZmine 3.9.0, following the steps: peak detection, ADAP chromatogram builder, local minimum feature resolver, 13C isotope filter, and feature list row filter. The processed MS/MS spectra were exported as MGF files and uploaded to GNPS (http://gnps.ucsd.edu, accessed on 26 March 2025). A spectral similarity network was built using the GNPS. The GNPS parameters were as follows: precursor ion mass tolerance, 0.02 Da; MS/MS fragment ion tolerance, 0.02 Da. The edges were filtered to obtain a cosine score above 0.6 and a minimum of five matched peaks, and the spectral network was visualized using Cytoscape 3.10.2. The MS/MS molecular network was downloaded from https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=cf83418bc24746bcb152ee2d4d66b6dd (accessed on 26 March 2025).
5. Conclusions
This study provided the first untargeted metabolite profiling of Q. mongolica pollen using LC–MS/MS-based molecular networking. A total of 69 flavonoid glycosides were tentatively annotated, highlighting the chemical diversity and bioactive potential of this material. These findings lay the groundwork for future investigations into the bioactive metabolites of Q. mongolica pollen and standardization studies.
The flavonoid composition and abundance in bee pollen are known to vary significantly depending on environmental conditions, including geographical origin, cultivation practices, and the timing of pollen collection [7,8,27,28]. In this study, the pollen analyzed was collected from Yangyang, Gangwon-do, Republic of Korea, an area characterized by distinct climatic conditions that could influence secondary metabolite biosynthesis pathways, ultimately affecting flavonoid diversity and abundance. Future research incorporating samples collected from different regions or seasons would provide deeper insights into the environmental influences on flavonoid profiles and help guide standardization efforts.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms26167930/s1.
Author Contributions
Conceptualization, S.B.K. and M.K.L.; methodology, Y.J. and S.B.K.; software, Y.J. and H.K.; validation, Y.J.; formal analysis, Y.J.; data curation, E.S.; writing—original draft preparation, Y.J.; writing—review and editing, S.B.K., H.K. and M.K.L.; visualization, Y.J.; supervision, S.B.K.; project administration, S.B.K.; funding acquisition, S.B.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a 2-Year Research Grant of Pusan National University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw NMR data for natural products were deposited in the Harvard Dataverse (https://dataverse.harvard.edu, accessed on 12 August 2025) and are accessible at DOI: [https://doi.org/10.7910/DVN/MYK0KV].
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kwak, J.-E.; Lee, J.-Y.; Baek, J.-Y.; Kim, S.W.; Ahn, M.-R. The Antioxidant and Anti-Inflammatory Properties of Bee Pollen from Acorn (Quercus acutissima Carr.) and Darae (Actinidia arguta). Antioxidants 2024, 13, 981. [Google Scholar] [CrossRef]
- Qiao, J.; Feng, Z.; Zhang, Y.; Xiao, X.; Dong, J.; Haubruge, E.; Zhang, H. Phenolamide and Flavonoid Glycoside Profiles of 20 Types of Monofloral Bee Pollen. Food Chem. 2023, 405, 134800. [Google Scholar] [CrossRef]
- El Ghouizi, A.; Bakour, M.; Laaroussi, H.; Ousaaid, D.; El Menyiy, N.; Hano, C.; Lyoussi, B. Bee Pollen as Functional Food: Insights into Its Composition and Therapeutic Properties. Antioxidants 2023, 12, 557. [Google Scholar] [CrossRef] [PubMed]
- Miłek, M.; Mołoń, M.; Kula-Maximenko, M.; Sidor, E.; Zaguła, G.; Dżugan, M. Chemical Composition and Bioactivity of Laboratory-Fermented Bee Pollen in Comparison with Natural Bee Bread. Biomolecules 2023, 13, 1025. [Google Scholar] [CrossRef]
- Bakour, M.; Laaroussi, H.; Ousaaid, D.; El Ghouizi, A.; Es-Safi, I.; Mechchate, H.; Lyoussi, B. Bee Bread as a Promising Source of Bioactive Molecules and Functional Properties: An up-to-Date Review. Antibiotics 2022, 11, 203. [Google Scholar] [CrossRef]
- Sen, N.B.; Vovk, I.; Kırmızıbekmez, H.; Guzelmeric, E. Phytochemical and Bioactivity Evaluation of Bee Pollen and Androecia of Castanea, Salix, and Quercus Species. Antioxidants 2024, 14, 40. [Google Scholar] [CrossRef] [PubMed]
- Rzepecka-Stojko, A.; Stojko, J.; Kurek-Górecka, A.; Górecki, M.; Kabała-Dzik, A.; Kubina, R.; Moździerz, A.; Buszman, E. Polyphenols from Bee Pollen: Structure, Absorption, Metabolism and Biological Activity. Molecules 2015, 20, 21732–21749, Erratum in: Molecules 2016, 21, 159. https://doi.org/10.3390/molecules21020159. [Google Scholar] [CrossRef] [PubMed]
- Miyata, R.; Hoshino, S.; Ahn, M.-R.; Kumazawa, S. Chemical Profiles of Korean Bee Pollens and Their Catechol-O-Methyltransferase Inhibitory Activities. J. Agric. Food Chem. 2022, 70, 1174–1181. [Google Scholar] [CrossRef]
- Rodríguez-Pólit, C.; Gonzalez-Pastor, R.; Heredia-Moya, J.; Carrera-Pacheco, S.E.; Castillo-Solis, F.; Vallejo-Imbaquingo, R.; Barba-Ostria, C.; Guamán, L.P. Chemical Properties and Biological Activity of Bee Pollen. Molecules 2023, 28, 7768. [Google Scholar] [CrossRef]
- Cornara, L.; Biagi, M.; Xiao, J.; Burlando, B. Therapeutic Properties of Bioactive Compounds from Different Honeybee Products. Front. Pharmacol. 2017, 8, 412. [Google Scholar] [CrossRef]
- Dias, M.C.; Pinto, D.C.G.A.; Silva, A.M.S. Plant Flavonoids: Chemical Characteristics and Biological Activity. Molecules 2021, 26, 5377. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.-Y.; Li, Q.; Bi, K.-S. Bioactive Flavonoids in Medicinal Plants: Structure, Activity and Biological Fate. Asian J. Pharm. Sci. 2018, 13, 12–23. [Google Scholar] [CrossRef]
- Joo, Y.; Seo, Y.H.; Lee, S.; Shin, E.; Yeon, S.W.; Kim, S.B.; Lee, M.K. Antioxidant and Tyrosinase-Inhibitory Activities and Biological Bioactivities of Flavonoid Derivatives from Quercus mongolica Pollen. Molecules 2025, 30, 794. [Google Scholar] [CrossRef]
- Serra Bonvehí, J.; Soliva Torrentó, M.; Centelles Lorente, E. Evaluation of Polyphenolic and Flavonoid Compounds in Honeybee-Collected Pollen Produced in Spain. J. Agric. Food Chem. 2001, 49, 1848–1853. [Google Scholar] [CrossRef]
- Hu, L.; Luo, Y.; Yang, J.; Cheng, C. Botanical Flavonoids: Efficacy, Absorption, Metabolism and Advanced Pharmaceutical Technology for Improving Bioavailability. Molecules 2025, 30, 1184. [Google Scholar] [CrossRef]
- Sajid, M.; Channakesavula, C.N.; Stone, S.R.; Kaur, P. Synthetic Biology towards Improved Flavonoid Pharmacokinetics. Biomolecules 2021, 11, 754. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lin, C.; Ma, X.; Tan, Y.; Wang, J.; Zeng, M. Functional Characterization of a Flavonoid Glycosyltransferase in Sweet Orange (Citrus sinensis). Front. Plant Sci. 2018, 9, 166. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J. Dietary Flavonoid Aglycones and Their Glycosides: Which Show Better Biological Significance? Crit. Rev. Food Sci. Nutr. 2017, 57, 1874–1905. [Google Scholar] [CrossRef]
- Maaiden, E.E.; Ullah, N.; Ezzariai, A.; Mazar, A.; Boukcim, H.; Hirich, A.; Nasser, B.; Qarah, N.; Kouisni, L.; Kharrassi, Y.E. Comparing Antioxidant and Cytoprotective Effects: Quercetin Glycoside vs. Aglycone from Ephedra alata. Phytomed. Plus 2024, 4, 100603. [Google Scholar] [CrossRef]
- Burlacu, E.; Nisca, A.; Tanase, C. A Comprehensive Review of Phytochemistry and Biological Activities of Quercus Species. Forests 2020, 11, 904. [Google Scholar] [CrossRef]
- Morales, D. Oak Trees (Quercus Spp.) as a Source of Extracts with Biological Activities: A Narrative Review. Trends Food Sci. Technol. 2021, 109, 116–125. [Google Scholar] [CrossRef]
- Di Marco, G.; D’Agostino, A.; Braglia, R.; Redi, E.L.; Iacobelli, S.; Gismondi, A.; Canini, A. Pollen Variability in Quercus L. Species and Relative Systematic Implications. Plant Physiol. Biochem. 2023, 204, 108079. [Google Scholar] [CrossRef]
- Lee, S.H.; Park, Y.D. Carbon Stock and Analysis on Factors Influencing in Quercus mongolica Stand at Mt. Gariwang, South Korea. Korean J. Agric. Sci. 2024, 51, 413–427. [Google Scholar] [CrossRef]
- Marzouk, M.M.; Hegazi, N.M.; El Shabrawy, M.O.A.; Farid, M.M.; Kawashty, S.A.; Hussein, S.R.; Saleh, N.A.M. Discriminative Metabolomics Analysis and Cytotoxic Evaluation of Flowers, Leaves, and Roots Extracts of Matthiola longipetala Subsp. Livida. Metabolites 2023, 13, 909. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Zhang, M.; Otsuki, K.; Li, W. Molecular Networking as a Natural Products Discovery Strategy. Acta Mater. Medica 2023, 2, 126–141. [Google Scholar] [CrossRef]
- Araújo, J.S.; Chambó, E.D.; de Carvalho Costa, M.A.P.; da Silva, S.M.P.C.; de Carvalho, C.A.L.; Estevinho, L.M. Chemical Composition and Biological Activities of Mono- and Heterofloral Bee Pollen of Different Geographical Origins. Int. J. Mol. Sci. 2017, 18, 921. [Google Scholar] [CrossRef] [PubMed]
- Prđun, S.; Svečnjak, L.; Valentić, M.; Marijanović, Z.; Jerković, I. Characterization of Bee Pollen: Physico-Chemical Properties, Headspace Composition and FTIR Spectral Profiles. Foods 2021, 10, 2103. [Google Scholar] [CrossRef]
- Fabre, N.; Rustan, I.; Hoffmann, E.; Quetin-Leclercq, J. Determination of Flavone, Flavonol, and Flavanone Aglycones by Negative Ion Liquid Chromatography Electrospray Ion Trap Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2001, 12, 707–715. [Google Scholar] [CrossRef]
- Cuyckens, F.; Claeys, M. Mass Spectrometry in the Structural Analysis of Flavonoids. J. Mass Spectrom. 2004, 39, 1–15. [Google Scholar] [CrossRef]
- Kachlicki, P.; Piasecka, A.; Stobiecki, M.; Marczak, Ł. Structural Characterization of Flavonoid Glycoconjugates and Their Derivatives with Mass Spectrometric Techniques. Molecules 2016, 21, 1494. [Google Scholar] [CrossRef]
- Li, Z.-H.; Guo, H.; Xu, W.-B.; Ge, J.; Li, X.; Alimu, M.; He, D.-J. Rapid Identification of Flavonoid Constituents Directly from PTP1B Inhibitive Extract of Raspberry (Rubus idaeus L.) Leaves by HPLC-ESI-QTOF-MS-MS. J. Chromatogr. Sci. 2016, 54, 805–810. [Google Scholar] [CrossRef]
- Cuyckens, F.; Claeys, M. Determination of the Glycosylation Site in Flavonoid Mono-O-Glycosides by Collision-Induced Dissociation of Electrospray-Generated Deprotonated and Sodiated Molecules. J. Mass Spectrom. 2005, 40, 364–372. [Google Scholar] [CrossRef]
- Abdallah, R.H.; Hassan, W.H.B.; Al-Massarani, S.M.; Abdel-Mageed, W.M.; Eldahmy, S.I.; Basudan, O.A.; Parveen, M.; El Senosy, E.; Abdelaziz, S. UPLC-ESI-MS/MS Profiling of Secondary Metabolites from Methanol Extracts of in Vivo and in Vitro Tissues of Daucus capillifolius Gilli (A Comparative Study). Molecules 2024, 29, 2694. [Google Scholar] [CrossRef]
- Avula, B.; Bae, J.-Y.; Wang, Y.-H.; Wang, M.; Osman, A.G.; Smith, K.; Yuk, J.; Ali, Z.; Plumb, R.; Isaac, G.; et al. Chemical Profiling and Characterization of Phenolic Acids, Flavonoids, Terpene Glycosides from Vangueria agrestis Using Ultra-High-Performance Liquid Chromatography/Ion Mobility Quadrupole Time-of-Flight Mass Spectrometry and Metabolomics Approach. Biomed. Chromatogr. 2020, 34, e4840. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Yang, Y.; Duan, Y.; Li, C.; Gao, H.; Liu, H.; Cui, Q.; Guo, Z.; Liu, X.; Wang, Z. Quality Marker Discovery and Quality Evaluation of Eucommia ulmoides Pollen Using UPLC-QTOF-MS Combined with a DPPH-HPLC Antioxidant Activity Screening Method. Molecules 2023, 28, 5288. [Google Scholar] [CrossRef] [PubMed]
- Hefny Gad, M.; Tuenter, E.; El-Sawi, N.; Younes, S.; El-Ghadban, E.; Demeyer, K.; Pieters, L.; Vander Heyden, Y.; Mangelings, D. Identification of Some Bioactive Metabolites in a Fractionated Methanol Extract from Ipomoea aquatica (Aerial Parts) through TLC, HPLC, UPLC-ESI-QTOF-MS and LC-SPE-NMR Fingerprints Analyses. Phytochem. Anal. 2018, 29, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yu, H.; Wu, H.; Pan, Y.; Wang, K.; Jin, Y.; Zhang, C. Characterization and Quantification by LC–MS/MS of the Chemical Components of the Heating Products of the Flavonoids Extract in Pollen Typhae for Transformation Rule Exploration. Molecules 2015, 20, 18352–18366. [Google Scholar] [CrossRef]
- Liu, M.; Dong, J.; Lin, Z.; Niu, Y.; Zhang, X.; Jiang, H.; Guo, N.; Li, W.; Wang, H.; Chen, S. Rapid Screening of Transferrin-Binders in the Flowers of Bauhinia Blakeana Dunn by on-Line High-Performance Liquid Chromatography–Diode-Array Detector–Electrospray Ionization–Ion-Trap–Time-of-Flight–Mass Spectrometry–Transferrin–Fluorescence Detection System. J. Chromatogr. A 2016, 1450, 17–28. [Google Scholar] [CrossRef]
- Parejo, I.; Jáuregui, O.; Viladomat, F.; Bastida, J.; Codina, C. Characterization of Acylated Flavonoid-O-Glycosides and Methoxylated Flavonoids from Tagetes Maxima by Liquid Chromatography Coupled to Electrospray Ionization Tandem Mass Spectrometry. Rapid Commun. Mass Spectrom. 2004, 18, 2801–2810. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).