Abstract
While most of the Alzheimer’s disease (AD) cases are sporadic and manifest after age 65 (late-onset AD, LOAD), a subset of patients develop symptoms earlier in life (early-onset, EOAD) due to mutations in the PSEN1, PSEN2, or APP genes with an autosomal-dominant inheritance pattern (AD-EOAD). In this study, we examined the association between age of onset (AoO) and first clinical manifestation (FCM) with the APOE and DAOA genotypes, previously described as modifiers of clinical phenotypes in LOAD and EOAD in 88 individuals clinically diagnosed with AD-EOAD due to the PSEN1 A431E variant (39 females, 49 males). We classified the population according to their genotype (APOEε2, APOEε3, and APOEε4 and DAOA G/G, G/A, and A/A) and FCM (cognitive, behavioral, motor, and memory impaired). Memory impairment was the most frequent symptom (51%), followed by motor disturbances (31.8%), cognitive symptoms other than memory (10.4%), and behavioral changes (6.8%). We found a significant association between APOE genotype and AoO (p < 0.001), with the APOEε4 allele being linked to a delayed onset (β = 4.04, SE = 1.11, p = 0.0003). Similarly, individuals with the DAOA rs2391191 A/A genotype showed a significantly later AoO compared to G/G carriers (β = 2.13, SE = 0.96, p = 0.0301). No significant association was found between APOE or DAOA genotypes and FCM. The findings suggest that both the APOEε4 allele and DAOA rs2391191 A/A genotype may act as genetic modifiers of AoO, delaying symptom onset in individuals with AD-EOAD. Further research is needed to elucidate the molecular pathways through which APOE and DAOA influence AD-EOAD progression.
1. Introduction
Alzheimer’s disease (AD) is the most prevalent form of dementia, accounting for approximately 60% to 80% of all diagnosed cases in the elderly population. In the United States alone, an estimated 6.9 million individuals are currently affected. Worldwide, more than 45 million people are living with dementia, and this number is projected to reach 135 million by 2050, primarily influenced by demographic changes and increased life expectancy [1,2]. AD is a progressive neurodegenerative disorder that leads to an irreversible deterioration in cognitive functions, memory, language, behavior, and judgment. Manifesting as cognitive decline, behavioral changes, and a gradual loss of independence, AD places a significant burden on patients, their families, and healthcare systems worldwide [3,4,5]. At the neuropathological level, AD is characterized by the extracellular deposition of amyloid-beta (Aβ) plaques and the intraneuronal accumulation of neurofibrillary tangles (NFTs) composed of hyperphosphorylated tau protein. These hallmark lesions are associated with widespread synaptic dysfunction, neuronal loss, and cortical atrophy. In addition to these classical features, increasing evidence points to the role of chronic neuroinflammation, cerebrovascular alterations, oxidative stress, and mitochondrial dysregulation as critical contributors to the multifaceted pathogenesis of the disease [6,7].
Although most cases of AD manifest after the age of 65—classified as late-onset AD (LOAD)—smaller subsets of individuals develop symptoms at an earlier age, known as early-onset AD (EOAD) [8]. EOAD typically manifests before age 65 and is frequently associated with a more aggressive clinical course, characterized by accelerated cognitive decline and atypical presentations. These often include focal cortical syndromes such as primary progressive aphasia and posterior cortical atrophy, which may complicate diagnosis and lead to initial misclassification. Pathologically, EOAD tends to exhibit severe tau deposition, as evidenced by higher tau-PET signals compared to LOAD [9,10,11,12]. Even though EOAD is frequently associated with Mendelian inheritance, only a small proportion of cases follow an autosomal-dominant pattern [13]. Autosomal-dominant EOAD (AD-EOAD, commonly known as Familial EOAD or Mendelian EOAD) accounts for approximately 10% of all EOAD cases; most of the cases result from variants in the PSEN1, PSEN2, or APP genes [14]. In contrast, LOAD is strongly influenced by the APOE gene, particularly the APOEε4 allele, which increases disease risk and lowers the age of onset (AoO) in a dose-dependent manner [15,16].
The APOE gene codes for apolipoprotein E (ApoE), a key regulator of lipid metabolism and neurological function, with significant implications for AD pathogenesis. ApoE is mainly synthesized by astrocytes in the brain. There are three major isoforms—ApoE2, ApoE3, and ApoE4—distinguished by amino acid substitutions at positions 112 and 158: ApoE2 (Cys112, Cys158), ApoE3 (Cys112, Arg158), and ApoE4 (Arg112, Arg158). These isoforms are encoded by the alleles APOEε2, APOEε3, and APOEε4, respectively, whose distribution and biological effects differ substantially. APOEε3 is the most common allele worldwide (~77%) and is considered neutral regarding AD risk. APOEε4, found in ~15% of the general population, produces an isoform with reduced lipid-binding stability and altered receptor interactions. In contrast, APOEε2, the rarest allele (~8%), encodes an isoform with enhanced lipid-binding capacity and greater efficiency in Aβ clearance, conferring a protective effect and often delaying AoO. ApoE facilitates lipid transport by interacting with lipoprotein receptors such as LDLr and LRP1, affecting neuronal function, synaptic plasticity, and membrane homeostasis. In the AD context, ApoE plays a crucial role in neural repair, neuroinflammation, and modulation of Aβ clearance, with its isoforms differentially influencing Aβ aggregation and clearance efficiency [17,18].
The APOEε4 allele is the most well-established genetic risk factor for LOAD, significantly increasing disease susceptibility in a dose-dependent manner. The presence of one ε4 allele increases the risk of developing AD by 2–4 times, while homozygosity (ε4/ε4) raises the risk by 8–10 times [15]. In contrast, the APOEε2 allele exerts a protective effect, reducing disease risk and delaying onset [19] relative to the most common APOEε3 allele. ApoE4 promotes amyloidogenic processing of the amyloid precursor protein (APP), leading to increased Aβ accumulation, impaired Aβ clearance, and exacerbated tau pathology, neuroinflammation, and blood–brain barrier dysfunction [20,21,22]. Understanding the multifaceted role of ApoE in AD pathogenesis is critical for the development of targeted therapeutic strategies to mitigate disease progression [22].
DAOA (also known as G72) encodes a protein that modulates the activity of D-amino acid oxidase (DAAO), an enzyme responsible for the degradation of D-serine. This amino acid serves as a potent co-agonist of N-methyl-D-aspartate receptors (NMDARs), which are critical for synaptic plasticity, learning, and memory [23,24].
The enzymes involved in the metabolism of D-amino acids have been associated with the pathophysiology of AD and schizophrenia [25,26]. In this context, the variant rs2391191 involves a guanine-to-adenine substitution at position 6231, resulting in an amino acid change from arginine to lysine at codon 30. This substitution has been reported to affect the structure and function of the DAOA protein [27].
The disruption of glutamate-mediated neurotransmission is considered a key mechanism in the pathogenesis of AD. L-serine is converted into D-serine by the enzyme serine racemase (SR), which is expressed in both astrocytes and presynaptic neurons. D-serine can be released into extracellular space by the alanine–serine–cysteine transporter (ASC-1) and ASCT-1 transporters. In contrast, L-serine is released by astrocytes and taken up by neurons via the same transporters, but in the reverse direction. D-serine is subsequently degraded by DAAO, an enzymatic activity that can be inhibited by DAOA, particularly when DAOA is localized to the outer mitochondrial membrane [28,29].
It has been proposed that the rs2391191 variant may indirectly reduce D-serine levels, thereby modulating the activation of NMDAR, which is known to be aberrantly upregulated in AD. This regulatory mechanism could potentially contribute to cognitive preservation and delay the onset of symptoms in EOAD [30].
Presenilin 1 (PS1, encoded by the gene PSEN1) is a crucial component of the γ-secretase complex, a multi-subunit protease involved in the intramembrane cleavage of several type I transmembrane proteins, most notably APP. Through this role, PS1 modulates the production of Aβ peptides [31,32]. Mutations in the PSEN1 gene are the most frequent cause of AD-EOAD, often leading to altered γ-secretase function, which increases the relative production of longer Aβ species [33,34]. The A431E variant is located within the ninth transmembrane domain of PS1 and has been identified in Mexican families. This variant is associated with an especially aggressive clinical course, including very early symptom onset, rapid cognitive decline, and frequent neuropsychiatric and motor disturbances [35]. Functional analyses suggest that the A431E variant significantly impacts the structural conformation and enzymatic activity of γ-secretase, resulting in elevated Aβ42 levels [36].
Since 1999, evidence has suggested that a significant proportion of individuals with AD-EOAD in the state of Jalisco, Mexico, carry the PSEN1 A431E pathogenic variant due to a founder effect. This phenomenon has been particularly observed in the mestizo population of Altos de Jalisco, a region characterized by mixed Spanish and indigenous ancestry [35,37].
The A431E variant in PSEN1 was first described by Rogaeva et al. [38] in five patients of unspecified origin. Subsequent investigations identified this variant in nine unrelated families native to Jalisco [37], strengthening the hypothesis of a common ancestral origin. In 2006, Murrell et al. reported 15 additional unrelated cases carrying the same variant [35], further reinforcing its association with a regional founder effect. The PSEN1 A431E variant is currently classified as pathogenic [35]. The clinical characterization of affected individuals revealed an AoO mean of 42.5 ± 3.9 years. Notably, there was marked clinical heterogeneity in the first clinical manifestations (FCM), with frequent reports of spastic paraparesis, language disturbances, and neuropsychiatric symptoms [36].
Previous studies have reported that the APOE genotype was associated with an effect on AoO [39,40,41] and clinical manifestations [42,43,44,45] in EOAD patients.
In the Colombian kindred with the PSEN1 E280A variant, studies have shown that the APOEε4 and APOEε2 alleles are associated with earlier and later AoO, respectively [40,46]. A larger study involving persons with various AD-EOAD mutations suggested only a subtle effect of earlier onset in association with the APOEε4 allele that was not statistically significant [47]. Interestingly, we previously published that patients with AD-EOAD and EOAD (with and without a known genetic cause) had a delay in the AoO due to the APOEε4 allele [48]. In the present study, we aimed to investigate the association between AoO and FCM with the APOE and DAOA genotypes in patients harboring the PSEN1 A431E pathogenic variant.
2. Results
2.1. Sociodemographic Information
A total of 88 index cases of AD-EOAD caused by the A431E variant in PSEN1 were evaluated. Of these cases, 49 (55.7%) were male and 39 (44.3%) were female. The average AoO was 41.81 years, ranging from 34 to 49 years (Table 1).

Table 1.
Clinical and demographic characteristics by APOE genotype in PSEN1 A431E variant carriers.
FCM were categorized into four main domains based on their frequency: cognitive (excluding memory), behavioral, motor, and memory-related symptoms. Within the cognitive domain, a total of nine cases (10.4%) were documented, including aphasia, anomia, dysgraphia, dyslalia, verbal perseveration, and disorientation. Behavioral symptoms were observed in six individuals (6.8%) and included irritability, depression, and aggressiveness. Motor symptoms were present in 28 cases (31.8%), predominantly characterized by gait disturbances, as well as dysarthria and apraxia. Finally, memory impairment was the most frequently reported symptom, present in 45 individuals (51%). This classification enabled a more structured analysis of symptom distribution and facilitated comparisons across domains in relation to genetic and clinical variables.
2.2. Age of Onset, DAOA, and APOE Genotype
The most frequent APOE genotype was ε3/ε3, found in 64 individuals (72.73%). The ε3/ε4 genotype was identified in 10 cases (11.36%), ε2/ε3 in 7 cases (7.96%), ε2/ε2 in 5 cases (5.69%), ε2/ε4 in 1 case (1.13%), and ε4/ε4 in 1 case (1.13%). In terms of allele frequency, ε3 was the most prevalent (82.38%), followed by ε2 (10.22%) and ε4 (7.4%). For analytical purposes, individuals were grouped based on the presence of the APOEε2 or ε4 alleles. The APOEε2+ subgroup included patients carrying at least one ε2 allele, specifically those with APOEε2/ε2 and APOEε2/ε3 genotypes. The APOEε4+ subgroup included those carrying at least one ε4 allele: APOEε2/ε4, APOEε3/ε4, and APOEε4/ε4 genotypes. This grouping approach allowed us to explore potential allele-specific effects on AoO while acknowledging the limitations imposed by the relatively small number of APOEε2 and APOEε4 homozygotes in the cohort. Regarding the DAOA rs2391191 genotypes, G/G was observed in 35 individuals (39.77%), G/A in 32 individuals (36.36%), and A/A in 21 individuals (23.87%). Allele frequency analysis showed that the G allele was the most prevalent (57.95%), followed by the A allele (42.05%).
We examined the distribution of APOE genotypes and their association with clinical characteristics in our PSEN1 variant carrier cohort (n = 88). The mean AoO significantly differed among APOE genotype groups (H = 18.0853, p = 0.00012), with APOEε2+ individuals presenting the earliest mean AoO (39.2 ± 3.1 years), followed by APOEε3 (41.6 ± 3.7 years), and APOEε4+ carriers showing the latest mean onset (45.8 ± 2.5 years).
A Mann–Whitney U test showed no significant difference in AoO between sexes (W = 903, p = 0.661). Likewise, the predominant FCM did not vary significantly across APOE genotypes (p = 0.7248), with memory impairment being the most frequent presentation in all groups. The results reveal a significant difference in the AoO across different APOE genotypes. A p-value less than 0.001 indicates a strong difference between AoO and APOE genotypes; however, no significant difference was observed between the FCM and APOE genotypes (Table 1).
We further assessed the potential influence of DAOA genotypes on the clinical phenotypes within the PSEN1 A431E variant carrier cohort. A statistically significant difference in AoO was detected among genotype groups (H = 10.334, p = 0.0057). Individuals with the A/A genotype exhibited a later mean AoO (44.1 ± 3.2 years) compared to G/G (41.4 ± 3.8 years) and G/A carriers (40.7 ± 3.9 years).
FCM did not differ significantly across genotype groups (p = 0.5284), with memory symptoms remaining the most prevalent initial presentation. The results reveal a significant difference in the AoO across different DAOA genotypes. A p-value of <0.05 indicates a strong difference between the AoO and DAOA genotypes; however, no significant association was observed between the FCM and DAOA genotypes (Table 2).

Table 2.
Clinical and demographic characteristics by DAOA genotype in PSEN1 A431E variant carriers.
2.3. Effect of APOE and DAOA Genotype on Age of Onset
Due to the significant findings from the non-parametric analysis, a simple linear regression was performed to quantify the impact of APOE and DAOA genotypes on AoO (Table 3 and Table 4). The results confirmed a significant association between APOE and DAOA genotypes and AoO, highlighting a potential role in modulating disease onset (Figure 1 and Figure 2). In contrast, no regression analysis was performed for the FCM, because Fisher’s exact test did not indicate a significant difference.

Table 3.
Simple linear regression of APOEε2 and APOEε4 alleles with AoO.

Table 4.
Simple linear regression of DAOA rs2391191 genotypes with AoO.
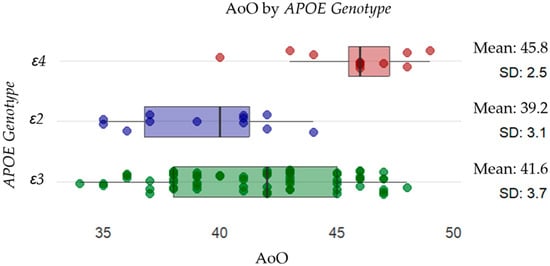
Figure 1.
Distribution of AoO across different APOE genotypes. Individual data points are displayed as scattered dots, with the mean AoO and standard deviation (SD) indicated for each genotype. The results suggest that individuals with the APOEε4 allele (red) (mean: 45.8, SD: 2.5) have a later onset than those with the APOEε2 allele (purple) (mean: 39.2, SD: 3.1) and APOEε3 (green) (mean: 41.6, SD: 3.7).
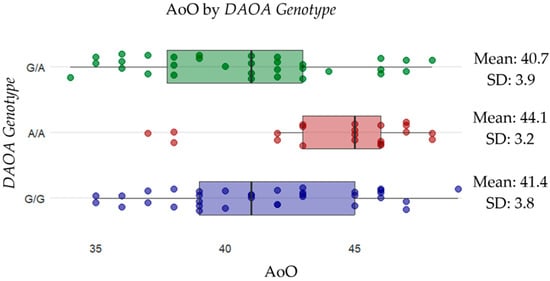
Figure 2.
Distribution of AoO across different DAOA genotypes. Scattered dots represent individual data points, with the mean AoO and standard deviation (SD) shown for each genotype. The results suggest that individuals with the A/A genotype (red) (mean: 44.1, SD: 3.2) have a later onset compared to G/A (purple) (mean: 39.2, SD: 3.1) and APOEε3 (green) (mean: 41.6, SD: 3.7).
To evaluate the impact of genetic variants on AoO in AD-EOAD, we performed a multiple linear regression including APOEε2, APOEε4, and genotypes of the DAOA rs2391191 variant as predictors. The model was statistically significant (F(4, 86) = 7.911, p = 1.88 × 10−5).
As shown in Table 5, the presence of the APOEε4 allele was significantly associated with a later AoO (β = 4.04, SE = 1.11, p = 0.000332), whereas the APOEε2 allele was not significantly associated with AoO (p = 0.14). Regarding DAOA, individuals homozygous for the A allele (A/A genotype) exhibited a significantly delayed onset compared to the reference genotype (G/G) (β = 2.13, SE = 0.96, p = 0.0301). The heterozygous G/A genotype did not show a statistically significant association (p = 0.39).

Table 5.
Multiple linear regression of APOE and DAOA with AoO.
As shown in Table 6, the presence of the APOEε4 allele was significantly associated with a later AoO (β = 6.24, SE = 1.83, p = 0.00105), whereas the APOEε2 allele did not show a significant association (p = 0.867). For DAOA, individuals with the A/A genotype exhibited a significantly delayed onset compared to the reference genotype G/G (β = 3.00, SE = 1.07, p = 0.0063). No significant association was observed for the heterozygous G/A genotype (p = 0.99). Interaction terms between APOE alleles and DAOA genotypes were included in the model but did not reach statistical significance.

Table 6.
Multiple linear regression model examining APOE–DAOA interaction effects on AoO.
These results suggest that both the APOEε4+ and the DAOA rs2391191 A/A genotype may independently modulate the clinical presentation of AD-EOAD by delaying the AoO (Figure 3).
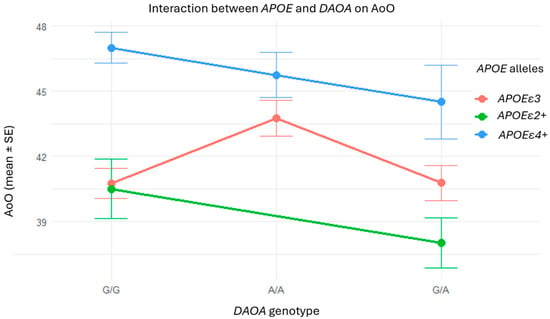
Figure 3.
Interaction between DAOA and APOE genotypes on AoO: no consistent additive or synergistic effects. Although patients with the A/A genotype or APOEε4+ individually tended to exhibit later onset, the visualization does not reveal any consistent additive or interaction patterns between these variants. The absence of clear trends supports the statistical results, indicating no synergistic effects between APOE and DAOA in this cohort.
3. Discussion
Understanding the genetic factors that influence the AoO in AD-EOAD is critical for unraveling disease mechanisms. While APOE and PSEN1 variants have well-documented effects on AD risk and progression, the interaction of these genes with other loci, such as DAOA, remains less clear.
Our findings reveal a significant association between APOE and DAOA genotypes and the AoO in individuals with AD-EOAD caused by the PSEN1 A431E variant. The presence of the APOEε4 allele was associated with a delayed clinical onset, a result that contrasts with well-established observations in LOAD, where APOEε4 is typically linked to earlier disease onset and increased severity [16,41,49,50,51,52].
Several studies have reported the heterogeneous effects of APOEε4 in EOAD and AD-EOAD populations [39,41,44,48,53,54], suggesting that its modulatory role may be context-dependent. This divergence from the LOAD findings supports the hypothesis that the influence of APOE on AD pathophysiology may vary according to age at onset and underlying genetic background.
The role of APOE in modulating the clinical trajectory of AD-EOAD has been a matter of ongoing debate. Initial studies, such as the one by Lendon et al. (1997), suggested that APOE genotypes had no significant impact on AoO in PSEN1 E280A carriers [55]. However, more recent work has challenged this view. Vélez et al. (2016), for example, reported a modifier effect of APOEε2, with significantly delayed onset among PSEN1 E280A carriers [40]. Similarly, Langella et al. (2023) showed that cognitive decline in AD-EOAD is accelerated in APOEε4 carriers and attenuated in APOEε2 carriers, underscoring the allele-specific influence of APOE on disease expression, even in the presence of fully penetrant monogenic mutations [56].
Our observation of a delayed onset in APOEε4 carriers with the PSEN1 A431E variant is particularly intriguing. It echoes the results by De Luca et al. (2016), who reported a paradoxical effect of APOEε4: accelerating disease onset in LOAD yet delaying it in certain EOAD contexts [39]. These unexpected results suggest that the modulatory role of APOE is not uniform across AD subtypes and may interact with specific genetic backgrounds—such as PSEN1 variants—to alter its phenotypic expression. Additionally, Smits et al. (2015) highlighted that APOEε4-negative individuals exhibited more rapid decline in non-memory cognitive domains, suggesting that APOE may influence not only AoO but also domain-specific trajectories of cognitive impairment [44]. Our results align with this growing recognition that APOE’s impact in AD extends beyond a unidimensional effect on AoO.
In the context of AD-EOAD caused by the PSEN1 A431E, the protective effect of APOEε2 appears to be non-significant, in contrast with observations in persons carrying the E280A PSEN1 variant which showed a delayed AoO associated with the APOEε2 allele [40]. The delayed onset observed in APOEε4 carriers challenges the conventional understanding of APOEε4 as a risk factor for accelerated cognitive decline. One possible explanation is that APOE may exert its effects later in the disease process, after significant neurodegeneration has already occurred. The delayed AoO in APOEε4 carriers may also reflect compensatory mechanisms, such as neuroinflammatory responses and altered lipid metabolism [56,57], which initially mitigate the pathological impact of the PSEN1 A431E variant.
The paradoxical findings of the present study can be explained by the concept of antagonistic pleiotropy. According to this model, the effects of a gene can be beneficial in some life stages and detrimental in others [58]. Research suggests that APOEε4 carriers may exhibit cognitive advantages in youth and early adulthood, including higher IQ scores [59], better performance on neuropsychological tests assessing attention and memory [60,61], and increased mental vitality. Furthermore, APOEε4 has been linked to personality traits such as sociability and positive emotionality, which may provide adaptive benefits during early life [62].
This compensatory mechanism enables APOEε4 carriers to exhibit relatively high cognitive performance early in life, even in the presence of subclinical neurocognitive changes associated with AD [63]. However, as APOEε4 carriers age, this compensatory process begins to fail. When the pathological burden of AD reaches a critical threshold, compensatory mechanisms are no longer sufficient to sustain cognitive function, resulting in a later onset of symptoms compared to non-carriers [64]. While APOEε4 enhances microglial activation, this effect becomes detrimental in LOAD, where prolonged or dysregulated activation impairs Aβ clearance and exacerbates neuroinflammation [65,66].
Our findings support the hypothesis that the DAOA rs2391191 variant may act as a genetic modifier, capable of influencing the phenotypic expression of AD-EOAD. Our findings contribute additional evidence to the role of common variants in shaping the heterogeneous clinical trajectories observed in individuals with AD-EOAD [40].
At the molecular level, DAOA may exert a modifying effect by indirectly regulating NMDAR signaling by modulating D-serine levels. Previous studies have reported that downregulation or reduced functional activity of DAOA—which may occur in individuals homozygous for the A allele—could lead to more stable D-serine levels, resulting in a more balanced activation of NMDARs. This mechanism may delay synaptic dysfunction in AD-EOAD (see Figure 4) [27,29,30,67,68].
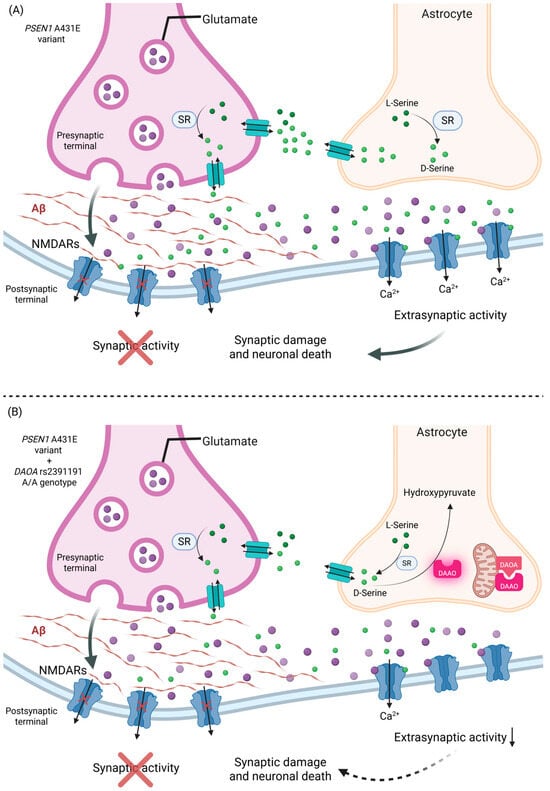
Figure 4.
Effect of the DAOA rs2391191 A/A genotype on glutamatergic signaling in the presence of the PSEN1 A431E variant. The PSEN1 A431E variant promotes the accumulation of Aβ at the synaptic cleft, blocking the normal synaptic activity and shifting the activation of NMDARs to the extrasynaptic space, where excitotoxic pathways associated with synaptic damage and neuronal death are enhanced. (A) In the absence of the DAOA rs2391191 A/A genotype, D-serine generated by SR is normally degraded due to DAAO inactivation by DAOA, enabling the activation of extrasynaptic NMDARs and favoring neuronal damage. (B) In the presence of the DAOA rs2391191 A/A genotype, it could decrease the affinity of DAOA for DAAO, allowing DAOA to remain active and degrade D-serine to hydroxypyruvate, reducing the activation of extrasynaptic NMDARs and attenuating excitotoxic damage. This mechanism could contribute to a later onset of symptoms in patients with AD-EOAD in A/A rs2391191 carriers. Created in BioRender. https://BioRender.com/93xxevk (accessed on 30 June 2025).
Moreover, the lack of an additive effect with APOEε4 suggests that DAOA operates through a distinct biological pathway, independent of lipid metabolism and cholesterol transport, mechanisms predominantly associated with ApoE.
Taken together, these findings support a model in which APOEε4 may be beneficial in early life but becomes detrimental in aging due to its effects on lipid metabolism, cellular stress responses, and Aβ aggregation. Further research is needed to dissect the precise molecular pathways through which APOE and DAOA influence AD-EOAD and to explore potential therapeutic strategies.
4. Materials and Methods
This study included 88 patients who were diagnosed clinically with AD-EOAD and confirmed to carry the PSEN1 A431E variant (39 females and 49 males). The patients were evaluated at the División de Genética at the Centro Médico Nacional de Occidente—Instituto Mexicano del Seguro Social (IMSS), in Guadalajara, Jalisco, Mexico, between 2012 and 2025. Genealogy and clinical data were also gathered from unaffected family members.
Primary caregivers of the patients were interviewed to determine AoO and to document the FCM. A thorough and structured clinical history was collected for each patient. Next, molecular analysis was performed on the probands to confirm the genetic diagnosis.
Written informed consent was obtained from the primary caregiver or legal representative of each participant prior to their inclusion in the study.
4.1. DNA Extraction and Sanger Sequencing
DNA was extracted from peripheral blood samples using the salting-out method [69]. For Sanger sequencing, the amplification was targeted specifically to exon 12 of the PSEN1 gene, where the variant A431E (c.1292C>A, rs63750083) is located. After amplifying the region of interest via PCR, the amplification product was purified using ExoSAP-IT™ Express reagent (Applied Biosystems™, Foster City, CA, USA) to remove any remaining primers and unincorporated nucleotides. The sequencing reaction was carried out using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems™, Foster City, CA, USA). Finally, the sequencing product was purified with the BigDye™ XTerminator Purification Kit (Applied Biosystems™, Foster City, CA, USA), according to the manufacturer’s specifications, to ensure a clear and contaminant-free sequence.
4.2. APOE and DAOA Genotyping by Real-Time PCR
APOE alleles were inferred from single-nucleotide polymorphisms (SNPs) rs7412 (C____904973_10) for APOEε2 allele and rs429358 (C___3084793_20) for APOEε4 allele, which were genotyped using real-time PCR. APOEε4 genotyping for these two SNPs was performed using a TaqMan™ assay (Applied Biosystems™, Foster City, CA, USA). Each reaction contained 20–50 ng of DNA, 2X TaqMan™ Genotyping Master Mix, and specific TaqMan™ probes labeled with VIC and FAM fluorophores to detect the rs7412 and rs429358 variants. The cycling conditions included an initial enzyme activation step at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 min.
Genotyping of the DAOA rs2391191 (C__16000591_10) variant was also performed using real-time PCR with TaqMan™ technology, following the same reaction setup and thermal cycling conditions as described for the APOE SNPs.
The fluorescence signals were analyzed using Applied Biosystems’ proprietary software. To assign a genotype to each patient, both the detection and intensity of the fluorophores were evaluated. For quality control, a subset of samples was genotyped in duplicate, and any ambiguous results were resolved by repeating the assay.
4.3. Dementia Diagnosis
The diagnosis of dementia was established through a multidisciplinary consensus involving a board-certified neurologist and two physicians specializing in dementia. The diagnostic process included a comprehensive neurological assessment, an in-depth clinical examination, and a detailed review of relevant medical and functional history before the dementia evaluation. Additionally, cognitive and behavioral changes were assessed through informant interviews to supplement clinical observations.
The classification of dementia followed internationally recognized guidelines, with cognitive impairment initially screened using the Mini-Mental State Examination (MMSE). For AD, the diagnosis was based on the criteria established by the National Institute of Neurological and Communicative Disorders and Stroke (NINCDS) in collaboration with the Alzheimer’s Disease and Related Disorders Association (ADRDA). These criteria ensure diagnostic accuracy by integrating clinical, cognitive, and functional assessments.
4.4. Ethics
This study was conducted in strict compliance with the Regulations of the General Health Law on Health Research [70], as well as the ethical principles outlined in the Declaration of Helsinki, last updated in October 2024 by the World Medical Association. Additionally, the study adhered to both national and international guidelines for best practices in clinical research [71].
According to the classification established in the Regulations of the General Health Law on Health Research [70], this study falls under type I research, meaning that it poses no risk to the participants. This study involved the collection of DNA samples through venipuncture, a standard, minimally invasive procedure. All samples were anonymized using a unique folio number and securely stored. The corresponding data were systematically recorded in a protected database, ensuring strict confidentiality and compliance with data protection regulations.
4.5. Statistical Analysis
Descriptive and comparative analyses of both sociodemographic and clinical data were conducted using RStudio v2024.12.1+563. Data visualization was performed through graphical representations generated in the same software.
To compare categorical variables, Fisher’s exact test or the chi-square test was applied, depending on data distribution and expected frequencies. To assess the normality of the age of symptom onset variable, a Shapiro–Wilk test was performed. The Kruskal–Wallis test was used to evaluate significant differences between the AoO, the FCM, and APOE and DAOA genotypes. To assess the extent of the effect of APOE and DAOA genotypes on the AoO, a simple linear regression analysis was performed. To further assess the extent of the effect of APOE and DAOA genotypes on AoO, a multiple linear regression analysis was conducted, including both genotypes as predictors.
5. Conclusions
This finding contrasts with previous evidence linking the APOEε4 allele to an increased risk and earlier onset of LOAD, indicating that its effect may differ in populations with a strong genetic background, such as this cohort. This result is particularly intriguing, because APOEε2 has been widely associated with a protective role against AD in sporadic cases.
The variability in AoO across genotypes underscores the complex role of APOE in modulating disease progression, potentially through mechanisms involving lipid metabolism, neuroinflammation, and amyloid processing. These findings suggest that APOEε4 may not always confer an earlier onset in certain genetic backgrounds, warranting further investigation into its potential modifying effects. However, these findings align with the hypothesis of antagonistic pleiotropy, where APOEε4 provides early-life advantages at the cost of late-life neurodegeneration. Moreover, the DAOA A/A genotype was found to delay the age of symptom onset by approximately 2 years, suggesting a possible protective influence. This effect may be mediated by DAOA’s role in modulating D-Serine levels, a key co-agonist of the NMDAR involved in glutamatergic neurotransmission, synaptic plasticity, and neurotoxicity.
Limitations and Future Directions
Despite the significance of our findings, several limitations must be acknowledged. First, our study is limited by the relatively small sample size, which may impact the generalizability of our results. Additionally, the cohort consists of individuals from a genetically homogeneous Mexican founder population, all carrying the PSEN1 A431E variant. While this homogeneity likely enhances our ability to detect genotype–phenotype associations by reducing background genetic noise, it also restricts the external validity of our results. Specifically, the observed associations may not be generalizable to individuals of different ethnic backgrounds, to sporadic forms of AD, or to carriers of other PSEN1 variants. For example, we observed that there is only one APOEε4/ε4 carrier in our cohort. This limits our ability to explore or interpret potential dose-dependent effects of the ε4 allele, which have been reported in other contexts to be associated with an earlier age at onset and increased disease severity. Larger cohorts are necessary to validate our findings and further investigate genotype-specific effects. However, as far as we know, this is the second largest population worldwide with AD-EOAD, which is considered an ultra-rare disease. Second, our study primarily focuses on AoO and does not comprehensively assess other clinical parameters, such as cognitive trajectories or biomarker progression. Longitudinal studies that incorporate multimodal biomarker assessments will be essential to elucidate the broader implications of the APOE and DAOA genotypes in AD-EOAD.
Additionally, while we propose potential mechanisms underlying the differential effects of APOE and DAOA genotypes in AD-EOAD, our study lacks the direct molecular or neuropathological data necessary to confirm these hypotheses. Future research should integrate functional studies examining these pathways, particularly in the context of PSEN1 variants, to better understand their combined impact on disease progression.
Author Contributions
Conceptualization, C.A.V.-G. and L.E.F.; methodology, C.A.V.-G., F.R.-L., and M.P.G.-A.; figures, A.G.-R.; writing—original draft preparation, C.A.V.-G., F.R.-L., and A.G.-R.; reviewing, L.E.F., S.D.-P., and J.M.R.; supervision L.E.F., M.P.G.-A., and S.D.-P.; investigation, C.A.V.-G.; medical diagnosis, L.E.F., S.D.-P., and J.M.R.; funding acquisition, L.E.F. and J.M.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by the National Institutes of Health/Fogarty International (R01 AG069013) and the Fundación IMSS with the Project Registration No. R-2022-1305-100.
Institutional Review Board Statement
Blood samples were obtained for diagnostic analysis after receiving ethical approval from the Ethics Committee of the Instituto Mexicano del Seguro Social (Project Registration No. R-2022-1305-100).
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
The data supporting the findings of this study can be obtained from the corresponding author upon reasonable request. Access to the data is restricted due to privacy and ethical considerations.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AD | Alzheimer’s Disease |
| Aβ | Amyloid-beta protein |
| NFTs | Neurofibrillary tangles |
| PET | Positron Emission Tomography |
| LOAD | Late-Onset Alzheimer’s Disease |
| EOAD | Early-Onset Alzheimer’s Disease |
| AD-EOAD | Autosomal-Dominant Early-Onset Alzheimer’s Disease |
| PSEN1 | Presenilin 1 gene |
| PS1 | Presenilin 1 |
| PSEN2 | Presenilin 2 gene |
| APP | Amyloid Precursor Protein gene |
| AoO | Age of Onset |
| APOE | Apolipoprotein E gene |
| ApoE | Apolipoprotein E |
| LDLr | Low-density lipoprotein receptor |
| LRP1 | LDL Receptor-Related Protein 1 |
| DAOA | D-Amino oxidase activator |
| DAAO | D-Amino oxidase |
| NMDA | N-methyl-D-aspartate |
| NMDAR | N-methyl-D-aspartate receptor |
| SR | Serine racemase |
| FCM | First Clinical Manifestation |
| SNP | Single-nucleotide polymorphism |
| SD | Standard deviation |
| IQ | Intelligence quotient |
| MMSE | Mini-mental state exam |
| NINCDS | National Institute of Neurological Disorders and Stroke |
| ADRDA | Alzheimer’s Disease and Related Disorders Association |
| IMSS | Instituto Mexicano del Seguro Social |
| ASC-1/ASCT-1 | Alanine–Serine–Cysteine Transporter 1 |
| PCR | Polymerase Chain Reaction |
References
- Alzheimer’s Association Report. 2024 Alzheimer’s Disease Facts and Figures. Alzheimers Dement. 2024, 20, 3708–3821. [Google Scholar] [CrossRef]
- Crous-Bou, M.; Minguillón, C.; Gramunt, N.; Molinuevo, J.L. Alzheimer’s Disease Prevention: From Risk Factors to Early Intervention. Alzheimers Res. Ther. 2017, 9, 71. [Google Scholar] [CrossRef]
- Pini, L.; Pievani, M.; Bocchetta, M.; Altomare, D.; Bosco, P.; Cavedo, E.; Galluzzi, S.; Marizzoni, M.; Frisoni, G.B. Brain Atrophy in Alzheimer’s Disease and Aging. Ageing Res. Rev. 2016, 30, 25–48. [Google Scholar] [CrossRef] [PubMed]
- Madnani, R.S. Alzheimer’s Disease: A Mini-Review for the Clinician. Front. Neurol. 2023, 14, 1178588. [Google Scholar] [CrossRef] [PubMed]
- DeTure, M.A.; Dickson, D.W. The Neuropathological Diagnosis of Alzheimer’s Disease. Mol. Neurodegener. 2019, 14, 32. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Sisodia, S.S.; Price, D.L. Neurofibrillary Tangles and β-Amyloid Deposits in Alzheimer’s Disease. Curr. Opin. Neurobiol. 1991, 1, 441–447. [Google Scholar] [CrossRef]
- Govindpani, K.; McNamara, L.G.; Smith, N.R.; Vinnakota, C.; Waldvogel, H.J.; Faull, R.L.; Kwakowsky, A. Vascular Dysfunction in Alzheimer’s Disease: A Prelude to the Pathological Process or a Consequence of It? J. Clin. Med. 2019, 8, 651. [Google Scholar] [CrossRef]
- Mendez, M.F. Early-Onset Alzheimer Disease. Neurol. Clin. 2017, 35, 263–281. [Google Scholar] [CrossRef]
- Filley, C.M.; Kelly, J.; Heaton, R.K. Neuropsychologic Features of Early- and Late-Onset Alzheimer’s Disease. Arch. Neurol. 1986, 43, 574–576. [Google Scholar] [CrossRef]
- Tanner, J.A.; Iaccarino, L.; Edwards, L.; Asken, B.M.; Gorno-Tempini, M.L.; Kramer, J.H.; Pham, J.; Perry, D.C.; Possin, K.; Malpetti, M.; et al. Amyloid, Tau and Metabolic PET Correlates of Cognition in Early and Late-Onset Alzheimer’s Disease. Brain 2022, 145, 4489–4505. [Google Scholar] [CrossRef]
- Ringman, J.M.; Monsell, S.; Ng, D.W.; Zhou, Y.; Nguyen, A.; Coppola, G.; Van Berlo, V.; Mendez, M.F.; Tung, S.; Weintraub, S.; et al. Neuropathology of Autosomal Dominant Alzheimer Disease in the National Alzheimer Coordinating Center Database. J. Neuropathol. Exp. Neurol. 2016, 75, 284–290. [Google Scholar] [CrossRef]
- Mendez, M.F. Early-Onset Alzheimer Disease and Its Variants. Contin. Lifelong Learn. Neurol. 2019, 25, 34–51. [Google Scholar] [CrossRef] [PubMed]
- Sirkis, D.W.; Bonham, L.W.; Johnson, T.P.; La Joie, R.; Yokoyama, J.S. Dissecting the Clinical Heterogeneity of Early-Onset Alzheimer’s Disease. Mol. Psychiatry 2022, 27, 2674–2688. [Google Scholar] [CrossRef] [PubMed]
- Lanoiselée, H.-M.; Nicolas, G.; Wallon, D.; Rovelet-Lecrux, A.; Lacour, M.; Rousseau, S.; Richard, A.-C.; Pasquier, F.; Rollin-Sillaire, A.; Martinaud, O.; et al. APP, PSEN1, and PSEN2 Mutations in Early-Onset Alzheimer Disease: A Genetic Screening Study of Familial and Sporadic Cases. PLoS Med. 2017, 14, e1002270. [Google Scholar] [CrossRef] [PubMed]
- Farrer, L.A. Effects of Age, Sex, and Ethnicity on the Association Between Apolipoprotein E Genotype and Alzheimer Disease: A Meta-Analysis. JAMA 1997, 278, 1349. [Google Scholar] [CrossRef]
- Sando, S.B.; Melquist, S.; Cannon, A.; Hutton, M.L.; Sletvold, O.; Saltvedt, I.; White, L.R.; Lydersen, S.; Aasly, J.O. APOE Ε4 Lowers Age at Onset and Is a High Risk Factor for Alzheimer’s Disease; A Case Control Study from Central Norway. BMC Neurol. 2008, 8, 9. [Google Scholar] [CrossRef]
- Tudorache, I.F.; Trusca, V.G.; Gafencu, A.V. Apolipoprotein E—A Multifunctional Protein with Implications in Various Pathologies as a Result of Its Structural Features. Comput. Struct. Biotechnol. J. 2017, 15, 359–365. [Google Scholar] [CrossRef]
- Hudry, E.; Klickstein, J.; Cannavo, C.; Jackson, R.; Muzikansky, A.; Gandhi, S.; Urick, D.; Sargent, T.; Wrobleski, L.; Roe, A.D.; et al. Opposing Roles of Apolipoprotein E in Aging and Neurodegeneration. Life Sci. Alliance 2019, 2, e201900325. [Google Scholar] [CrossRef]
- Corder, E.H.; Saunders, A.M.; Risch, N.J.; Strittmatter, W.J.; Schmechel, D.E.; Gaskell, P.C.; Rimmler, J.B.; Locke, P.A.; Conneally, P.M.; Schmader, K.E. Protective Effect of Apolipoprotein E Type 2 Allele for Late Onset Alzheimer Disease. Nat. Genet. 1994, 7, 180–184. [Google Scholar] [CrossRef]
- Keene, C.D.; Cudaback, E.; Li, X.; Montine, K.S.; Montine, T.J. Apolipoprotein E Isoforms and Regulation of the Innate Immune Response in Brain of Patients with Alzheimer’s Disease. Curr. Opin. Neurobiol. 2011, 21, 920–928. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, L.-M.; Wu, J. Cross-Talk between Apolipoprotein E and Cytokines. Mediators Inflamm. 2011, 2011, 949072. [Google Scholar] [CrossRef]
- Valdez-Gaxiola, C.A.; Rosales-Leycegui, F.; Gaxiola-Rubio, A.; Moreno-Ortiz, J.M.; Figuera, L.E. Early- and Late-Onset Alzheimer’s Disease: Two Sides of the Same Coin? Diseases 2024, 12, 110. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-H.; Yang, H.-T.; Chiu, C.-C.; Lane, H.-Y. Blood Levels of D-Amino Acid Oxidase vs. D-Amino Acids in Reflecting Cognitive Aging. Sci. Rep. 2017, 7, 14849. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-H.; Chiu, C.-C.; Huang, C.-H.; Yang, H.-T.; Lane, H.-Y. pLG72 Levels Increase in Early Phase of Alzheimer’s Disease but Decrease in Late Phase. Sci. Rep. 2019, 9, 13221. [Google Scholar] [CrossRef] [PubMed]
- Piubelli, L.; Murtas, G.; Rabattoni, V.; Pollegioni, L. The Role of D-Amino Acids in Alzheimer’s Disease. J. Alzheimers Dis. 2021, 80, 475–492. [Google Scholar] [CrossRef]
- Seckler, J.M.; Lewis, S.J. Advances in D-Amino Acids in Neurological Research. Int. J. Mol. Sci. 2020, 21, 7325. [Google Scholar] [CrossRef]
- Su, W.; Zhu, T.; Xu, L.; Wei, Y.; Zeng, B.; Zhang, T.; Cui, H.; Wang, J.; Jia, Y.; Wang, J.; et al. Effect of DAOA Genetic Variation on White Matter Alteration in Corpus Callosum in Patients with First-Episode Schizophrenia. Brain Imaging Behav. 2021, 15, 1748–1759. [Google Scholar] [CrossRef]
- Cheng, L.; Hattori, E.; Nakajima, A.; Woehrle, N.S.; Opal, M.D.; Zhang, C.; Grennan, K.; Dulawa, S.C.; Tang, Y.-P.; Gershon, E.S.; et al. Expression of the G72/G30 Gene in Transgenic Mice Induces Behavioral Changes. Mol. Psychiatry 2014, 19, 175–183. [Google Scholar] [CrossRef]
- Cheng, Y.-J.; Lin, C.-H.; Lane, H.-Y. D-Amino Acids and pLG72 in Alzheimer’s Disease and Schizophrenia. Int. J. Mol. Sci. 2021, 22, 10917. [Google Scholar] [CrossRef]
- Pollegioni, L.; Piubelli, L.; Molla, G.; Rosini, E. D-Amino Acid Oxidase-pLG72 Interaction and D-Serine Modulation. Front. Mol. Biosci. 2018, 5, 3. [Google Scholar] [CrossRef]
- De Strooper, B.; Saftig, P.; Craessaerts, K.; Vanderstichele, H.; Guhde, G.; Annaert, W.; Von Figura, K.; Van Leuven, F. Deficiency of Presenilin-1 Inhibits the Normal Cleavage of Amyloid Precursor Protein. Nature 1998, 391, 387–390. [Google Scholar] [CrossRef]
- Haass, C.; Selkoe, D.J. Soluble Protein Oligomers in Neurodegeneration: Lessons from the Alzheimer’s Amyloid β-Peptide. Nat. Rev. Mol. Cell Biol. 2007, 8, 101–112. [Google Scholar] [CrossRef]
- Wolfe, M.S. Dysfunctional γ-Secretase in Familial Alzheimer’s Disease. Neurochem. Res. 2019, 44, 5–11. [Google Scholar] [CrossRef]
- Wu, L.; Rosa-Neto, P.; Hsiung, G.-Y.R.; Sadovnick, A.D.; Masellis, M.; Black, S.E.; Jia, J.; Gauthier, S. Early-Onset Familial Alzheimer’s Disease (EOFAD). Can. J. Neurol. Sci. J. Can. Sci. Neurol. 2012, 39, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Murrell, J.; Ghetti, B.; Cochran, E.; Macias-Islas, M.A.; Medina, L.; Varpetian, A.; Cummings, J.L.; Mendez, M.F.; Kawas, C.; Chui, H.; et al. The A431E Mutation in PSEN1 Causing Familial Alzheimer’s Disease Originating in Jalisco State, Mexico: An Additional Fifteen Families. Neurogenetics 2006, 7, 277–279. [Google Scholar] [CrossRef] [PubMed]
- Dumois-Petersen, S.; Gallegos-Arreola, M.P.; Magaña-Torres, M.T.; Perea-Díaz, F.J.; Ringman, J.M.; Figuera, L.E. Autosomal Dominant Early Onset Alzheimer’s Disease in the Mexican State of Jalisco: High Frequency of the Mutation PSEN1 c. 1292C >A and Phenotypic Profile of Patients. Am. J. Med. Genet. C Semin. Med. Genet. 2020, 184, 1023–1029. [Google Scholar] [CrossRef]
- Yescas, P.; Huertas-Vazquez, A.; Villarreal-Molina, M.T.; Rasmussen, A.; Tusié-Luna, M.T.; López, M.; Canizales-Quinteros, S.; Alonso, M.E. Founder Effect for the Ala431Glu Mutation of the Presenilin 1 Gene Causing Early-Onset Alzheimer’s Disease in Mexican Families. Neurogenetics 2006, 7, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Rogaeva, E. The Solved and Unsolved Mysteries of the Genetics of Early-Onset Alzheimer ’s Disease. NeuroMolecular Med. 2002, 2, 1–10. [Google Scholar] [CrossRef]
- De Luca, V.; Orfei, M.D.; Gaudenzi, S.; Caltagirone, C.; Spalletta, G. Inverse Effect of the APOE Epsilon4 Allele in Late- and Early-Onset Alzheimer’s Disease. Eur. Arch. Psychiatry Clin. Neurosci. 2016, 266, 599–606. [Google Scholar] [CrossRef]
- Vélez, J.I.; Lopera, F.; Sepulveda-Falla, D.; Patel, H.R.; Johar, A.S.; Chuah, A.; Tobón, C.; Rivera, D.; Villegas, A.; Cai, Y.; et al. APOE*E2 Allele Delays Age of Onset in PSEN1 E280A Alzheimer’s Disease. Mol. Psychiatry 2016, 21, 916–924. [Google Scholar] [CrossRef]
- Polsinelli, A.J.; Lane, K.A.; Manchella, M.K.; Logan, P.E.; Gao, S.; Apostolova, L.G. APOE Ε4 Is Associated with Earlier Symptom Onset in LOAD but Later Symptom Onset in EOAD. Alzheimers Dement. 2023, 19, 2212–2217. [Google Scholar] [CrossRef]
- Van Der Vlies, A.E.; Pijnenburg, Y.A.L.; Koene, T.; Klein, M.; Kok, A.; Scheltens, P.; Van Der Flier, W.M. Cognitive Impairment in Alzheimer’s Disease Is Modified by APOE Genotype. Dement. Geriatr. Cogn. Disord. 2007, 24, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Van Der Vlies, A.E.; Koedam, E.L.G.E.; Pijnenburg, Y.A.L.; Twisk, J.W.R.; Scheltens, P.; Van Der Flier, W.M. Most Rapid Cognitive Decline in APOE Ε4 Negative Alzheimer’s Disease with Early Onset. Psychol. Med. 2009, 39, 1907–1911. [Google Scholar] [CrossRef] [PubMed]
- Smits, L.L.; Pijnenburg, Y.A.L.; Van Der Vlies, A.E.; Koedam, E.L.G.E.; Bouwman, F.H.; Reuling, I.E.W.; Scheltens, P.; Van Der Flier, W.M. Early Onset APOE E4-Negative Alzheimer’s Disease Patients Show Faster Cognitive Decline on Non-Memory Domains. Eur. Neuropsychopharmacol. 2015, 25, 1010–1017. [Google Scholar] [CrossRef]
- Dong, L.; Li, J.; Liu, C.; Mao, C.; Wang, J.; Lei, D.; Huang, X.; Chu, S.; Hou, B.; Feng, F.; et al. Effects of ApoE Genotype on Clinical Phenotypes in Early-onset and Late-onset Alzheimer’s Disease in China: Data from the PUMCH Dementia Cohort. Brain Behav. 2021, 11, e2373. [Google Scholar] [CrossRef]
- Pastor, P.; Roe, C.M.; Villegas, A.; Bedoya, G.; Chakraverty, S.; García, G.; Tirado, V.; Norton, J.; Ríos, S.; Martínez, M.; et al. Apolipoprotein Eε4 modifies Alzheimer’s disease onset in an E280A PS1 kindred. Ann. Neurol. 2003, 54, 163–169. [Google Scholar] [CrossRef]
- Ryman, D.C.; Acosta-Baena, N.; Aisen, P.S.; Bird, T.; Danek, A.; Fox, N.C.; Goate, A.; Frommelt, P.; Ghetti, B.; Langbaum, J.B.S.; et al. Symptom Onset in Autosomal Dominant Alzheimer Disease: A Systematic Review and Meta-Analysis. Neurology 2014, 83, 253–260. [Google Scholar] [CrossRef]
- Valdez-Gaxiola, C.A.; Maciel-Cruz, E.J.; Hernández-Peña, R.; Dumois-Petersen, S.; Rosales-Leycegui, F.; Gallegos-Arreola, M.P.; Moreno-Ortiz, J.M.; Figuera, L.E. Potential Modifying Effect of the APOEε4 Allele on Age of Onset and Clinical Manifestations in Patients with Early-Onset Alzheimer’s Disease with and without a Pathogenic Variant in PSEN1 in a Sample of the Mexican Population. Int. J. Mol. Sci. 2023, 24, 15687. [Google Scholar] [CrossRef]
- Dhana, K.; Aggarwal, N.T.; Rajan, K.B.; Barnes, L.L.; Evans, D.A.; Morris, M.C. Impact of the Apolipoprotein E Ε4 Allele on the Relationship Between Healthy Lifestyle and Cognitive Decline: A Population-Based Study. Am. J. Epidemiol. 2021, 190, 1225–1233. [Google Scholar] [CrossRef]
- Liu, C.-C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer Disease: Risk, Mechanisms and Therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef]
- Raber, J.; Huang, Y.; Ashford, J.W. ApoE Genotype Accounts for the Vast Majority of AD Risk and AD Pathology. Neurobiol. Aging 2004, 25, 641–650. [Google Scholar] [CrossRef]
- Coon, K.D.; Myers, A.J.; Craig, D.W.; Webster, J.A.; Pearson, J.V.; Lince, D.H.; Zismann, V.L.; Beach, T.G.; Leung, D.; Bryden, L.; et al. A High-Density Whole-Genome Association Study Reveals That APOE Is the Major Susceptibility Gene for Sporadic Late-Onset Alzheimer’s Disease. J. Clin. Psychiatry 2007, 68, 613–618. [Google Scholar] [CrossRef]
- Van Der Flier, W.M.; Pijnenburg, Y.A.; Fox, N.C.; Scheltens, P. Early-Onset versus Late-Onset Alzheimer’s Disease: The Case of the Missing APOE Ɛ4 Allele. Lancet Neurol. 2011, 10, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Almkvist, O.; Johansson, C.; Laffita-Mesa, J.; Thordardottir, S.; Graff, C. APOE Ε4 Influences Cognitive Decline Positively in APP and Negatively in PSEN1 Mutation Carriers with Autosomal-dominant Alzheimer’s Disease. Eur. J. Neurol. 2022, 29, 3580–3589. [Google Scholar] [CrossRef] [PubMed]
- Lendon, C.L.; Martinez, A.; Behrens, I.M.; Kosik, K.S.; Madrigal, L.; Norton, J.; Neuman, R.; Myers, A.; Busfield, F.; Wragg, M.; et al. E280A PS-1 mutation causes Alzheimer’s disease but age of onset is not modified by ApoE alleles. Human Mutation. 1997, 3, 186–195. [Google Scholar] [CrossRef]
- Langella, S.; Barksdale, N.G.; Vasquez, D.; Aguillon, D.; Chen, Y.; Su, Y.; Acosta-Baena, N.; Acosta-Uribe, J.; Baena, A.Y.; Garcia-Ospina, G.; et al. Effect of Apolipoprotein Genotype and Educational Attainment on Cognitive Function in Autosomal Dominant Alzheimer’s Disease. Nat. Commun. 2023, 14, 5120. [Google Scholar] [CrossRef]
- Cordy, J.M.; Hussain, I.; Dingwall, C.; Hooper, N.M.; Turner, A.J. Exclusively Targeting β-Secretase to Lipid Rafts by GPI-Anchor Addition up-Regulates β-Site Processing of the Amyloid Precursor Protein. Proc. Natl. Acad. Sci. USA 2003, 100, 11735–11740. [Google Scholar] [CrossRef]
- Mitteldorf, J. What Is Antagonistic Pleiotropy? Biochem. Mosc. 2019, 84, 1458–1468. [Google Scholar] [CrossRef]
- Yu, Y.W.-Y.; Lin, C.-H.; Chen, S.-P.; Hong, C.-J.; Tsai, S.-J. Intelligence and Event-Related Potentials for Young Female Human Volunteer Apolipoprotein E Ε4 and Non-Ε4 Carriers. Neurosci. Lett. 2000, 294, 179–181. [Google Scholar] [CrossRef]
- Mondadori, C.R.A.; De Quervain, D.J.-F.; Buchmann, A.; Mustovic, H.; Wollmer, M.A.; Schmidt, C.F.; Boesiger, P.; Hock, C.; Nitsch, R.M.; Papassotiropoulos, A.; et al. Better Memory and Neural Efficiency in Young Apolipoprotein E 4 Carriers. Cereb. Cortex 2007, 17, 1934–1947. [Google Scholar] [CrossRef]
- Wright, R.O.; Hu, H.; Silverman, E.K.; Tsaih, S.W.; Schwartz, J.; Bellinger, D.; Palazuelos, E.; Weiss, S.T.; Hernandez-Avila, M. Apolipoprotein E Genotype Predicts 24-Month Bayley Scales Infant Development Score. Pediatr. Res. 2003, 54, 819–825. [Google Scholar] [CrossRef]
- Keltikangas-Järvinen, L.; Räikkönen, K.; Lehtimäki, T. Dependence between Apolipoprotein E Phenotypes and Temperament in Children, Adolescents, and Young Adults. Psychosom. Med. 1993, 55, 155–163. [Google Scholar] [CrossRef]
- Tuminello, E.R.; Han, S.D. The Apolipoprotein E Antagonistic Pleiotropy Hypothesis: Review and Recommendations. Int. J. Alzheimer’s Dis. 2011, 2011, 726197. [Google Scholar] [CrossRef]
- Han, S.D.; Bondi, M.W. Revision of the Apolipoprotein E Compensatory Mechanism Recruitment Hypothesis. Alzheimers Dement. 2008, 4, 251–254. [Google Scholar] [CrossRef]
- Fernandez, C.G.; Hamby, M.E.; McReynolds, M.L.; Ray, W.J. The Role of APOE4 in Disrupting the Homeostatic Functions of Astrocytes and Microglia in Aging and Alzheimer’s Disease. Front. Aging Neurosci. 2019, 11, 14. [Google Scholar] [CrossRef]
- Lee, S.; Devanney, N.A.; Golden, L.R.; Smith, C.T.; Schwartz, J.L.; Walsh, A.E.; Clarke, H.A.; Goulding, D.S.; Allenger, E.J.; Morillo-Segovia, G.; et al. APOE Modulates Microglial Immunometabolism in Response to Age, Amyloid Pathology, and Inflammatory Challenge. Cell Rep. 2023, 42, 112196. [Google Scholar] [CrossRef] [PubMed]
- Madeira, C.; Lourenco, M.V.; Vargas-Lopes, C.; Suemoto, C.K.; Brandão, C.O.; Reis, T.; Leite, R.E.P.; Laks, J.; Jacob-Filho, W.; Pasqualucci, C.A.; et al. D-Serine Levels in Alzheimer’s Disease: Implications for Novel Biomarker Development. Transl. Psychiatry 2015, 5, e561. [Google Scholar] [CrossRef] [PubMed]
- Sacchi, S.; Cappelletti, P.; Pirone, L.; Smaldone, G.; Pedone, E.; Pollegioni, L. Elucidating the Role of the pLG72 R30K Substitution in Schizophrenia Susceptibility. FEBS Lett. 2017, 591, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A Simple Salting out Procedure for Extracting DNA from Human Nucleated Cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef]
- Cámara de Diputados. Reglamento de La Ley General de Salud en Materia de Investigación Para La Salud. DOF 02-04-2014. Available online: https://www.diputados.gob.mx/LeyesBiblio/regley/Reg_LGS_MIS.pdf (accessed on 9 July 2025).
- World Medical Association. Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects. 2024. Available online: https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/ (accessed on 9 July 2025).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).