Bioinformatic Analysis of the Leptin–Ob-R Interface: Structural Modeling, Thermodynamic Profiling, and Stability in Diverse Microenvironments
Abstract
1. Introduction
2. Results and Discussion
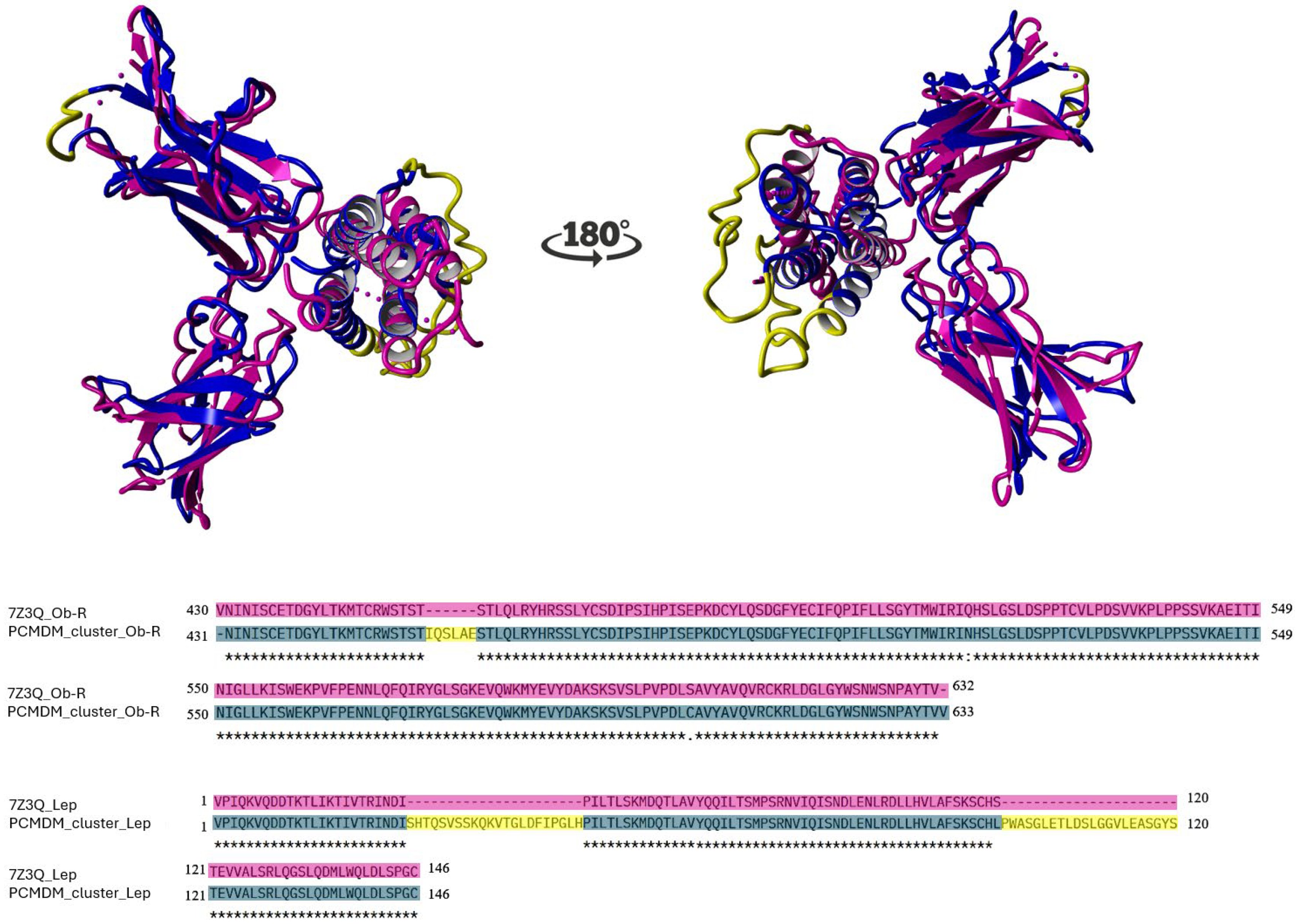
2.1. Sequence Conservation and Structural Consistency
2.2. Interface Variation Across Species
2.3. Analysis of the Stability of Leptin–CRH2 Complexes
2.4. Effects of Physicochemical Conditions on Leptin–CRH2 Complex Stability
2.4.1. Effect of pH on Binding Stability
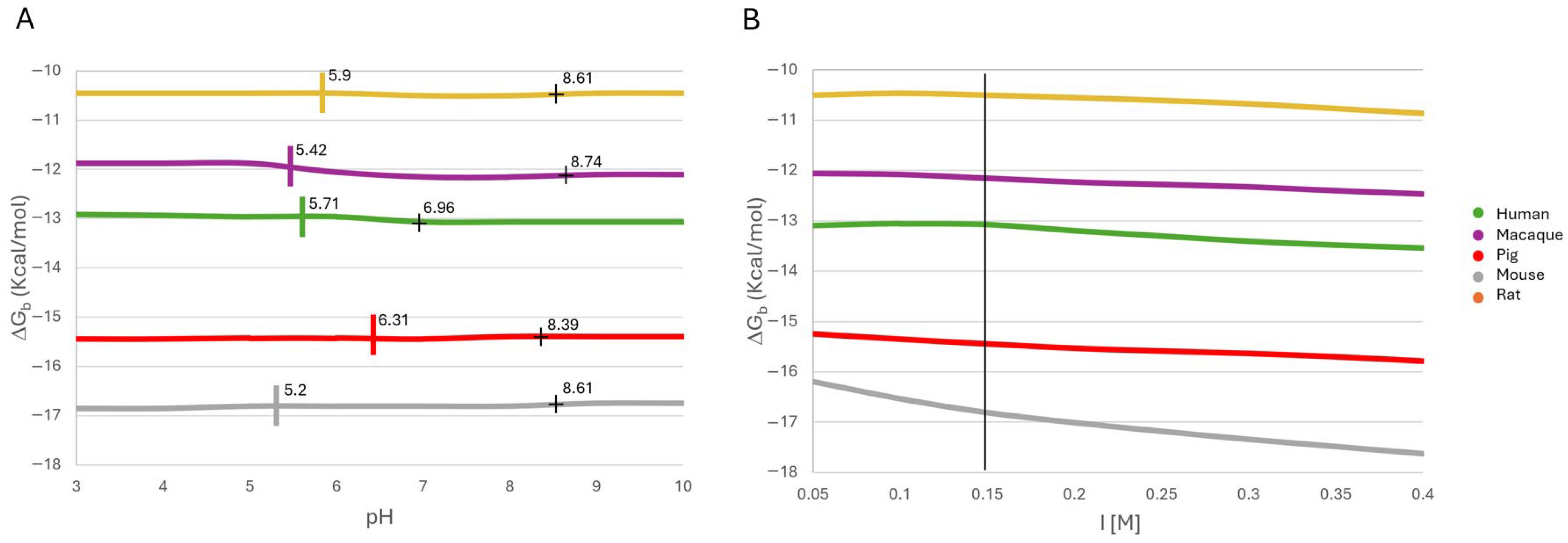
2.4.2. Effect of Ionic Strength on Binding Stability
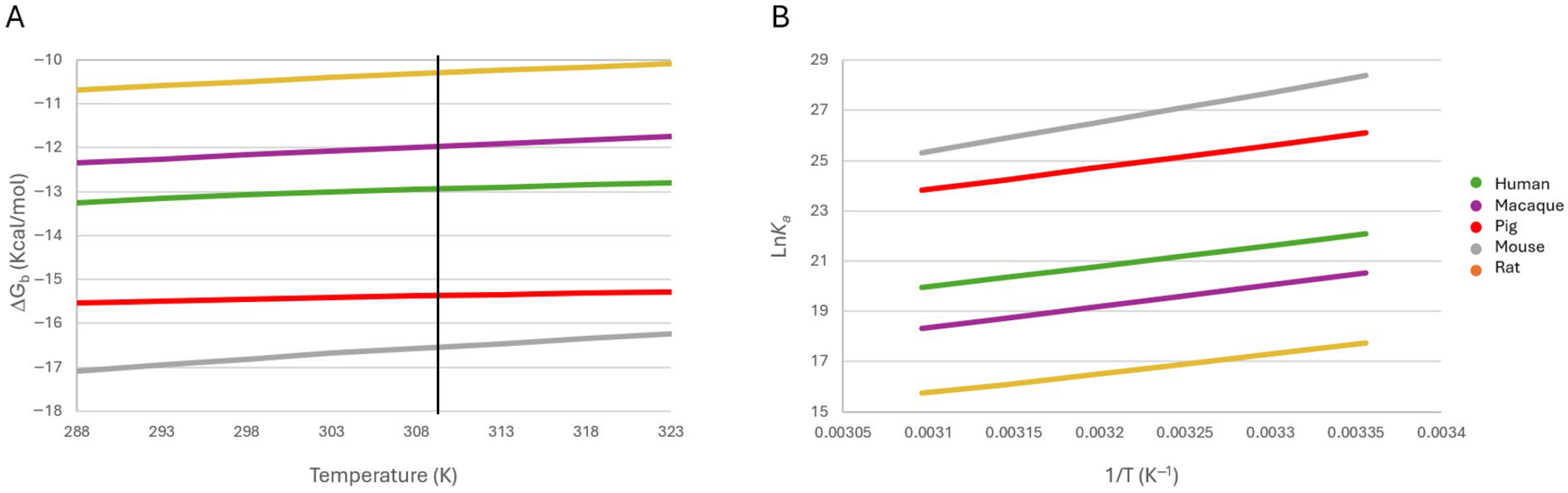
2.4.3. Effect of Temperature on Binding Stability
2.5. Physiological and Pathological Implications
2.6. Thermodynamic Analysis
3. Materials and Methods
3.1. Selection and Preparation of Homologous Sequences
3.2. Model Construction and Structural Optimization
3.3. Interface Characterization
3.4. Binding Energy Prediction and Thermodynamic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sáinz, N.; González-Navarro, C.J.; Martínez, J.A.; Moreno-Aliaga, M.J. Leptin Signaling as a Therapeutic Target of Obesity. Expert Opin. Ther. Targets 2015, 19, 893–909. [Google Scholar] [CrossRef]
- La Cava, A. Leptin in Inflammation and Autoimmunity. Cytokine 2017, 98, 51–58. [Google Scholar] [CrossRef]
- Wauman, J.; Zabeau, L.; Tavernier, J. The Leptin Receptor Complex: Heavier Than Expected? Front. Endocrinol. 2017, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, I.; Bado, A.; Vissuzaine, C.; Buyse, M.; Kermorgant, S.; Laigneau, J.P.; Attoub, S.; Lehy, T.; Henin, D.; Mignon, M.; et al. Leptin Secretion and Leptin Receptor in the Human Stomach. Gut 2000, 47, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Solberg, R.; Aas, V.; Thoresen, G.H.; Kase, E.T.; Drevon, C.A.; Rustan, A.C.; Reseland, J.E. Leptin Expression in Human Primary Skeletal Muscle Cells Is Reduced during Differentiation. J. Cell. Biochem. 2005, 96, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Masuzaki, H.; Ogawa, Y.; Isse, N.; Satoh, N.; Okazaki, T.; Shigemoto, M.; Mori, K.; Tamura, N.; Hosoda, K.; Yoshimasa, Y. Human Obese Gene Expression. Adipocyte-Specific Expression and Regional Differences in the Adipose Tissue. Diabetes 1995, 44, 855–858. [Google Scholar] [CrossRef]
- Moharana, K.; Zabeau, L.; Peelman, F.; Ringler, P.; Stahlberg, H.; Tavernier, J.; Savvides, S.N. Structural and Mechanistic Paradigm of Leptin Receptor Activation Revealed by Complexes with Wild-Type and Antagonist Leptins. Structure 2014, 22, 866–877. [Google Scholar] [CrossRef]
- Tartaglia, L.A.; Dembski, M.; Weng, X.; Deng, N.; Culpepper, J.; Devos, R.; Richards, G.J.; Campfield, L.A.; Clark, F.T.; Deeds, J.; et al. Identification and Expression Cloning of a Leptin Receptor, OB-R. Cell 1995, 83, 1263–1271. [Google Scholar] [CrossRef]
- Zhang, F.; Basinski, M.B.; Beals, J.M.; Briggs, S.L.; Churgay, L.M.; Clawson, D.K.; DiMarchi, R.D.; Furman, T.C.; Hale, J.E.; Hsiung, H.M.; et al. Crystal Structure of the Obese Protein Ieptin-E100. Nature 1997, 387, 206–209. [Google Scholar] [CrossRef]
- Haglund, E.; Sułkowska, J.I.; He, Z.; Feng, G.-S.; Jennings, P.A.; Onuchic, J.N. The Unique Cysteine Knot Regulates the Pleotropic Hormone Leptin. PLoS ONE 2012, 7, e45654. [Google Scholar] [CrossRef]
- Haglund, E.; Sulkowska, J.I.; Noel, J.K.; Lammert, H.; Onuchic, J.N.; Jennings, P.A. Pierced Lasso Bundles Are a New Class of Knot-like Motifs. PLOS Comput. Biol. 2014, 10, e1003613. [Google Scholar] [CrossRef] [PubMed]
- Haglund, E.; Pilko, A.; Wollman, R.; Jennings, P.A.; Onuchic, J.N. Pierced Lasso Topology Controls Function in Leptin. J. Phys. Chem. B 2017, 121, 706–718. [Google Scholar] [CrossRef] [PubMed]
- Haglund, E.; Nguyen, L.; Schafer, N.P.; Lammert, H.; Jennings, P.A.; Onuchic, J.N. Uncovering the Molecular Mechanisms behind Disease-Associated Leptin Variants. J. Biol. Chem. 2018, 293, 12919–12933. [Google Scholar] [CrossRef] [PubMed]
- Peelman, F.; Iserentant, H.; Smet, A.-S.D.; Vandekerckhove, J.; Zabeau, L.; Tavernier, J. Mapping of Binding Site III in the Leptin Receptor and Modeling of a Hexameric Leptin·Leptin Receptor Complex *. J. Biol. Chem. 2006, 281, 15496–15504. [Google Scholar] [CrossRef]
- Peelman, F.; Beneden, K.V.; Zabeau, L.; Iserentant, H.; Ulrichts, P.; Defeau, D.; Verhee, A.; Catteeuw, D.; Elewaut, D.; Tavernier, J. Mapping of the Leptin Binding Sites and Design of a Leptin Antagonist *. J. Biol. Chem. 2004, 279, 41038–41046. [Google Scholar] [CrossRef]
- Mancour, L.V.; Daghestani, H.N.; Dutta, S.; Westfield, G.H.; Schilling, J.; Oleskie, A.N.; Herbstman, J.F.; Chou, S.Z.; Skiniotis, G. Ligand-Induced Architecture of the Leptin Receptor Signaling Complex. Mol. Cell 2012, 48, 655–661. [Google Scholar] [CrossRef]
- Saxton, R.A.; Caveney, N.A.; Moya-Garzon, M.D.; Householder, K.D.; Rodriguez, G.E.; Burdsall, K.A.; Long, J.Z.; Garcia, K.C. Structural Insights into the Mechanism of Leptin Receptor Activation. Nat. Commun. 2023, 14, 1797. [Google Scholar] [CrossRef]
- Hiroike, T.; Higo, J.; Jingami, H.; Toh, H. Homology Modeling of Human Leptin/Leptin Receptor Complex. Biochem. Biophys. Res. Commun. 2000, 275, 154–158. [Google Scholar] [CrossRef]
- Iserentant, H.; Peelman, F.; Defeau, D.; Vandekerckhove, J.; Zabeau, L.; Tavernier, J. Mapping of the Interface between Leptin and the Leptin Receptor CRH2 Domain. J. Cell Sci. 2005, 118, 2519–2527. [Google Scholar] [CrossRef]
- Heintzman, D.R.; Fisher, E.L.; Rathmell, J.C. Microenvironmental Influences on T Cell Immunity in Cancer and Inflammation. Cell. Mol. Immunol. 2022, 19, 316–326. [Google Scholar] [CrossRef]
- Gomez-Pinilla, F.; Myers, S.K. Traumatic Brain Injury from a Peripheral Axis Perspective: Uncovering the Roles of Liver and Adipose Tissue in Temperature Regulation. Prog. Neurobiol. 2025, 247, 102733. [Google Scholar] [CrossRef]
- Möller, J.; Schroer, M.A.; Erlkamp, M.; Grobelny, S.; Paulus, M.; Tiemeyer, S.; Wirkert, F.J.; Tolan, M.; Winter, R. The Effect of Ionic Strength, Temperature, and Pressure on the Interaction Potential of Dense Protein Solutions: From Nonlinear Pressure Response to Protein Crystallization. Biophys. J. 2012, 102, 2641–2648. [Google Scholar] [CrossRef]
- Honrath, B.; Krabbendam, I.E.; Culmsee, C.; Dolga, A.M. Small Conductance Ca2+-Activated K+ Channels in the Plasma Membrane, Mitochondria and the ER: Pharmacology and Implications in Neuronal Diseases. Neurochem. Int. 2017, 109, 13–23. [Google Scholar] [CrossRef]
- de Almeida, A.N.S.; de Souza Ferreira, S.L.; Balata, P.M.M.; da Cunha, D.A.; Pernambuco, L.; da Silva, H.J. Thermography in Complementary Assessments of Head and Neck Muscles: A Scoping Review. J. Oral. Rehabil. 2022, 49, 1188–1196. [Google Scholar] [CrossRef] [PubMed]
- Bedford, J.M. Effects of Elevated Temperature on the Epididymis and Testis: Experimental Studies. In Temperature and Environmental Effects on the Testis; Zorgniotti, A.W., Ed.; Springer: Boston, MA, USA, 1991; pp. 19–32. ISBN 978-1-4684-5913-5. [Google Scholar]
- Chen, W. Thermometry and Interpretation of Body Temperature. Biomed. Eng. Lett. 2019, 9, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Geneva, I.I.; Cuzzo, B.; Fazili, T.; Javaid, W. Normal Body Temperature: A Systematic Review. Open Forum Infect. Dis. 2019, 6, ofz032. [Google Scholar] [CrossRef]
- Chiche, J.; Ilc, K.; Laferrière, J.; Trottier, E.; Dayan, F.; Mazure, N.M.; Brahimi-Horn, M.C.; Pouysségur, J. Hypoxia-Inducible Carbonic Anhydrase IX and XII Promote Tumor Cell Growth by Counteracting Acidosis through the Regulation of the Intracellular pH. Cancer Res. 2008, 69, 358–368. [Google Scholar] [CrossRef]
- Pérez-Pérez, A.; Sánchez-Jiménez, F.; Vilariño-García, T.; Sánchez-Margalet, V. Role of Leptin in Inflammation and Vice Versa. Int. J. Mol. Sci. 2020, 21, 5887. [Google Scholar] [CrossRef]
- Inoue, W.; Luheshi, G.N. Acute Starvation Alters Lipopolysaccharide-Induced Fever in Leptin-Dependent and -Independent Mechanisms in Rats. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2010, 299, R1709–R1719. [Google Scholar] [CrossRef]
- Luheshi, G.N. Leptin and Cytolines:Actions and Interactions in Fever and Appetite Control. In NeuroImmune Biology; Berczi, I., Gorczynski, R.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2001; Volume 1, pp. 283–291. [Google Scholar]
- Londraville, R.L.; Prokop, J.W.; Duff, R.J.; Liu, Q.; Tuttle, M. On the Molecular Evolution of Leptin, Leptin Receptor, and Endospanin. Front. Endocrinol. 2017, 8, 58. [Google Scholar] [CrossRef]
- Tsirigotaki, A.; Dansercoer, A.; Verschueren, K.H.G.; Marković, I.; Pollmann, C.; Hafer, M.; Felix, J.; Birck, C.; Van Putte, W.; Catteeuw, D.; et al. Mechanism of Receptor Assembly via the Pleiotropic Adipokine Leptin. Nat. Struct. Mol. Biol. 2023, 30, 551–563. [Google Scholar] [CrossRef] [PubMed]
- López-Hidalgo, M.; Caro-Gómez, L.A.; Romo-Rodríguez, R.; Herrera-Zuñiga, L.D.; Anaya-Reyes, M.; Rosas-Trigueros, J.L.; Benítez-Cardoza, C.G. Atomistic Mechanism of Leptin and Leptin-Receptor Association. J. Biomol. Struct. Dyn. 2023, 41, 2231–2248. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.; Sander, C. Protein Structure Comparison by Alignment of Distance Matrices. J. Mol. Biol. 1993, 233, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Kufareva, I.; Abagyan, R. Methods of Protein Structure Comparison. In Homology Modeling: Methods and Protocols; Orry, A.J.W., Abagyan, R., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 231–257. ISBN 978-1-61779-588-6. [Google Scholar]
- Vangone, A.; Bonvin, A.M. Contacts-Based Prediction of Binding Affinity in Protein-Protein Complexes. eLife 2015, 4, e07454. [Google Scholar] [CrossRef]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A Web Server for Predicting the Binding Affinity of Protein–Protein Complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef]
- Peelman, F.; Zabeau, L.; Moharana, K.; Savvides, S.N.; Tavernier, J. 20 YEARS OF LEPTIN: Insights into Signaling Assemblies of the Leptin Receptor. J. Endocrinol. 2014, 223, T9–T23. [Google Scholar] [CrossRef]
- Reyes-Espinosa, F.; Juárez-Saldivar, A.; Palos, I.; Herrera-Mayorga, V.; García-Pérez, C.; Rivera, G. In Silico Analysis of Homologous Heterodimers of Cruzipain-Chagasin from Structural Models Built by Homology. Int. J. Mol. Sci. 2019, 20, 1320. [Google Scholar] [CrossRef]
- Hajjar, S.; Zhou, X. pH Sensing at the Intersection of Tissue Homeostasis and Inflammation. Trends Immunol. 2023, 44, 807–825. [Google Scholar] [CrossRef]
- Kato, Y.; Ozawa, S.; Miyamoto, C.; Maehata, Y.; Suzuki, A.; Maeda, T.; Baba, Y. Acidic Extracellular Microenvironment and Cancer. Cancer Cell Int. 2013, 13, 89. [Google Scholar] [CrossRef]
- Rahman, M.A.; Yadab, M.K.; Ali, M.M. Emerging Role of Extracellular pH in Tumor Microenvironment as a Therapeutic Target for Cancer Immunotherapy. Cells 2024, 13, 1924. [Google Scholar] [CrossRef]
- Lee, M.J.; Yaffe, M.B. Protein Regulation in Signal Transduction. Cold Spring Harb. Perspect. Biol. 2016, 8, a005918. [Google Scholar] [CrossRef]
- Jeon, S.; Jeon, Y.; Lim, J.-Y.; Kim, Y.; Cha, B.; Kim, W. Emerging Regulatory Mechanisms and Functions of Biomolecular Condensates: Implications for Therapeutic Targets. Signal Transduct. Target Ther. 2025, 10, 4. [Google Scholar] [CrossRef]
- Hu, H.-J.; Song, M. Disrupted Ionic Homeostasis in Ischemic Stroke and New Therapeutic Targets. J. Stroke Cerebrovasc. Dis. 2017, 26, 2706–2719. [Google Scholar] [CrossRef]
- Yu, S.P.; Canzoniero, L.M.T.; Choi, D.W. Ion Homeostasis and Apoptosis. Curr. Opin. Cell Biol. 2001, 13, 405–411. [Google Scholar] [CrossRef]
- Miotto, M.; Olimpieri, P.P.; Di Rienzo, L.; Ambrosetti, F.; Corsi, P.; Lepore, R.; Tartaglia, G.G.; Milanetti, E. Insights on Protein Thermal Stability: A Graph Representation of Molecular Interactions. Bioinformatics 2019, 35, 2569–2577. [Google Scholar] [CrossRef]
- Faust, O.; Rajendra Acharya, U.; Ng, E.Y.K.; Hong, T.J.; Yu, W. Application of Infrared Thermography in Computer Aided Diagnosis. Infrared Phys. Technol. 2014, 66, 160–175. [Google Scholar] [CrossRef] [PubMed]
- Lubkowska, A.; Chudecka, M. Thermal Characteristics of Breast Surface Temperature in Healthy Women. Int. J. Environ. Res. Public Health 2021, 18, 1097. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, L.d.B.; Prodonoff, J.S.; Favero de Aguiar, C.; Correa-da-Silva, F.; Castoldi, A.; Bakker, N.v.T.; Davanzo, G.G.; Castelucci, B.; Pereira, J.A.d.S.; Curtis, J.; et al. Leptin Signaling Suppression in Macrophages Improves Immunometabolic Outcomes in Obesity. Diabetes 2022, 71, 1546–1561. [Google Scholar] [CrossRef]
- Park, J.; Euhus, D.M.; Scherer, P.E. Paracrine and Endocrine Effects of Adipose Tissue on Cancer Development and Progression. Endocr. Rev. 2011, 32, 550–570. [Google Scholar] [CrossRef]
- Lin, T.-C.; Hsiao, M. Leptin and Cancer: Updated Functional Roles in Carcinogenesis, Therapeutic Niches, and Developments. Int. J. Mol. Sci. 2021, 22, 2870. [Google Scholar] [CrossRef]
- Dessie, G.; Ayelign, B.; Akalu, Y.; Shibabaw, T.; Molla, M.D. Effect of Leptin on Chronic Inflammatory Disorders: Insights to Therapeutic Target to Prevent Further Cardiovascular Complication. Diabetes Metab. Syndr. Obes. 2021, 14, 3307–3322. [Google Scholar] [CrossRef]
- Hachemi, I.; U-Din, M. Brown Adipose Tissue: Activation and Metabolism in Humans. Endocrinol. Metab. 2023, 38, 214–222. [Google Scholar] [CrossRef]
- Llanos, A.A.M.; Aremu, J.B.; Cheng, T.-Y.D.; Chen, W.; Chekmareva, M.A.; Cespedes Feliciano, E.M.; Qin, B.; Lin, Y.; Omene, C.; Khoury, T.; et al. Greater Body Fatness Is Associated with Higher Protein Expression of LEPR in Breast Tumor Tissues: A Cross-Sectional Analysis in the Women’s Circle of Health Study. Front. Endocrinol. 2022, 13, 879164. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher Sequence Analysis Tools Framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo—Distance Constraints Applied on Model Quality Estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef]
- Krissinel, E. On the Relationship between Sequence and Structure Similarities in Proteomics. Bioinformatics 2007, 23, 717–723. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Secondary-Structure Matching (SSM), a New Tool for Fast Protein Structure Alignment in Three Dimensions. Acta Cryst. D 2004, 60, 2256–2268. [Google Scholar] [CrossRef]
- Krieger, E.; Joo, K.; Lee, J.; Lee, J.; Raman, S.; Thompson, J.; Tyka, M.; Baker, D.; Karplus, K. Improving Physical Realism, Stereochemistry and Side-Chain Accuracy in Homology Modeling: Four Approaches That Performed Well in CASP8. Proteins 2009, 77, 114–122. [Google Scholar] [CrossRef]
- Land, H.; Humble, M.S. YASARA: A Tool to Obtain Structural Guidance in Biocatalytic Investigations. In Protein Engineering: Methods and Protocols; Bornscheuer, U.T., Höhne, M., Eds.; Springer: New York, NY, USA, 2018; pp. 43–67. ISBN 978-1-4939-7366-8. [Google Scholar]
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Buß, O.; Rudat, J.; Ochsenreither, K. FoldX as Protein Engineering Tool: Better Than Random Based Approaches? Comput. Struct. Biotechnol. J. 2018, 16, 25–33. [Google Scholar] [CrossRef]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX Web Server: An Online Force Field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]


| Species | *ΔGb (kcal/mol) | *Kd M (× 10−10) | *Ka M−1(× 1010) | $ΔH (kcal/mol) | $-TΔS (kcal/mol) |
|---|---|---|---|---|---|
| Human | −13.17 | 2.54 | 0.392 | −16.28 | 3.11 |
| Mouse | −16.81 | 4612.39 | 216.807 | −23.45 | 6.64 |
| Rat | −10.50 | 0.01 | 0.005 | −15.36 | 4.87 |
| Pig | −15.44 | 462.98 | 21.599 | −17.31 | 1.87 |
| Macaque | −12.152 | 0.12 | 0.082 | −16.93 | 4.78 |
| Human | Temperature (°C) | Ka [M]* | pH | Ka [M]# | I [M] | Ka [M]$ |
|---|---|---|---|---|---|---|
| 25 | 3.93 × 109 | 3 | 3.02 × 109 | 0.05 | 4.09 × 109 | |
| 30 | 2.46 × 109 | 4 | 3.12 × 109 | 0.1 | 3.83 × 109 | |
| 35 | 1.57 × 109 | 5 | 3.27 × 109 | 0.15 | 3.93 × 109 | |
| 40 | 1.03 × 109 | 6 | 3.25 × 109 | 0.2 | 4.81 × 109 | |
| 45 | 6.86 × 108 | 7 | 3.93 × 109 | 0.25 | 5.80 × 109 | |
| 8 | 3.93 × 109 | 0.3 | 6.91 × 109 | |||
| 9 | 3.93 × 109 | 0.35 | 7.84 × 109 | |||
| 10 | 3.93 × 109 | 0.4 | 8.69 × 109 | |||
| Mouse | Temperature (°C) | Ka [M]* | pH | Ka [M]# | I [M] | Ka [M]$ |
| 25 | 2.17 × 1012 | 3 | 2.33 × 1012 | 0.05 | 7.69 × 1011 | |
| 30 | 1.09 × 1012 | 4 | 2.33 × 1012 | 0.1 | 1.36 × 1012 | |
| 35 | 5.76 × 1011 | 5 | 2.17 × 1012 | 0.15 | 2.17 × 1012 | |
| 40 | 3.16 × 1011 | 6 | 2.17 × 1012 | 0.2 | 3.02 × 1012 | |
| 45 | 1.76 × 1011 | 7 | 2.17 × 1012 | 0.25 | 4.08 × 1012 | |
| 8 | 2.17 × 1012 | 0.3 | 5.31 × 1012 | |||
| 9 | 1.97 × 1012 | 0.35 | 6.81 × 1012 | |||
| 10 | 1.97 × 1012 | 0.4 | 8.66 × 1012 | |||
| Rat | Temperature (°C) | Ka [M]* | pH | Ka [M]# | I [M] | Ka [M]$ |
| 25 | 5.09 × 107 | 3 | 4.71 × 107 | 0.05 | 5.13 × 107 | |
| 30 | 3.25 × 107 | 4 | 4.71 × 107 | 0.1 | 4.80 × 107 | |
| 35 | 2.11 × 107 | 5 | 4.71 × 107 | 0.15 | 5.09 × 107 | |
| 40 | 1.42 × 107 | 6 | 4.71 × 107 | 0.2 | 5.53 × 107 | |
| 45 | 9.74 × 106 | 7 | 5.09 × 107 | 0.25 | 6.10 × 107 | |
| 8 | 5.09 × 107 | 0.3 | 6.82 × 107 | |||
| 9 | 4.72 × 107 | 0.35 | 8.02 × 107 | |||
| 10 | 4.72 × 107 | 0.4 | 9.42 × 107 | |||
| Pig | Temperature (°C) | Ka [M]* | pH | Ka [M]# | I [M] | Ka [M]$ |
| 25 | 2.16 × 1011 | 3 | 2.16 × 1011 | 0.05 | 1.54 × 1011 | |
| 30 | 1.31 × 1011 | 4 | 2.16 × 1011 | 0.1 | 1.85 × 1011 | |
| 35 | 8.29 × 1010 | 5 | 2.09 × 1011 | 0.15 | 2.16 × 1011 | |
| 40 | 5.23 × 1010 | 6 | 2.09 × 1011 | 0.2 | 2.51 × 1011 | |
| 45 | 3.37 × 1010 | 7 | 2.16 × 1011 | 0.25 | 2.74 × 1011 | |
| 8 | 1.99 × 1011 | 0.3 | 2.99 × 1011 | |||
| 9 | 1.99 × 1011 | 0.35 | 3.32 × 1011 | |||
| 10 | 1.99 × 1011 | 0.4 | 3.83 × 1011 | |||
| Macaque | Temperature (°C) | Ka [M]* | pH | Ka [M]# | I [M] | Ka [M]$ |
| 25 | 8.28 × 108 | 3 | 5.25 × 108 | 0.05 | 7.07 × 108 | |
| 30 | 5.13 × 108 | 4 | 5.25 × 108 | 0.1 | 7.34 × 108 | |
| 35 | 3.24 × 108 | 5 | 5.25 × 108 | 0.15 | 8.28 × 108 | |
| 40 | 2.08 × 108 | 6 | 7.03 × 108 | 0.2 | 9.45 × 108 | |
| 45 | 1.35 × 108 | 7 | 8.28 × 108 | 0.25 | 1.02 × 109 | |
| 8 | 8.33 × 108 | 0.3 | 1.11 × 109 | |||
| 9 | 7.66 × 108 | 0.35 | 1.25 × 109 | |||
| 10 | 7.66 × 108 | 0.4 | 1.42 × 109 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortega-López, G.I.; Reyes-Espinosa, F.; López-Y-López, V.E.; Benítez-Cardoza, C.G. Bioinformatic Analysis of the Leptin–Ob-R Interface: Structural Modeling, Thermodynamic Profiling, and Stability in Diverse Microenvironments. Int. J. Mol. Sci. 2025, 26, 6955. https://doi.org/10.3390/ijms26146955
Ortega-López GI, Reyes-Espinosa F, López-Y-López VE, Benítez-Cardoza CG. Bioinformatic Analysis of the Leptin–Ob-R Interface: Structural Modeling, Thermodynamic Profiling, and Stability in Diverse Microenvironments. International Journal of Molecular Sciences. 2025; 26(14):6955. https://doi.org/10.3390/ijms26146955
Chicago/Turabian StyleOrtega-López, Gabriel I., Francisco Reyes-Espinosa, Víctor Eric López-Y-López, and Claudia G. Benítez-Cardoza. 2025. "Bioinformatic Analysis of the Leptin–Ob-R Interface: Structural Modeling, Thermodynamic Profiling, and Stability in Diverse Microenvironments" International Journal of Molecular Sciences 26, no. 14: 6955. https://doi.org/10.3390/ijms26146955
APA StyleOrtega-López, G. I., Reyes-Espinosa, F., López-Y-López, V. E., & Benítez-Cardoza, C. G. (2025). Bioinformatic Analysis of the Leptin–Ob-R Interface: Structural Modeling, Thermodynamic Profiling, and Stability in Diverse Microenvironments. International Journal of Molecular Sciences, 26(14), 6955. https://doi.org/10.3390/ijms26146955








