Abscisic Acid Enhances Ex Vitro Acclimatization Performance in Hop (Humulus lupulus L.)
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification of PYLs in H. lupulus
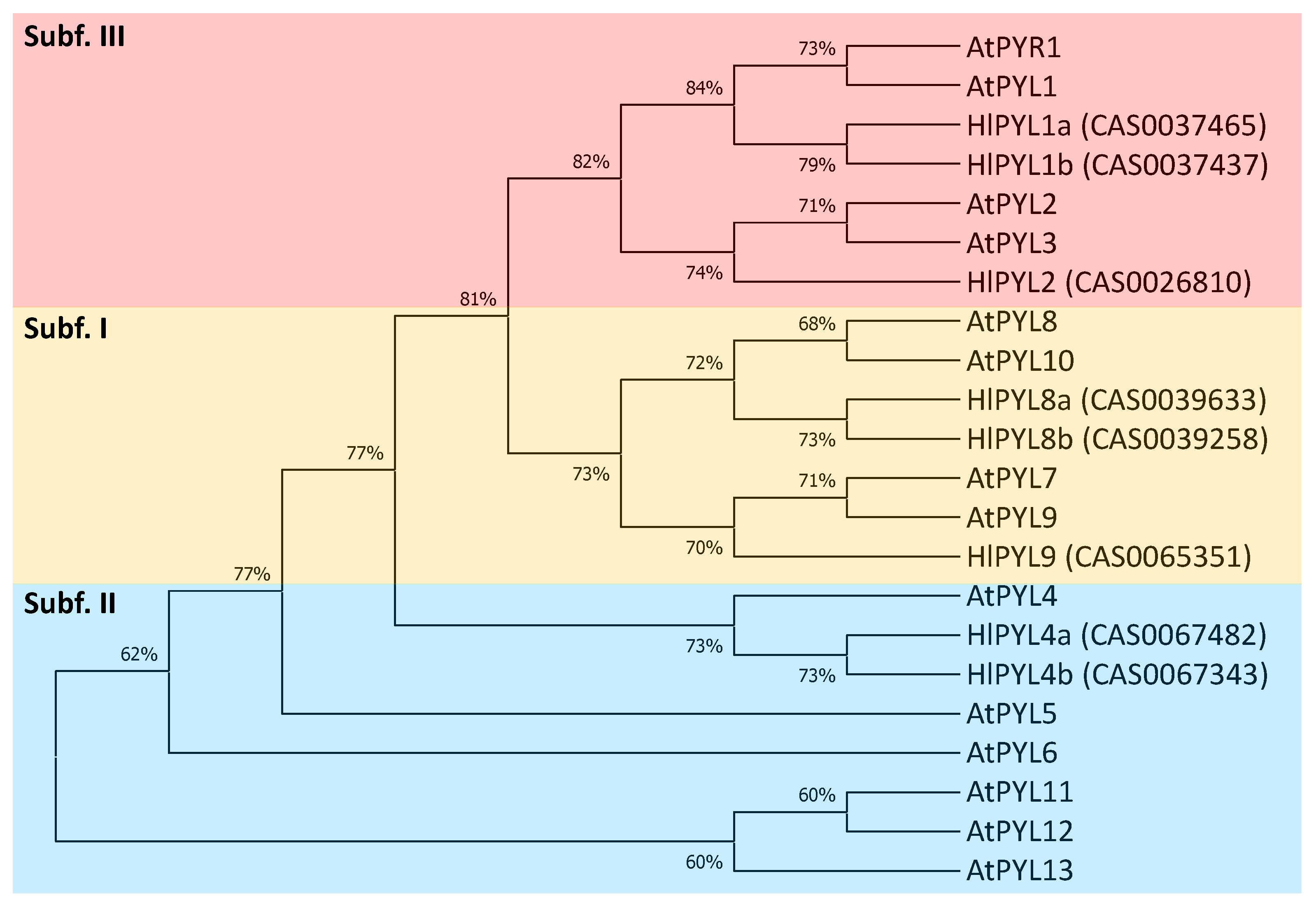
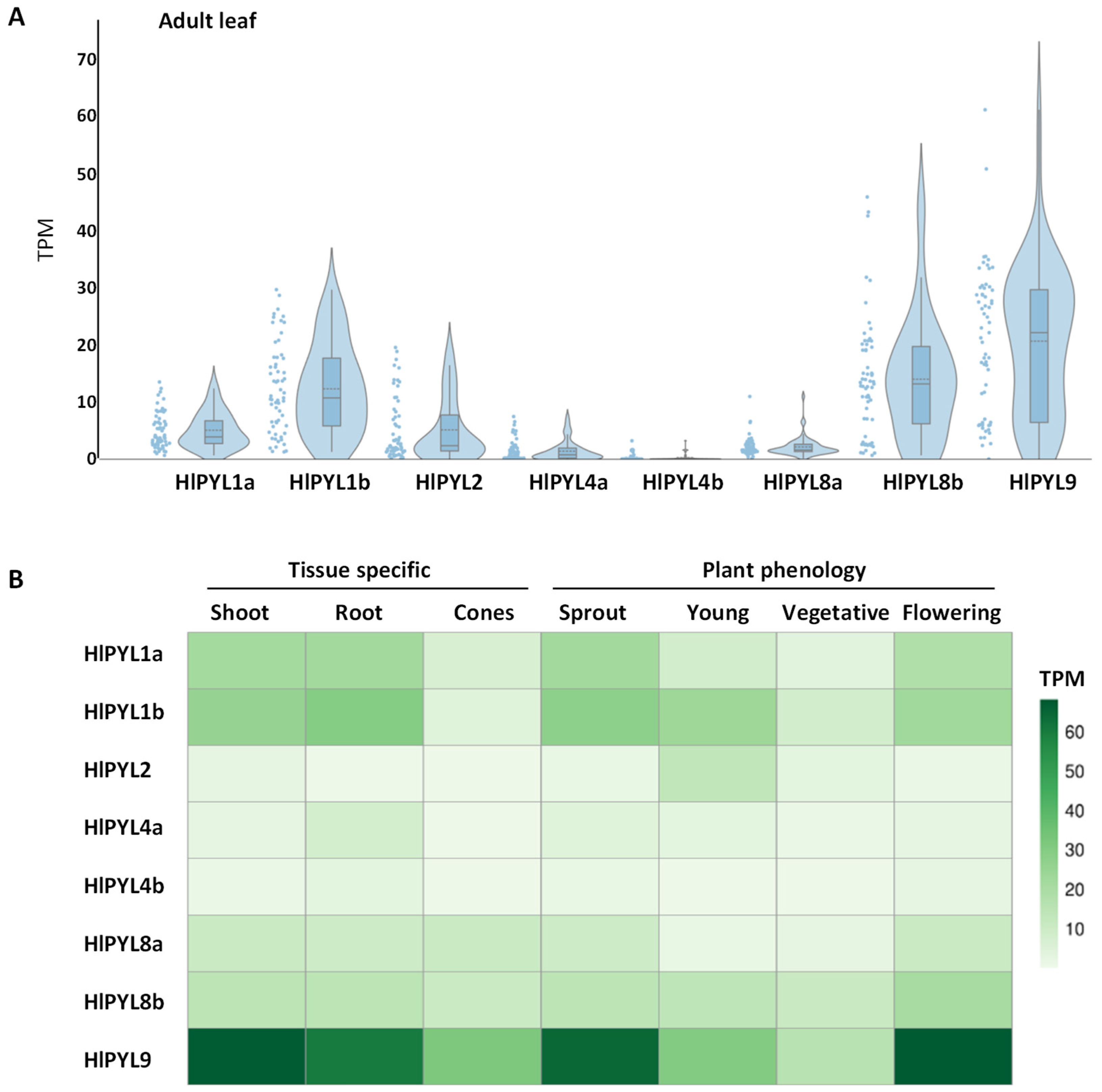
2.2. Phylogeny, Gene Structure Analysis, and Tissue-Specific Expression Pattern of HlPYLs
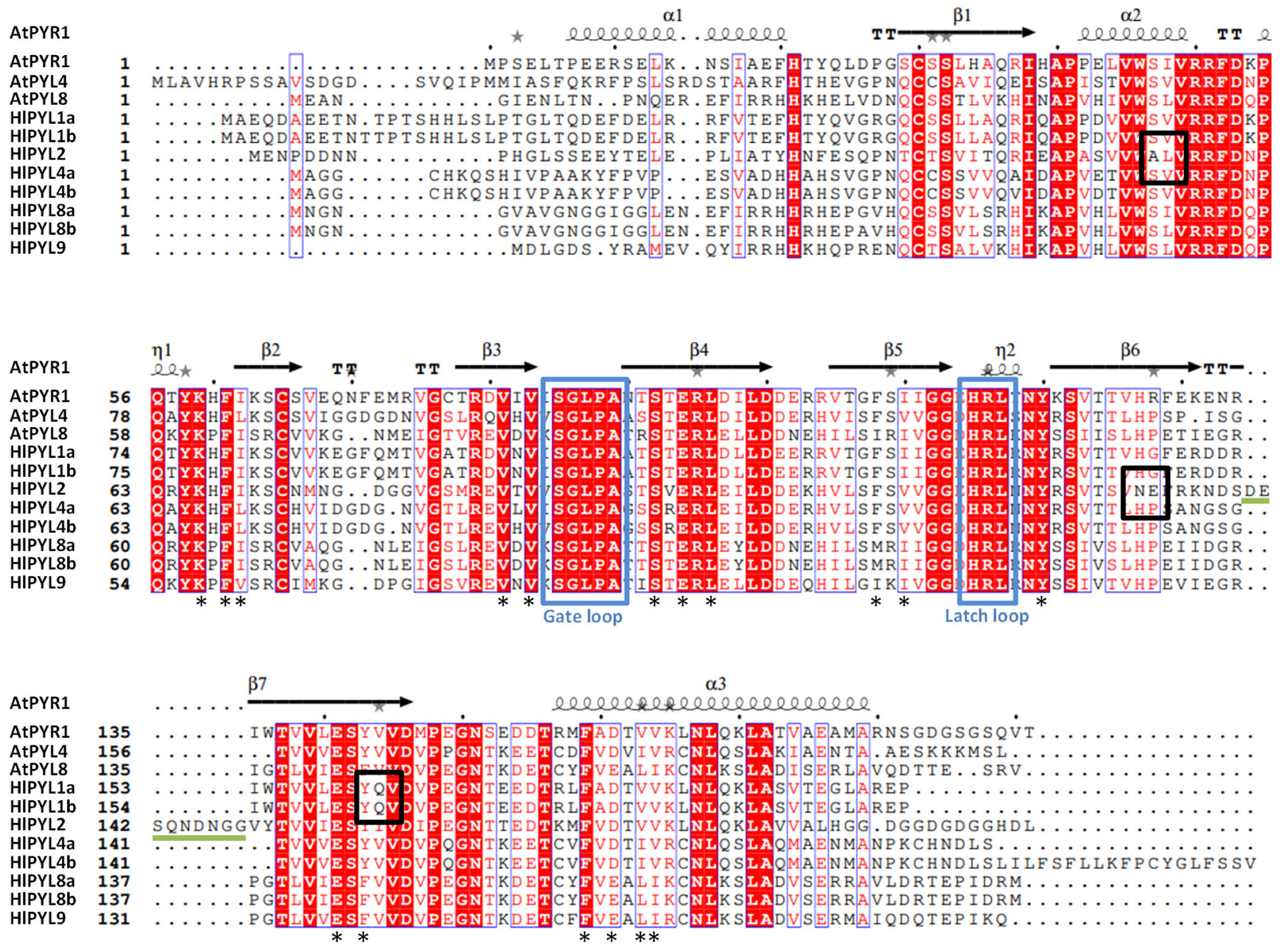
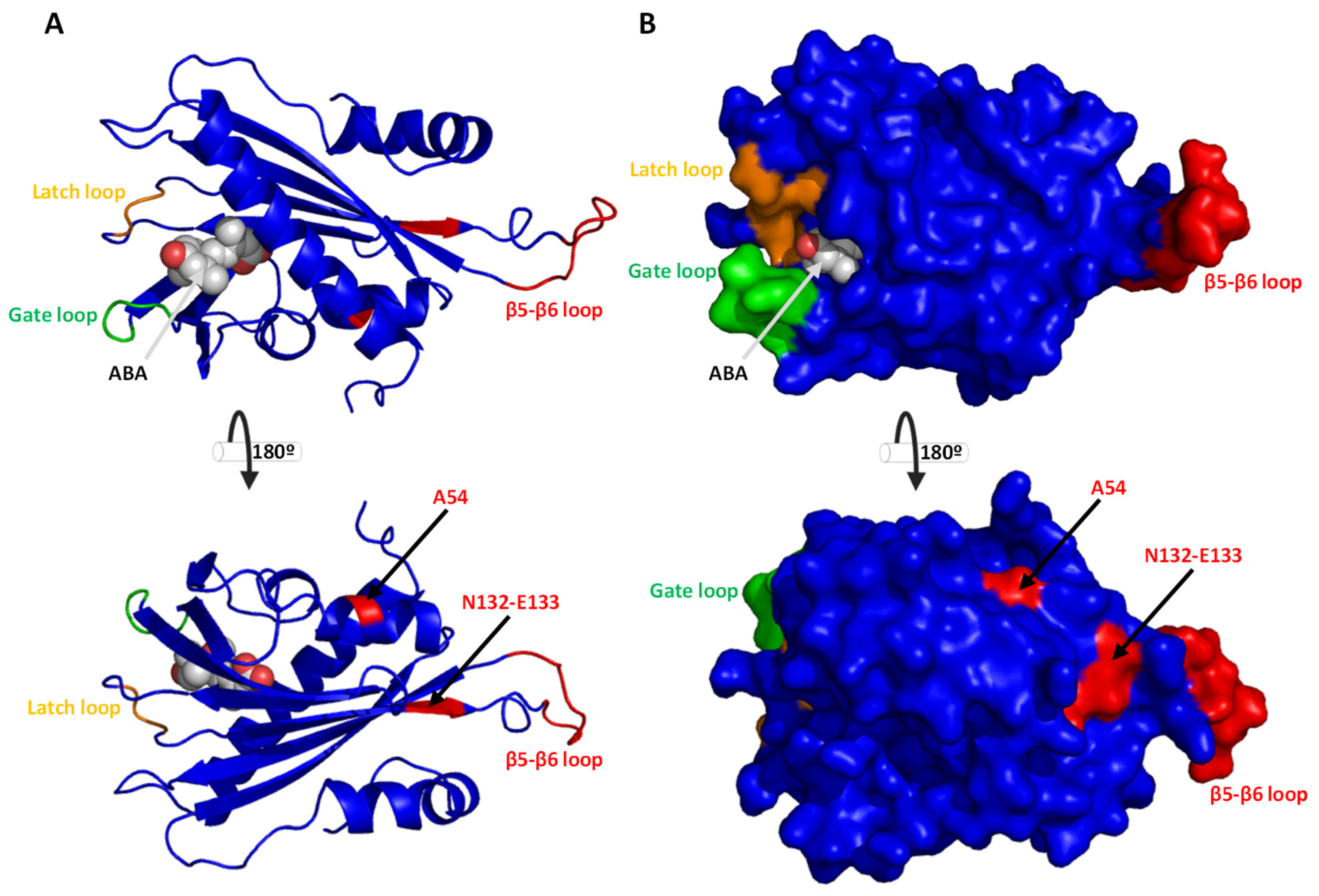
2.3. Conserved Motif, Protein Alignments, and 3D Topology Analysis of PYLs in Hop
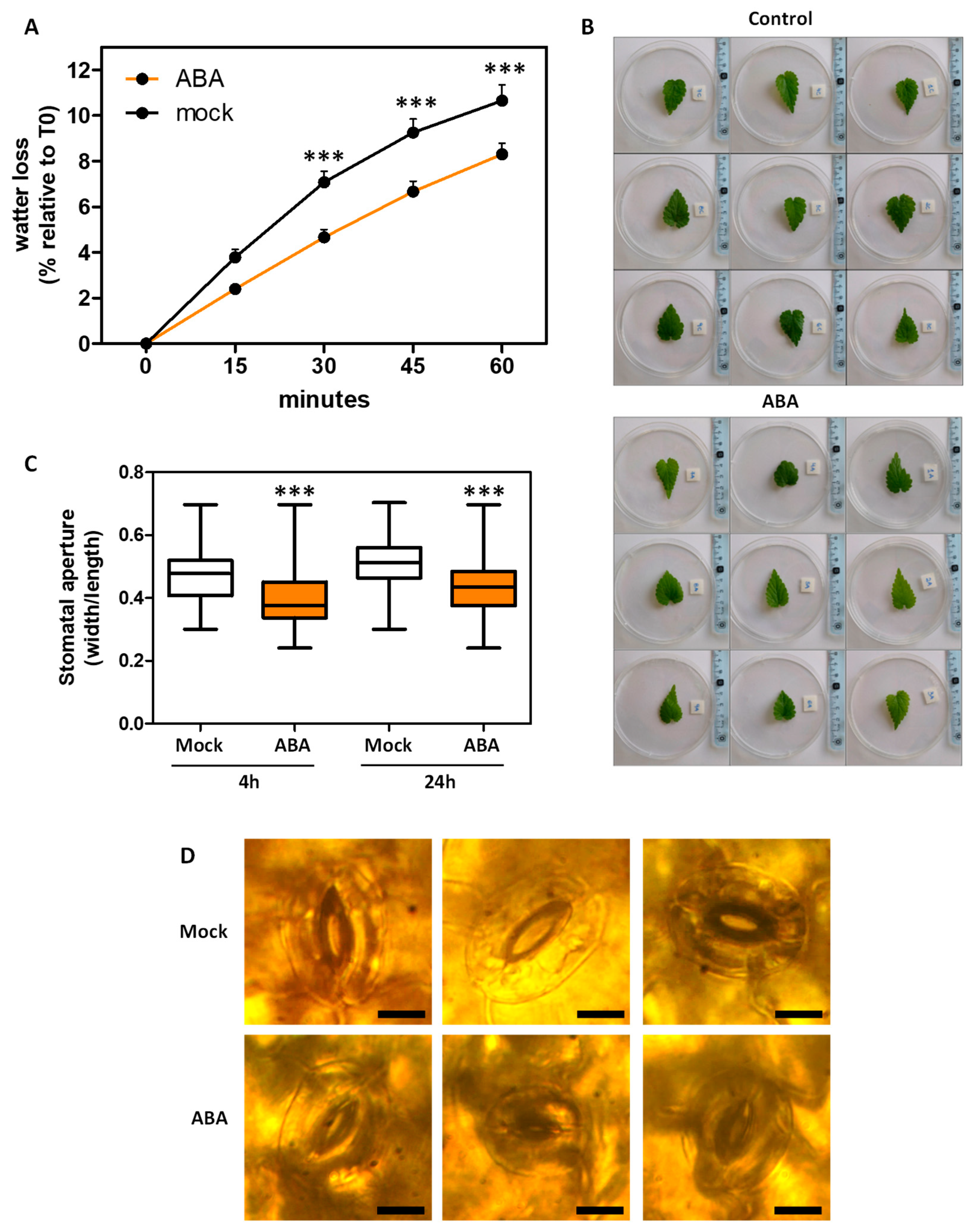
2.4. ABA Effects in Plant Ex Vitro Acclimatization and Transpiration
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification of PYL Genes in Humulus lupulus L.
4.2. Phylogenetic and Gene Structure Analysis of HlPYLs
4.3. Analysis of ABA Receptor Expression in Hop Public Transcriptomic Data
4.4. Conserved Motifs Analysis, Protein Alignments, and Homology Modeling of HlPYLs 3D Structure
4.5. Plant Material, ABA Treatment, and Acclimatization to Ex Vitro Conditions
4.6. Leaf Transpiration and Stomatal Aperture Assays
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zanoli, P.; Zavatti, M. Pharmacognostic and pharmacological profile of Humulus lupulus L. J. Ethnopharmacol. 2008, 116, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Agehara, S.; Acosta-Rangel, A.; Gallardo, M.; Vallad, G. Selection and Preparation of Planting Material for Successful Hop Production in Florida: HS1381, 9/2020. EDIS 2020, 2020. [Google Scholar] [CrossRef]
- Di Sario, L.; Zubillaga, M.F.; Moreno, C.F.Z.; Pizzio, G.A.; Boeri, P.A. Micropropagation of Mapuche hop and evaluation of synthetic seed storage conditions. Plant Cell Tissue Organ Cult. 2025, 160, 1–12. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.; Bressan, R.A.; Song, C.; Zhu, J.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Xu, X.; Liu, H.; Praat, M.; Pizzio, G.A.; Jiang, Z.; Driever, S.M.; Wang, R.; Van De Cotte, B.; Villers, S.L.Y.; Gevaert, K.; et al. Stomatal opening under high X temperatures is controlled by the OST1-regulated TOT3–AHA1 module. Nat. Plants 2025, 11, 105–117. [Google Scholar] [CrossRef]
- Tombesi, S.; Nardini, A.; Frioni, T.; Soccolini, M.; Zadra, C.; Farinelli, D.; Poni, S.; Palliotti, A. Stomatal closure is induced by hydraulic signals and maintained by ABA in drought-stressed grapevine. Sci. Rep. 2015, 5, 12449. [Google Scholar] [CrossRef]
- Falchi, R.; Petrussa, E.; Braidot, E.; Sivilotti, P.; Boscutti, F.; Vuerich, M.; Calligaro, C.; Filippi, A.; Herrera, J.C.; Sabbatini, P. Analysis of Non-Structural Carbohydrates and Xylem Anatomy of Leaf Petioles Offers New Insights in the Drought Response of Two Grapevine Cultivars. Int. J. Mol. Sci. 2020, 21, 1457. [Google Scholar] [CrossRef]
- Saleh, B.; Alshehadah, E.; Slaman, H. Abscisic Acid (ABA) and Salicylic Acid (SA) Content in Relation to Transcriptional Patterns in Grapevine (Vitis vinifera L.) under Salt Stress. J. Plant Biochem. Physiol. 2020, 8, 245. [Google Scholar]
- Lamers, J.; Zhang, Y.; van Zelm, E.; Leong, C.K.; Meyer, A.J.; de Zeeuw, T.; Verstappen, F.; Veen, M.; Deolu-Ajayi, A.O.; Gommers, C.M.M.; et al. Abscisic acid signaling gates salt-induced responses of plant roots. Proc. Natl. Acad. Sci. USA 2025, 122, e2406373122. [Google Scholar] [CrossRef]
- Alonso, R.; Berli, F.J.; Bottini, R.; Piccoli, P. Acclimation mechanisms elicited by sprayed abscisic acid, solar UV-B and water deficit in leaf tissues of field-grown grapevines. Plant Physiol. Biochem. 2015, 91, 56–60. [Google Scholar] [CrossRef]
- Murcia, G.; Fontana, A.; Pontin, M.; Baraldi, R.; Bertazza, G.; Piccoli, P.N. ABA and GA3 regulate the synthesis of primary and secondary metabolites related to alleviation from biotic and abiotic stresses in grapevine. Phytochemistry 2017, 135, 34–52. [Google Scholar] [CrossRef]
- Pizzio, G.A.; Mayordomo, C.; Illescas-Miranda, J.; Coego, A.; Bono, M.; Sanchez-Olvera, M.; Martin-Vasquez, C.; Samantara, K.; Merilo, E.; Forment, J.; et al. Basal ABA signaling balances transpiration and photosynthesis. Physiol. Plant. 2024, 176, e14494. [Google Scholar] [CrossRef]
- Kirsch, F.; Klahn, S.; Hagemann, M. Salt-Regulated Accumulation of the Compatible Solutes Sucrose and Glucosylglycerol in Cyanobacteria and Its Biotechnological Potential. Front. Microbiol. 2019, 10, 2139. [Google Scholar] [CrossRef]
- Ruiz-Partida, R.; Rosario, S.; Lozano-Juste, J. An Update on Crop ABA Receptors. Plants 2021, 10, 1087. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Fung, P.; Nishimura, N. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.; Rodrigues, A.; Saez, A.; Rubio, S.; Antoni, R.; Dupeux, F.; Park, S.Y.; Márquez, J.A.; Cutler, S.R.; Rodriguez, P.L. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009, 60, 575–588. [Google Scholar] [CrossRef]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef]
- Vlad, F.; Rubio, S.; Rodrigues, A.; Sirichandra, C.; Belin, C.; Robert, N.; Leung, J.; Rodriguez, P.L.; Laurière, C.; Merlot, S. Protein Phosphatases 2C Regulate the Activation of the Snf1-Related Kinase OST1 by Abscisic Acid in Arabidopsis. Plant Cell 2009, 21, 3170–3184. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Bono, M.; Rivera-Moreno, M.; Albert, A.; Rodriguez, P.L. Phosphorylation/dephosphorylation-mediated regulation of ABI1/2 activity and stability for fine-tuning ABA signaling. Mol. Plant 2025, 18, 1103–1105. [Google Scholar] [CrossRef]
- Gong, L.; Alabdallah, N.M.; Altihani, F.A.; Al-Balawi, S.M.; Anazi, H.K.; Alharbi, B.M.; Hasan, M.M. TOT3-AHA1 module: Its role in fine-tuning stomatal responses. Front. Plant Sci. 2025, 16, 1582196. [Google Scholar] [CrossRef]
- Pizzio, G.A.; Rodriguez, P.L. Dual regulation of SnRK2 signaling by Raf-like MAPKKKs. Mol. Plant 2022, 15, 1260–1262. [Google Scholar] [CrossRef]
- Xu, K.; Wang, P. Genome-wide analysis of SnRK2 gene family and functional characterization of CpSnRK2.6 in drought and salt resistance in Cucurbita pepo L. Plant Sci. 2025, 359, 112638. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhang, X.; Cao, M.; Yin, L.; Gao, A.; An, K.; Gao, S.; Guo, S.; Yin, H. Genome-Wide Identification, Expression and Interaction Analyses of PP2C Family Genes in Chenopodium quinoa. Genes 2023, 15, 41. [Google Scholar] [CrossRef]
- Bono, M.; Ferrer-Gallego, R.; Pou, A.; Rivera-Moreno, M.; Benavente, J.L.; Mayordomo, C.; Deis, L.; Carbonell-Bejerano, P.; Pizzio, G.A.; Navarro-Payá, D.; et al. Chemical activation of ABA signaling in grapevine through the iSB09 and AMF4 ABA receptor agonists enhances water use efficiency. Physiol. Plant. 2024, 176, e14635. [Google Scholar] [CrossRef] [PubMed]
- Lei, P.; Wei, X.; Gao, R.; Huo, F.; Nie, X.; Tong, W.; Song, W. Genome-wide identification of PYL gene family in wheat: Evolution, expression and 3D structure analysis. Genomics 2021, 113, 854–866. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.; He, A.; Yang, H.; Song, L.; Li, H.; Wu, C.; Ruan, J. Genome-wide identification, abiotic stress, and expression analysis of PYL family in Tartary buckwheat (Fagopyrum tataricum (L.) Gaertn.) during grain development. BMC Plant Biol. 2024, 24, 725. [Google Scholar] [CrossRef]
- Gul, S.; Gul, H.; Shahzad, M.; Ullah, I.; Shahzad, A.; Khan, S.U. Comprehensive analysis of potato (Solanum tuberosum) PYL genes highlights their role in stress responses. Funct. Plant Biol. 2024, 51, FP24094. [Google Scholar] [CrossRef]
- Padgitt-Cobb, L.K.; Kingan, S.B.; Wells, J.; Elser, J.; Kronmiller, B.; Moore, D.; Concepcion, G.; Peluso, P.; Rank, D.; Jaiswal, P.; et al. A draft phased assembly of the diploid Cascade hop (Humulus lupulus) genome. Plant Genome 2021, 14, e20072. [Google Scholar] [CrossRef]
- Gonzalez-Guzman, M.; Rodriguez, L.; Lorenzo-Orts, L.; Pons, C.; Sarrion-Perdigones, A.; Fernandez, M.A.; Peirats-Llobet, M.; Forment, J.; Moreno-Alvero, M.; Cutler, S.R.; et al. Tomato PYR/PYL/RCAR abscisic acid receptors show high expression in root, differential sensitivity to the abscisic acid agonist quinabactin, and the capability to enhance plant drought resistance. J. Exp. Bot. 2014, 65, 4451–4464. [Google Scholar] [CrossRef]
- Long, M.; Betrán, E.; Thornton, K.; Wang, W. The origin of new genes: Glimpses from the young and old. Nat. Rev. Genet. 2003, 11, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Alvero, M.; Yunta, C.; Gonzalez-Guzman, M.; Lozano-Juste, J.; Benavente, J.L.; Arbona, V.; Menéndez, M.; MartinezRipoll, M.; Infantes, L.; Gomez-Cadenas, A.; et al. Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor. Mol. Plant 2017, 10, 1250–1253. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Di Sario, L.; Boeri, P.; Matus, J.T.; Pizzio, G.A. Plant Biostimulants to Enhance Abiotic Stress Resilience in Crops. Int. J. Mol. Sci. 2025, 26, 1129. [Google Scholar] [CrossRef]
- Pizzio, G.A. Potential Implications of the Phytohormone Abscisic Acid in Human Health Improvement at the Central Nervous System. Ann. Epidemiol. Public Health 2022, 5, 1090. [Google Scholar]
- Miao, C.; Xiao, L.; Hua, K.; Zou, C.; Zhao, Y.; Bressan, R.A.; Zhu, J. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. USA 2018, 115, 6058–6063. [Google Scholar] [CrossRef]
- Mosquna, A.; Peterson, F.C.; Park, S.Y.; Lozano-Juste, J.; Volkman, B.F.; Cutler, S.R. Potent and selective activation of abscisic acid receptors in vivo by mutational stabilization of their agonist-bound conformation. Proc. Natl. Acad. Sci. USA 2011, 108, 20838–20843. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Duan, C.; Chen, P.; Li, Q.; Dai, S.; Sun, L.; Ji, K.; Sun, Y.; Xu, W.; et al. The expression profiling of the CsPYL, CsPP2C and CsSnRK2 gene families during fruit development and drought stress in cucumber. J. Plant Physiol. 2012, 169, 1874–1882. [Google Scholar] [CrossRef]
- Zhou, L.; Li, R.; Yang, X.; Peng, Y.; Wang, Y.; Xu, Q.; Yang, Y.; Iqbal, A.; Su, X.; Zhou, Y. Interaction of R2R3-MYB transcription factor EgMYB111 with ABA receptors enhances cold tolerance in oil palm. Int. J. Biol. Macromol. 2025, 305, 141223. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, J.; Poree, F.; Schaeufele, R.; Helmke, H.; Frackenpohl, J.; Lehr, S.; von Koskull-Döring, P.; Christmann, A.; Schnyder, H.; et al. Abscisic Acid Receptors and Coreceptors Modulate Plant Water Use Efficiency and Water Productivity. Plant Physiol. 2019, 180, 1066–1080. [Google Scholar] [CrossRef]
- Arbona, V.; Zandalinas, S.I.; Manzi, M.; González-Guzmán, M.; Rodriguez, P.L.; Gómez-Cadenas, A. Depletion of Abscisic Acid Levels in Roots of Flooded Carrizo Citrange (Poncirus trifoliata L. Raf. × Citrus sinensis L. Osb.) Plants Is a Stress-Specific Response Associated to the Differential Expression of PYR/PYL/RCAR Receptors. Plant Mol. Biol. 2017, 93, 623–640. [Google Scholar] [CrossRef]
- Dias, M.C.; Correia, C.; Moutinho-Pereira, J.; Oliveira, H.; Santos, C. Study of the effects of foliar application of ABA during acclimatization. Plant Cell Tissue Organ Cult. 2014, 117, 213–224. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Santiago, A.; Orduña, L.; Fernández, J.D.; Vidal, Á.; de Martín-Agirre, I.; Lisón, P.; Vidal, E.A.; Navarro-Payá, D.; Matus, J.T. The Plantae Visualization Platform: A comprehensive web-based tool for the integration, visualization, and analysis of omic data across plant and related species. bioRxiv 2024. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. Clustvis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Motif Search. Available online: https://www.genome.jp/tools/motif/ (accessed on 1 October 2023).
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, G.; Haas, J.; Schwede, T. QMEANDisCo—Distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef]
- Liu, T.; Wang, P.; Wang, Z.; Dun, W.; Li, J.; Yu, R. SPY Interacts with Tubulin and Regulates Abscisic Acid-Induced Stomatal Closure in Arabidopsis. Plant Direct 2025, 9, e70063. [Google Scholar] [CrossRef]
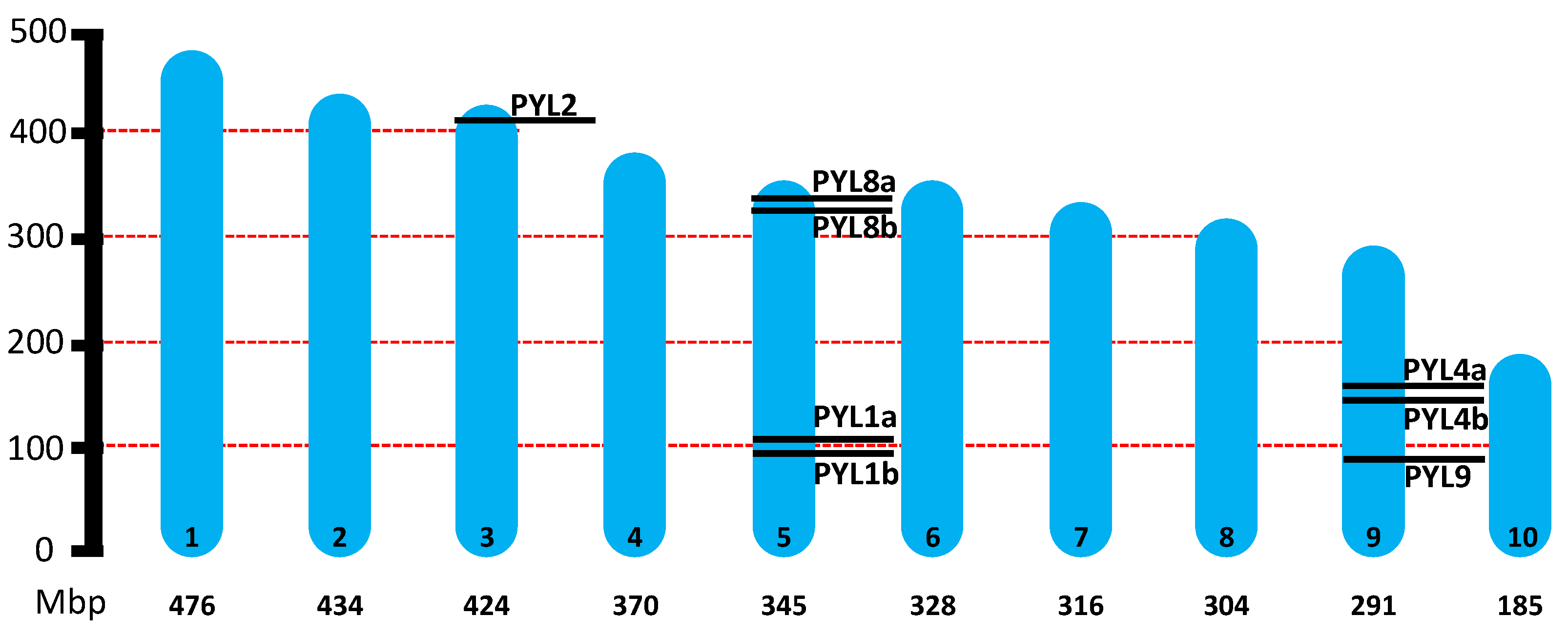



| Gene ID | Gene Name | Genome Location (Start..Stop) | ORF Length (bp) | Protein Size (aa) |
|---|---|---|---|---|
| HUMLU_CAS0037465 | HlPYL1a | CH5/Scaffold 24 (101098846..101099927) | 600 | 200 |
| HUMLU_CAS0037437 | HlPYL1b | CH5/Scaffold 24 (97872371..97873468) | 603 | 201 |
| HUMLU_CAS0026810 | HlPYL2 | CH3/Scaffold 1533 (420945668..420946409) | 615 | 204 |
| HUMLU_CAS0067482 | HlPYL4a | CH9/Scaffold 49 (153780561..153781466) | 579 | 192 |
| HUMLU_CAS0067343 | HlPYL4b | CH9/Scaffold 49 (147637069..147638078) | 633 | 211 |
| HUMLU_CAS0039633 | HlPYL8a | CH5/Scaffold 24 (335968664..335971620) | 579 | 192 |
| HUMLU_CAS0039258 | HlPYL8b | CH5/Scaffold 24 (323689689..323692723) | 579 | 192 |
| HUMLU_CAS0065351 | HlPYL9 | CH9/Scaffold 49 (93847944..93850402) | 558 | 185 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Sario, L.; Navarro-Payá, D.; Zubillaga, M.F.; Matus, J.T.; Boeri, P.A.; Pizzio, G.A. Abscisic Acid Enhances Ex Vitro Acclimatization Performance in Hop (Humulus lupulus L.). Int. J. Mol. Sci. 2025, 26, 6923. https://doi.org/10.3390/ijms26146923
Di Sario L, Navarro-Payá D, Zubillaga MF, Matus JT, Boeri PA, Pizzio GA. Abscisic Acid Enhances Ex Vitro Acclimatization Performance in Hop (Humulus lupulus L.). International Journal of Molecular Sciences. 2025; 26(14):6923. https://doi.org/10.3390/ijms26146923
Chicago/Turabian StyleDi Sario, Luciana, David Navarro-Payá, María F. Zubillaga, José Tomás Matus, Patricia A. Boeri, and Gastón A. Pizzio. 2025. "Abscisic Acid Enhances Ex Vitro Acclimatization Performance in Hop (Humulus lupulus L.)" International Journal of Molecular Sciences 26, no. 14: 6923. https://doi.org/10.3390/ijms26146923
APA StyleDi Sario, L., Navarro-Payá, D., Zubillaga, M. F., Matus, J. T., Boeri, P. A., & Pizzio, G. A. (2025). Abscisic Acid Enhances Ex Vitro Acclimatization Performance in Hop (Humulus lupulus L.). International Journal of Molecular Sciences, 26(14), 6923. https://doi.org/10.3390/ijms26146923







