The Potential of Universal Primers for Barcoding of Subtropical Crops: Actinidia, Feijoa, Citrus, and Tea
Abstract
1. Introduction
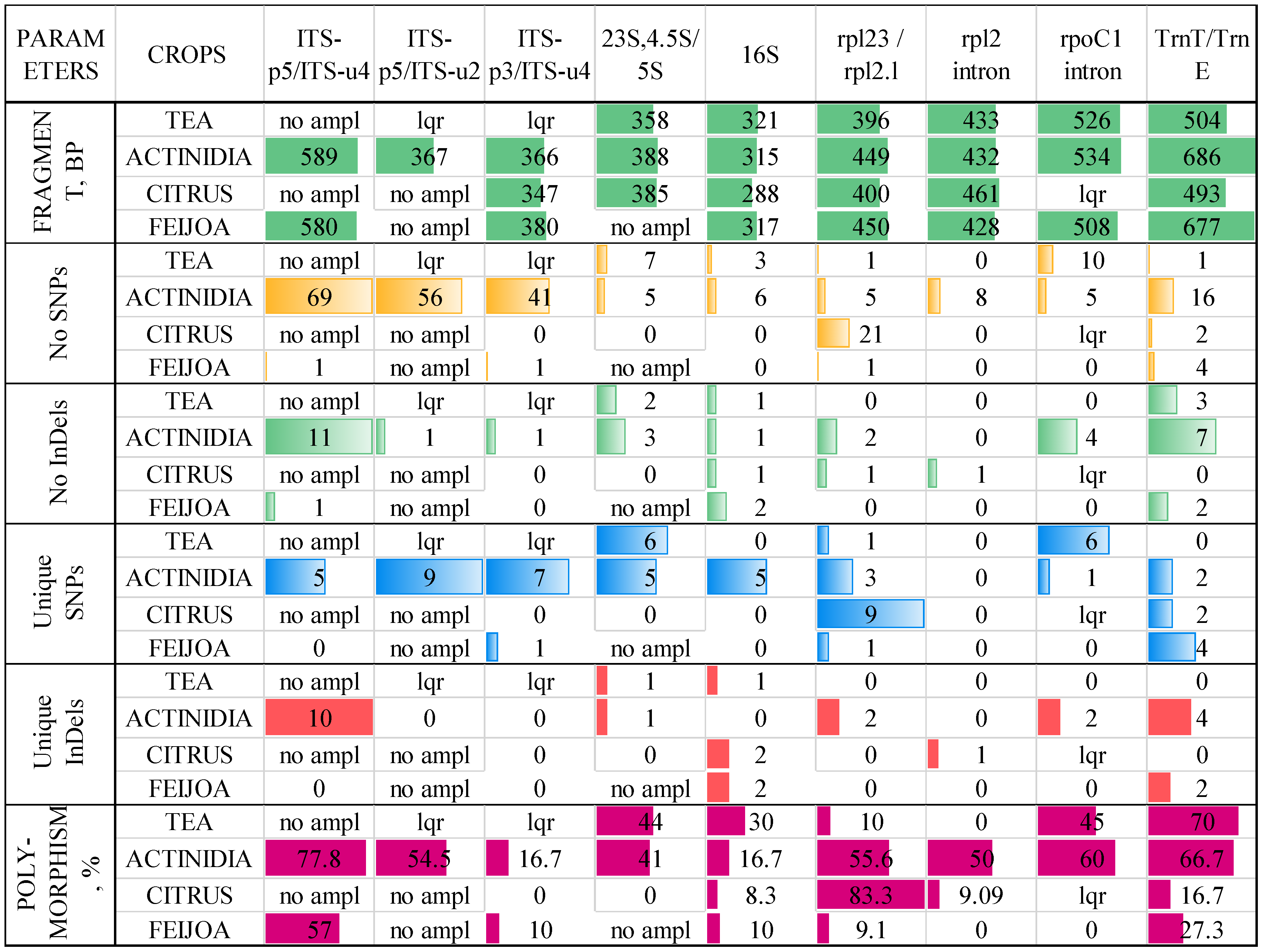
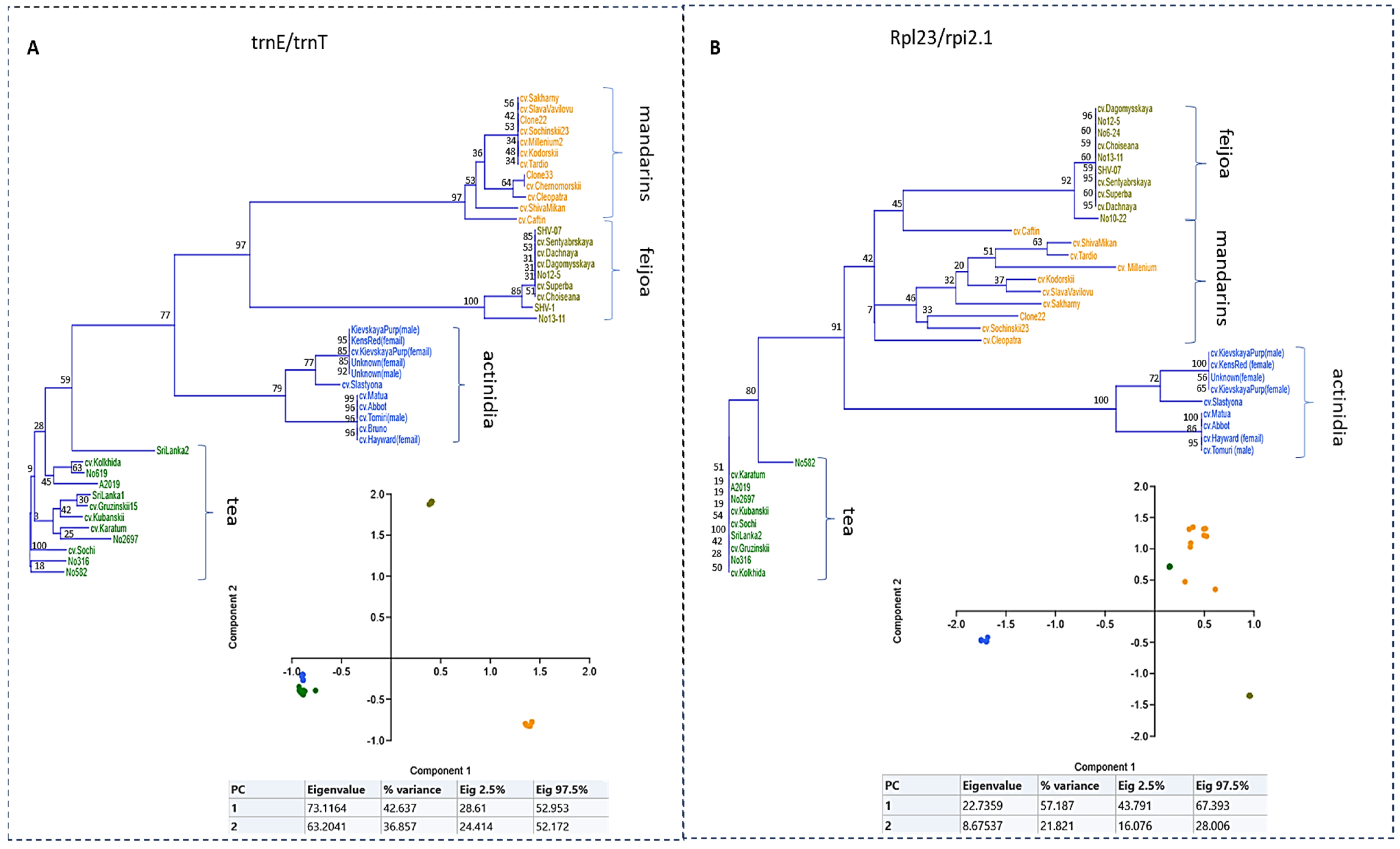
2. Results
3. Discussion
4. Materials and Methods
4.1. Plant Materials and DNA Extraction
4.2. DNA Primers and PCR Conditions
4.3. Sanger Sequencing and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seyis, F.; Yurteri, E.; Ozcan, A. Tea (Camellia sinensis) Cultivation and Breeding in Turkey: Past and Present Status. Ekin J. Crop Breed. Genet. 2019, 5, 111–119. [Google Scholar]
- Maharramov, M.A.; Jahangirov, M.M.; Maharramova, S.I. Azerbaijan Tea (Camellia sinensis L.): Chemical Components, Pharmacology and the Dynamics of the Amino Acids. In Tea Chemistry and Pharmacology; Gonçalo, J., Ed.; IntechOpen: Norderstedt, Germany, 2020; pp. 367–638. [Google Scholar]
- Zhang, Q.; Li, T.; Wang, Q.; LeCompte, J.; Harkess, R.L.; Bi, G. Screening Tea Cultivars for Novel Climates: Plant Growth and Leaf Quality of Camellia sinensis Cultivars Grown in Mississippi, United States. Front. Plant Sci. 2020, 11, 280. [Google Scholar] [CrossRef] [PubMed]
- Ryndin, A.V.; Kulyan, R.V.; Slepchenko, N.A. Subtropical and flower crops breeding at the Subtropical Scientific Centre. Vavilov J. Genet. Breed. 2021, 25, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Daraselia, M.K.; Vorontsov, V.V.; Gvasalia, V.P. Tea Culture in the USSR; Metsniereba: Tbilisi, Georgia, 1989. [Google Scholar]
- Samarina, L.S.; Matskiv, A.O.; Koninskaya, N.G.; Shkhalakhova, R.M.; Gvasaliya, M.V.; Tsaturyan, G.A.; Ryndin, A.V.; Pchikhachev, E.K.; Manakhova, K.A.; Shumeev, A.N.; et al. Genetic diversity and genome size variability in germplasm collection of tea plant (Camellia sinensis L. Kuntze) in Russia. Front. Plant Sci. 2022, 12, 800141. [Google Scholar] [CrossRef] [PubMed]
- Kulyan, R.; Samarina, L.; Shkhalakhova, R.; Kuleshov, A.; Ukhatova, Y.; Antonova, O.; Koninskaya, N.; Matskiv, A.; Malyarovskaya, V.; Ryndin, A. InDel and SCoT Markers for Genetic Diversity Analysis in a Citrus Collection from the Western Caucasus. Int. J. Mol. Sci. 2023, 24, 8276. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Jones, L.; Brennan, G.L.; Lowe, A.; Creer, S.; Ford, C.R.; de Vere, N. Shifts in honeybee foraging reveal historical changes in floral resources. Commun. Biol. 2021, 4, 37. [Google Scholar] [CrossRef]
- Mohammed Abubakar, B.; MohdSalleh, F.; Shamsir Omar, M.S.; Wagiran, A. DNA Barcoding and Chromatography Fingerprints for the Authentication of Botanicals in Herbal Medicinal Products. Evid Based Complement Altern. Med. 2017, 2017, 1352948. [Google Scholar] [CrossRef]
- Xiang, X.G.; Hu, H.; Wang, W.; Jin, X.H. DNA barcoding of the recently evolved genus Holcoglossum (Orchidaceae: Aeridinae): A test of DNA barcode candidates. Mol. Ecol. Resour. 2011, 11, 1012–1021. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef]
- CBOL Plant Working Group. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [PubMed]
- Haidera, N.; Wilkinson, M.J. A Set of Plastid DNA-Specific Universal Primers for Flowering Plants. Russ. J. Genet. 2011, 47, 1066–1077. [Google Scholar] [CrossRef]
- Corvalán, L.; de Melo-Ximenes, A.A.; Carvalho, L.R.; Silva-Neto, C.D.; Diniz-Filho, J.A.F.; Telles, M.P.C.; Nunes, R. Is There a Key Primer for Amplification of Core Land Plant DNA Barcode Regions (rbcL and matK)? Ecol. Evol. 2025, 15, e70961. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hu, Y.; He, M.; Zhang, B.; Wu, W.; Cai, P.; Huo, D.; Hong, Y. Comparative chloroplast genomes: Insights into the evolution of the chloroplast genome of Camellia sinensis and the phylogeny of Camellia. BMC Genom. 2021, 22, 138. [Google Scholar] [CrossRef]
- Whitworth, T.L.; Dawson, R.D.; Magalon, H.; Baudry, E. DNA barcoding cannot reliably identify species of the blowfly genus Protocalliphora (Diptera: Calliphoridae). Proc. R. Soc. B Biol. Sci. 2007, 274, 1731–1739. [Google Scholar] [CrossRef]
- Dong, W.; Cheng, T.; Li, C.; Xu, C.; Long, P.; Chen, C.; Zhou, S. Discriminating plants using the DNA barcode rbcLb: An appraisal based on a large data set. Mol. Ecol. Resour. 2013, 14, 336–343. [Google Scholar] [CrossRef]
- Hidayat, T.; Abdullah, F.I.; Kuppusamy, C.; Samad, A.A.; Wagiran, A. Molecular Identification of Malaysian Pineapple Cultivar Based on Internal Transcribed Spacer Region. APCBEE Procedia 2012, 4, 146–151. [Google Scholar] [CrossRef][Green Version]
- Dissanayake, U.H.K.; Senevirathna, R.W.K.M.; Ranaweera, L.T.; Wijesundara, W.W.M.U.K.; Jayarathne, H.S.M.; Weebadde, C.K.; Sooriyapathirana, S.D.S.S. Characterization of Cassava (Manihot esculenta Crantz) Cultivars in Sri Lanka using Morphological, Molecular and Organoleptic Parameters. Trop. Agric. Res. 2019, 30, 51–70. [Google Scholar] [CrossRef]
- Dhivya, S.; Ashutosh, S.; Gowtham, I.; Baskar, V.; Harini, A.B.; Mukunthakumar, S.; Sathishkumar, R. Molecular identification and evolutionary relationships between the subspecies of Musa by DNA barcodes. BMC Genom. 2020, 21, 659. [Google Scholar] [CrossRef]
- Hürkan, K. Employing Barcode High-Resolution Melting Technique for Authentication of Apricot Cultivars. J. Agric. Sci. 2022, 28, 251–258. [Google Scholar] [CrossRef]
- Nguyen, T.P.; Do, K.T. Identification of Mango (Mangifera indica L.) cultivars in the Mekong Delta using ISSR markers and DNA barcodes. J. Appl. Biol. Biotechnol. 2025, 13, 68–75. [Google Scholar] [CrossRef]
- Omari Alzahrani, F.; Dguimi, H.M.; Alshaharni, M.O.; Albalawi, D.; Zaoui, S. Employing plant DNA barcodes for pomegranate species identification in Al-Baha Region, Saudi Arabia. J. Umm Al-Qura Univ. Appl. Sci. 2024, 10, 136–144. [Google Scholar] [CrossRef]
- Castro, C.; Hernandez, A.; Alvarado, L.; Flores, D. DNA Barcodes in Fig Cultivars (Ficus carica L.) Using ITS Regions of Ribosomal DNA, the psbA-trnH Spacer and the matK Coding Sequence. Am. J. Plant Sci. 2015, 6, 95–102. [Google Scholar] [CrossRef]
- Li, D.Z.; Gao, L.M.; Li, H.T.; Wang, H.; Ge, X.J.; Liu, J.Q.; Chen, Z.D.; Zhou, S.L.; Chen, S.L.; Yang, J.B.; et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Li, C.; Sun, J.; Zuo, Y.; Shi, S.; Cheng, T.; Guo, J.; Zhou, S. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015, 5, 8348. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Liu, J.; Yu, J.; Wang, L.; Zhou, S. Highly Variable Chloroplast Markers for Evaluating Plant Phylogeny at Low Taxonomic Levels and for DNA Barcoding. PLoS ONE 2012, 7, e35071. [Google Scholar] [CrossRef]
- Galimberti, A.; Labra, M.; Sandionigi, A.; Bruno, A.; Mezzasalma, V.; De Mattia, F. DNA Barcoding for Minor Crops and Food Traceability. Adv. Agric. 2014, 2014, 831875. [Google Scholar] [CrossRef]
- Sayed, H.A.; Mostafa, S.; Haggag, I.M.; Hassan, N.A. DNA Barcoding of Prunus Species Collection Conserved in the National Gene Bank of Egypt. Mol. Biotechnol. 2023, 65, 410–418. [Google Scholar] [CrossRef]
- Pang, X.; Liu, C.; Shi, L.; Liu, R.; Liang, D.; Li, H.; Cherny, S.S.; Chen, S. Utility of the trnH–psbA Intergenic Spacer Region and Its Combinations as Plant DNA Barcodes: A Meta-Analysis. PLoS ONE 2012, 7, e48833. [Google Scholar] [CrossRef]
- Feng, S.; Jiao, K.; Zhu, Y.; Wang, H.; Jiang, M.; Wang, H. Molecular identification of species of Physalis (Solanaceae) using a candidate DNA barcode: The chloroplast psbA–trnH intergenic region. Genome 2018, 61, 15–20. [Google Scholar] [CrossRef]
- Awad, M.; Fahmy, R.M.; Mosa, K.A.; Helmy, M.; El-Feky, F.A. Identification of effective DNA barcodes for Triticum plants through chloroplast genome-wide analysis. Comput. Biol. Chem. 2017, 71, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. Isolation of Plant DNA from Fresh Tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]

| # | Species | Genotype | Origin |
|---|---|---|---|
| 1 | Camellia sinensis (L.) Kuntze | #619 | Tetraploid, local form, ɣ-irradiation |
| 2 | Camellia sinensis (L.) Kuntze | #582 | Triploid, local form, ɣ-irradiation |
| 3 | Camellia sinensis (L.) Kuntze | #2697 | Aneuploid, local form, ɣ-irradiation |
| 4 | Camellia sinensis (L.) Kuntze | cv. Sochi | Diploid, local cultivar, clonal selection |
| 5 | Camellia sinensis (L.) Kuntze | cv. Kubanskii | Diploid, local cultivar, clonal selection |
| 6 | Camellia sinensis (L.) Kuntze | cv. Kolkhida | Diploid, local cultivar, clonal selection |
| 7 | Camellia sinensis (L.) Kuntze | #316 | Aneuploid, local form, ɣ-irradiation |
| 8 | Camellia sinensis (L.) Kuntze | cv. Gruzinskii15 | Diploid, clonal selection |
| 9 | Camellia sinensis (L.) Kuntze | SriLanka1 | Diploid, clonal selection |
| 10 | Camellia sinensis (L.) Kuntze | SriLanka2 | Diploid, clonal selection |
| 11 | Camellia sinensis (L.) Kuntze | cv. Karatum | Triploid, local cultivar |
| 12 | Camellia sinensis (L.) Kuntze | A2019 | Diploid, local breeding line |
| 13 | Actinidia chinensis var. deliciosa (A.Chev.) A. Chev. | Tomuri (male) | New Zealand |
| 14 | Actinidia chinensis var. deliciosa (A.Chev.) A. Chev. | Hayward (female) | New Zealand |
| 15 | Actinidia arguta (Siebold & Zucc.) Planch. ex Miq. | Kievskaya Purpurnaya (female) | Ukraine |
| 16 | Actinidia arguta (Siebold & Zucc.) Planch. ex Miq. | Unknown (male) | Russia |
| 17 | Actinidia kolomikta (Maxim. & Rupr.) Maxim. | Slastyona (female) | Russia |
| 18 | Actinidia arguta (Siebold & Zucc.) Planch. ex Miq. | Unknown (female) | Russia |
| 19 | Actinidia arguta (Siebold & Zucc.) Planch. ex Miq. | Ken’s Red (female) | New Zealand |
| 20 | Actinidia arguta (Siebold & Zucc.) Planch. ex Miq. | Kievskaya Purpurnaya (male) | Ukraine |
| 21 | Actinidia chinensis var. deliciosa (A.Chev.) A. Chev | Bruno (female) | New Zealand |
| 22 | Actinidia chinensis var. deliciosa (A.Chev.) A. Chev | Abbott | New Zealand |
| 23 | Actinidia chinensis var. deliciosa (A.Chev.) A. Chev | Allison | New Zealand |
| 24 | Actinidia chinensis var. deliciosa (A.Chev.) A. Chev | Matua | New Zealand |
| 25 | Citrus reshni hort. ex. Tanaka | Cleopatra | India |
| 26 | Citrus reticulata Blanco. | Tardio | Italy |
| 27 | Citrus unshiu Marcow. × C. × leiocarpa | Chernomorskii | Local cultivar |
| 28 | Citrus unshiu Marcow. | Sochinskii23 | Local cultivar |
| 29 | Citrus unshiu Marcow. | Clone22 | Local cultivar |
| 30 | Citrus reticulata var. austera Swingle | Caftin | USA |
| 31 | Citrus unshiu Marcow. | Slava Vavilovu | Local cultivar |
| 32 | Citrus unshiu Marcow. | Sakharny | Local cultivar |
| 33 | Citrus unshiu Marcow. | Kodorskii | Local cultivar |
| 34 | Citrus unshiu Marcow. | Clone33 | Local cultivar |
| 35 | Citrus leiocarpa hort. ex Tanaka | Shiva Mikan | India |
| 36 | Citrus unshiu Marcow. | Millenium2 | Local cultivar |
| 37 | Feijoa sellowiana (O. Berg) Burret | #6-24 | Local breeding line |
| 38 | Feijoa sellowiana (O. Berg) Burret | #13-11 | Local breeding line |
| 39 | Feijoa sellowiana (O. Berg) Burret | #10-22 | Local breeding line |
| 40 | Feijoa sellowiana (O. Berg) Burret | Choiseana | USA |
| 41 | Feijoa sellowiana (O. Berg) Burret | Dachnaya | Local cultivar |
| 42 | Feijoa sellowiana (O. Berg) Burret | Superba | USA |
| 43 | Feijoa sellowiana (O. Berg) Burret | Sentyabrskaya | Local cultivar |
| 44 | Feijoa sellowiana (O. Berg) Burret | SHV-07 | Local breeding line |
| 45 | Feijoa sellowiana (O. Berg) Burret | #12-5 | Local breeding line |
| 46 | Feijoa sellowiana (O. Berg) Burret | Dagomysskaya | Local cultivar |
| 47 | Feijoa sellowiana (O. Berg) Burret | SHV-1 | Local breeding line |
| Primers Code | Primer Sequences 5′–3′ | Product Length, bp | Amplified Region | Reference (doi) |
|---|---|---|---|---|
| ITS-P5/ITS-U4 | F-ccttatcayttagaggaaggag; R-rgtttcttttcctccgctta | 700–800 | nuclear genome, ITS-region | [14] |
| ITS-P5/ITS-U2 | F-ccttatcayttagaggaaggag; R-gcgttcaaagaytcgatgrttc | No data | nuclear genome, ITS-region | [14] |
| ITS-P3/ITS-U4 | F-ygactctcggcaacggata; R-rgtttcttttcctccgctta | 410–480 | nuclear genome, ITS-region | [14] |
| 23S,4.5S/5S | F-tctcctccgacttccctag; R-accatgaacgaggaaaggc | 400–430 | chloroplast genome | [14] |
| 16S | F-attgcgtcgttgtgcctgg; R-gatacgttgttaggtgctcc | 350–370 | chloroplast genome | [14] |
| PetB/PetD | F-tagggggaattacacttac; R-cattaacatgaatacggcag | 490–500 | chloroplast genome | [14] |
| rpl23/rpl2.L | F-gaagaagcttgtacagtttgg; R-tttacttacggcgacgaag | 490–500 | chloroplast genome | [14] |
| rpl2 intron | F-attgagttcagtagttcctc; R-ccaaactgtacaagcttcttc | 430–520 | chloroplast genome | [14] |
| RPOC1 intron | F-gagtaacatgaagctcag; R-gtttcctttcatccggct | 540–670 | chloroplast genome | [14] |
| TRNK intron | F-gtctacatcatcggtagag; R-caacccaatcgctcttttg | 430–500 | chloroplast genome | [14] |
| TRNE-UUC/TRNT-GUU | F-tcctgaaccactagacgatg; R-atggcgttactctaccactg | 834 | chloroplast genome | [17] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samarina, L.S.; Koninskaya, N.G.; Shkhalakhova, R.M.; Simonyan, T.A.; Tsaturyan, G.A.; Shurkina, E.S.; Kulyan, R.V.; Omarova, Z.M.; Tutberidze, T.V.; Ryndin, A.V.; et al. The Potential of Universal Primers for Barcoding of Subtropical Crops: Actinidia, Feijoa, Citrus, and Tea. Int. J. Mol. Sci. 2025, 26, 6921. https://doi.org/10.3390/ijms26146921
Samarina LS, Koninskaya NG, Shkhalakhova RM, Simonyan TA, Tsaturyan GA, Shurkina ES, Kulyan RV, Omarova ZM, Tutberidze TV, Ryndin AV, et al. The Potential of Universal Primers for Barcoding of Subtropical Crops: Actinidia, Feijoa, Citrus, and Tea. International Journal of Molecular Sciences. 2025; 26(14):6921. https://doi.org/10.3390/ijms26146921
Chicago/Turabian StyleSamarina, Lidiia S., Natalia G. Koninskaya, Ruset M. Shkhalakhova, Taisiya A. Simonyan, Gregory A. Tsaturyan, Ekaterina S. Shurkina, Raisa V. Kulyan, Zuhra M. Omarova, Tsiala V. Tutberidze, Alexey V. Ryndin, and et al. 2025. "The Potential of Universal Primers for Barcoding of Subtropical Crops: Actinidia, Feijoa, Citrus, and Tea" International Journal of Molecular Sciences 26, no. 14: 6921. https://doi.org/10.3390/ijms26146921
APA StyleSamarina, L. S., Koninskaya, N. G., Shkhalakhova, R. M., Simonyan, T. A., Tsaturyan, G. A., Shurkina, E. S., Kulyan, R. V., Omarova, Z. M., Tutberidze, T. V., Ryndin, A. V., & Orlov, Y. L. (2025). The Potential of Universal Primers for Barcoding of Subtropical Crops: Actinidia, Feijoa, Citrus, and Tea. International Journal of Molecular Sciences, 26(14), 6921. https://doi.org/10.3390/ijms26146921









