Abstract
The human placenta is a rapidly developing transient organ that is key to pregnancy success. Early development of the conceptus occurs in a low oxygen environment before oxygenated maternal blood begins to flow into the placenta at ~10–12 weeks’ gestation. This process is likely to substantially affect overall placental gene expression. Transcript variability underlying gene expression has yet to be profiled. In this study, accurate transcript expression profiles were identified for 84 human placental chorionic villus tissue samples collected across 6–23 weeks’ gestation. Differential gene expression (DGE), differential transcript expression (DTE) and differential transcript usage (DTU) between 6–10 weeks’ and 11–23 weeks’ gestation groups were assessed. In total, 229 genes had significant DTE yet no significant DGE. Integration of DGE and DTE analyses found that differential expression patterns of individual transcripts were commonly masked upon aggregation to the gene-level. Of the 611 genes that exhibited DTU, 534 had no significant DGE or DTE. The four most significant DTU genes ADAM10, VMP1, GPR126, and ASAH1, were associated with hypoxia-responsive pathways. Transcript usage is a likely regulatory mechanism in early placentation. Identification of functional roles will facilitate new insight in understanding the origins of pregnancy complications.
1. Introduction
The placenta undergoes rapid growth and development across a short lifespan during gestation [1]. The myriad functions of the placenta include transport of nutrients, gases, and wastes between the maternal and fetal circulations, mediation of maternal immune tolerance, regulation of maternal insulin sensitivity and protection of the fetus against xenobiotics [2]. Successful initiation of placentation involves trophoblast differentiation along both villus and extravillous pathways. Extravillous cytotrophoblasts (EVTs) form a cytotrophoblastic shell in the first few weeks’ post conception which completely encircles the conceptus. EVTs invade, colonize, and occlude the uteroplacental arteries during the first trimester of development [3]. Occluding EVTs in the spiral arterioles, so-called trophoblast plugs, begin to be dislodged at around 10–12 weeks’ gestation, facilitating the flow of oxygen-rich maternal blood into the placental intervillous space [1,4]. Thus, there is a transition between two biologically distinct developmental environments over early to mid gestation, with a physiological low oxygen environment up to ~10 weeks’ gestation, and a physiologically “normal” oxygen environment developing after this time. The shift from a low to an increasingly normal physiological oxygen environment late in the first trimester [4] is a critical time in placental development, with deficiency in EVT invasion associated with complications ranging from miscarriage to preeclampsia [5,6]. Transcriptional dynamics across gestation have previously been profiled in gene expression studies, including between mid gestation and term [7], first and third trimester [8,9] and more recently between 6–10 weeks’ and 11–23 weeks’ gestation [10]. Significant changes in microRNAs (miRNAs) have also been reported between the first and third trimesters [11] and between 6–10 weeks’ and 11–23 weeks’ gestation with placenta-specific miRNA clusters reflected in maternal plasma [12]. Roles of long non-coding RNA (lncRNA) in placental development have also been reviewed including their role as cis regulators of autosomal gene expression [13], and in pregnancy complications and immune system regulation [14].
Approximately 95% of all multi-exon genes have more than one alternatively spliced form which serves to increase protein diversity [15,16]. Profiling at the transcript-level enables detection of differential transcript expression (DTE) within a gene that may be masked upon aggregation to the gene-level [17]. Most previous studies have used differential gene expression (DGE) analysis to characterize the placental transcriptome, despite evidence that individual transcript isoforms perform their own distinct functions in human placenta [18,19,20,21]. While differential expression analysis is an indispensable tool to comprehensively profile transcriptional dynamics, it is limited in that it ignores underlying changes in transcript usage. Indeed, the presence of variable isoform usage may hinder detection of significant changes in gene expression [22]. Differential transcript usage (DTU) analysis captures the changes in transcript proportions which can uncover the contribution of individual transcripts to overall gene expression [23,24]. To date, relatively few studies on placenta have investigated the impact of transcript usage on the placental transcriptome and these have been primarily focused on preeclampsia and intrauterine growth restriction (IUGR) [25,26] and EVT differentiation [27]. Assessing DTU in early gestation may offer a unique perspective on the regulation of placental differentiation and growth. Changes in protein abundance altered by isoform usage and intron retention have been previously identified in human breast adenocarcinoma cells using RNA-seq integrated with mass spectrometry [28], highlighting the functional significance of DTU. In placenta, several genes have already been shown in previous studies to exhibit alternative expression of transcript isoforms throughout development including PlGF [19,29], VEGF [30], IGF2 [20], and FLT1 [21,31], suggesting modulation of gene expression through usage of specific transcripts.
Profiling individual transcript expression and DTU across 6–23 weeks’ gestation can provide novel insights on the effects of varying oxygen tension during this dynamically changing period in placental development. At present, the complexities of variable transcript usage in human placenta across early gestation remain understudied, attributable to the paucity of samples reported in the literature and compounded by the difficulties in sampling the placenta during ongoing pregnancy. In order to investigate the effects of variable transcript expression and usage, a previously studied gene expression dataset of 84 chorionic villus tissue samples (6–23 weeks’ gestation) [10] was used to identify transcript expression profiles. In this study DGE, DTE and DTU analyses were performed to identify changes in the expression and usage of individual transcripts with respect to gene expression. Gene ontology (GO) enrichment analysis of significant genes highlighted enrichment of biological and molecular processes driven by DGE, DTE, and DTU. Profiling at a resolution of individual transcripts allowed identification of potentially functional impacts resulting from transcript variability in samples between 6–10 weeks’ and 11–23 weeks’ gestation.
2. Results
After initial filtering of the data, a total of 14,687 transcripts were detected. These belonged to 10,078 unique genes, of which 3028 had more than one detectable isoform. Of the 14,687 transcripts, the 10 with the highest expression across all 84 samples, in order of expression normalized by transcript length, were MTRNR2L12-001, MTRNRL2-001, CGA-001, MTRNR2L8-001, CSH1-001, MTRNR2L1-001, CSH2-001, WDR74-005, KISS1-001, and PSG3-001 (Table S1). Principal component analysis of transcript expression showed that 26.22% of variance within the data was explained by PC1 indicating that gestational age represents the largest source of variability within the data (Figure 1).
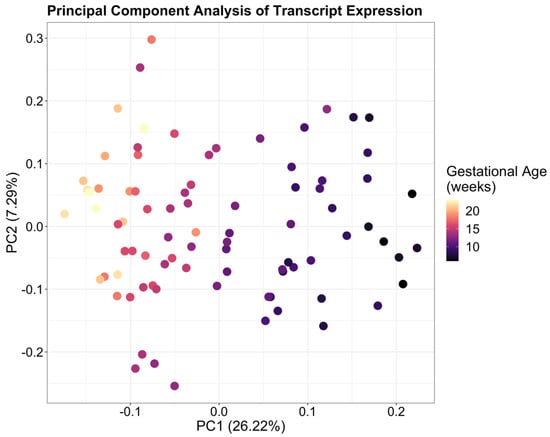
Figure 1.
Principal Component Analysis of transcript-level expression in placenta showing a clear association between gestational age across 6–23 weeks’ gestation and PC1. Samples in the earlier weeks of gestation are localized on the right with mid-gestation samples on the left, indicating that gestational age represents the largest source of variability within the data.
2.1. Changes in the Placental Transcriptome from Early to Mid Gestation Are Highly Enriched for Genes Involved in Cell Migration and Transmembrane Signaling
A total of 1642 genes were significantly different in at least one of three analyses, DGE, DTE and/or DTU. Comparison between analyses revealed the number of genes common to the three approaches (Figure 2). DTE analysis identified a total of 1011 differentially expressed (DE) transcripts, from 861 genes (Figure S1); DGE analysis identified 879 DE genes; and 611 genes showed observable changes in transcript usage (DTU; FDR < 0.05). There were also 682 genes common to more than one analysis with 27 genes observed in all three (Figure 2). Of the 1642 genes identified across all analyses, 10 were long non-coding RNA (lncRNA). The 10 lncRNAs identified were AC079612.1, AC080112.2, C8orf31, and LINC01554 in DGE; MSC-AS1, LINC02860, C1QTNF1-AS1 and MIRLET7BHG in both DGE and DTE; AC110619.1 in DTU; and LINC01118 identified in DGE, DTE, and DTU analyses. A total of 2358 transcripts, from the 1395 genes were significant in DTE and DTU analyses. Of the 1395 genes, 632 had one detectable transcript after filtering for low read counts, while 461 genes expressed two transcripts, and 302 expressed three or more transcripts. Of the 2358 transcripts, 1987 were protein coding and 367 were non-coding. The three most abundant types of non-coding transcripts consisted of processed transcripts with no open reading frame (ORF) (n = 139), transcripts containing retained introns (n = 132), and transcripts targeted by nonsense-mediated decay (NMD) (n = 86). Two protein coding transcripts were from immunoglobulin genes and two unprocessed pseudogenes were also in the set.

Figure 2.
Overlap of significant genes from DGE, DTE, and DTU analyses between 6–10 weeks’ and 11–23 weeks’ gestation placenta. Statistical significance was determined using an FDR < 0.05 and |logFC| > 1 for both DGE and DTE analysis, and an FDR < 0.05 for DTU.
Changing expression patterns of PGF isoforms, previously shown to associate with preeclampsia [29], identified six isoforms of PGF (Figure S2), three of which were protein coding and three of which that were non-coding with retained introns. IGF2 exhibited dominant expression of a single transcript (IGF2-003) along with four other minor transcripts, to make a total of five isoforms (Figure S2). The FLT1 gene contains 30 exons with 5 isoforms (9 isoforms in GRCh38) and has also been shown to be involved in preeclampsia [31]. Three transcripts (FLT1-001, FLT1-201, FLT1-203) were identified in placenta across 6–23 weeks’ gestation, with dominant expression of FLT1-203 (Figure S2). Two isoforms of VEGFB were also identified (VEGFB-001, VEGFB-005) in placenta during early to mid-gestation.
GO enrichment analysis of the 1642 significant genes identified Biological Processes such as cellular responses to cell migration, cellular responses to chemical stimulus and organic substances, and defense response. The top Molecular Functions included transmembrane signaling receptor activity, glycosaminoglycan binding, and receptor ligand activity, with cellular components enriched within the plasma membrane (Table 1). As a change in oxygen tension occurs from about 10 weeks’ gestation, significantly enriched GO terms (FDR > 0.05) were queried for any processes relating to oxygen tension or hypoxia response. Terms containing the expressions “oxygen” and “hypoxia” were assessed returning two biological processes which were response to oxygen-containing compound and reactive oxygen species metabolic process. GO enrichment was then performed for each separate analysis and the top biological processes and molecular functions identified. The top enriched terms in DGE were metal ion transmembrane transporter activity, gated channel activity and adaptive immune response. In DTE, they were cell surface receptor signaling pathway, cell communication, and multicellular organismal process. For DTU, top enriched terms were organic anion transport, signaling receptor activator activity, and active transmembrane transporter activity.

Table 1.
Top 20 enriched GO terms from gene ontology analysis using the 1642 genes that were significant in DGE, DTE, and DTU.
Of the 27 genes overlapping all three analyses (Table S2), the largest number of transcripts were found in FN1 (n = 9), CD36 (n = 6) and FOS (n = 5). Multiple constituent transcripts with DTE were identified for 17 genes, with ADGRD1 (formerly GPR133), ANGPTL1, GPRC5C, and CD36 expressing the greatest numbers of changing transcripts. Of the 27 genes, 25 expressed at least two transcripts with changing proportions between 6–10 weeks’ and 11–23 weeks’ gestation. Both CD36 and NRP2, which had significant DTU at the gene-level, had only one transcript each (CD36-016, NRP2-005) with significantly changing proportions (Figure S3). Of note, CD36 had six transcripts that were all statistically significant in DTE (FDR < 0.05, |logFC| > 1), with one transcript (CD36-016) statistically significant for changing proportions in transcript-level DTU (FDR < 0.05; Table S2). One lncRNA (LINC01118) was statistically significant in all three analyses and had detectable expression for three isoforms, two of which were significant in DGE, DTE and DTU analysis (LINC01118-001, LINC01118-002). Apart from one lncRNA, all other genes in the set were protein coding. However, transcript level annotations revealed that five genes expressed transcripts with a retained intron (ANKRD33, CD36, FN1, GPRC5C, PEAK1), five expressed processed transcripts with no ORF (CD36, F3, FOS, SBSPON, SLC30A2) and one gene (HPGD) expressed a transcript targeted by NMD. Protein coding transcripts were the vast majority, comprising 46 of the 52 transcripts expressed from the set of 27 genes.
2.2. Variable Transcript Expression Masks Differential Gene Expression
A greater number of DE transcripts were found to be more highly expressed in 11–23 weeks’ (upregulated) compared to transcripts more highly expressed in 6–10 weeks’ gestation (down-regulated), with 795 transcripts upregulated and 216 transcripts down-regulated, indicating a possible increase in transcriptome complexity with increasing differentiation of the developing tissue. These changes are consistent with those observed in DGE analysis, for which 678 genes were upregulated while 201 were down-regulated. GO analysis of genes in DTE also found that, as with the statistically significant genes in DGE, growth related processes were highly enriched in 6–10 weeks’ gestation, while immune related functions were enriched in 11–23 weeks’ gestation. Transcripts from 861 individual genes were identified using DTE (Figure 2), with 749 expressing only a single DE transcript while 112 genes expressed two or more DE transcripts. The genes with the greatest number of DE transcripts were NUCB2 and the previously mentioned CD36, which each expressed six DE transcripts, representing complex expression patterns within these genes (Figure S4).
The top 10 most significantly upregulated transcripts in the later gestational group, ordered by FDR, were WNT10A-001, MSC-001, C8orf4-001, DPYSL3-001, NALCN-001, SVEP-003, ANGPTL1-003, PREX2-001, ALPP-001 and TMEM176B-004 (Table S3). The WNT gene family transcript WNT10A-001 and musculin transcript MSC-001 had the highest logFC (logFC > 3) of all upregulated transcripts. The top 10 most significantly down-regulated transcripts were HBZ-001, HBE1-201, CLDN10-002, CLDN10-001, SLC16A3-018, TUBB3-001, CGB7-001, CLDN6-201, GOLT1A-001, and C7orf71-001 (Table S3). Both HBZ-001 and HBE1-201 transcripts, members of the hemoglobin subunit family of genes, had the largest changes in expression (|logFC| > 6) of all transcripts, corresponding to the assumed change in oxygen tension between gestational groups. Multiple highly significant transcripts from the claudin gene family, which mediate angiogenesis and trophoblast invasion [32], were found to be down-regulated in mid gestation (CLDN10-002, CLDN10-001, CLDN6-201, CLDN7-003, and CLDN3-001), with only CLDN1-001 found to be upregulated.
Significant DTE in the absence of any significant DGE was found for 229 genes. However, when comparing between DGE and DTE analyses, the use of discrete FDR and logFC thresholds may artificially reduce the overlap between DE genes and transcripts. With the thresholds used here (FDR < 0.05, |logFC| > 1), 179 genes were considered unique to DTE, and 247 genes were considered unique to DGE analysis (Figure 2). To identify transcripts that appeared to be genuinely lacking in any significant DGE, but were still significant in DTE analysis, genes with the greatest disparities in gene- and transcript-level results were filtered for by comparing the logFC and FDR from both DGE and DTE. Only genes which were non-significant in DGE (FDR > 0.05) and which showed a discrepancy (>1) between logFC estimates were considered as truly unique to DTE. Under these stringent criteria, only 15 uniquely DE transcripts remained (Figure 3) with no gene-level differential expression (Figure S5). Six of these transcripts (ADAM10-015, VMP1-004, MTUS1-005, GPR126-009, RPS24-001, GDPD5-012, and TFPI2-002) were also identified in DTU analysis (FDR < 0.05). Of the 15 transcripts, 7 were upregulated and 8 were down-regulated at 11–23 weeks’ compared to 6–10 weeks’ gestation with four clusters identified (Figure 3). Hierarchical clustering identified two groups which contained only upregulated transcripts which were ITSN1-001, GDPD5-012, and MGAT1-004 in the larger cluster and TFPI2-002 and FLT1-001 in the smaller cluster. One contained only down-regulated transcripts PSG9-008, HBA2-002, and SLC16A3-018, and one contained the transcripts CALM1-005, MTUS1-005, VMP1-004, AZIN1-012, ADAM10-015, RPS24-001, and PSG5-005, which were DE in either direction. Genes in this set also exhibited transcript expression masking paradigms including collapsing of expression (FN1), dominant transcript expression (PSG9) (Figure S6) and cancellation of expression (VMP1) [17] (Figure S7).

Figure 3.
Expression of the top 15 transcripts in DTE analysis that exhibited the greatest difference in expression compared to DGE analysis. Rows (transcripts) are horizontally clustered using the Ward D2 method with gaps distinguishing each cluster, and columns (samples) are ordered by gestational age. A vertical gap in the heatmap separates the counts into the two sample groups at 6–10 weeks’ and 11–23 weeks’ gestation.
2.3. Global Differences in Transcript Usage Were Observed from Early to Mid Gestation
DTU analysis was performed comparing data at 6–10 weeks’ and 11–23 weeks’ gestation to identify genes exhibiting changes in transcript proportions across the 10–11 weeks’ transition period. A total of 2307 genes were analyzed that satisfied the initial filtering criteria, of which 611 were statistically significant for gene-level DTU (Table S4). Changes in individual transcript proportions between sample groups were quantified by differences between median proportions of transcripts compared between 6–10 weeks’ and 11–23 weeks’ gestation (Figure 4). A total of 582 transcripts were found to decrease in proportion from 6–10 weeks’ to 11–23 weeks’ gestation, while 598 transcripts were shown to increase from 6–10 weeks’ to 11–23 weeks’ gestation. Two patterns of DTU were observed. The first is when two transcripts from the same gene show changes of similar magnitude, but in opposite directions. Whilst this may be expected to indicate a switch between two dominant isoforms, a more common pattern was a large magnitude change in one of the two isoforms, as seen for ADAM10 (Figure 5).
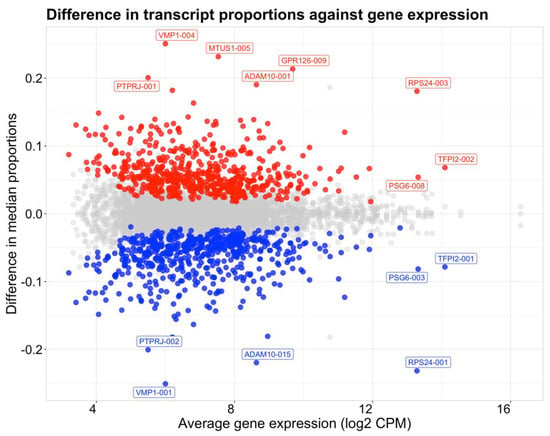
Figure 4.
Scatterplot showing median transcript proportion differences compared to average gene expression (log2 CPM) between placental villus tissue at 11–23 weeks’ and 6–10 weeks’ gestation. Significant transcripts (FDR < 0.05) with a difference in median proportions between sample groups outside the interquartile range were highlighted and colored by the direction of change. Red points indicate an increase in proportion of a transcript from 6–10 weeks’ to 11–23 weeks’ gestation and blue points indicate a decrease from 6–10 to 11–23 gestational weeks. Transcripts with the largest changes in proportion (>0.2) or changing transcripts with the highest average gene expression (>12) are labelled.

Figure 5.
Placental villus ADAM10 gene expression, transcript expression, and transcript usage between 6–10 weeks’ and 11–23 weeks’ gestation with intron-exon structures of each transcript. (A) Structures of ADAM10 isoforms. Coding sequences in each transcript are opaque with non-coding regions transparent. All transcripts are transcribed on the reverse strand. Each transcript is positioned relative to the gene region start site. (B) Gene expression of ADAM10 in log2 CPM across 6–23 weeks’ gestation. (C) Boxplot of ADAM10 transcript proportions of total gene expression between 6–10 weeks’ and 11–23 weeks’ gestation. Transcripts with significant changing proportions are indicated (*). (D) Transcript expression in log2 TPM scaled by transcript length of ADAM10 across 6–23 weeks’ gestation, showing a near 32-fold (log2 5-fold) decrease in expression of the non-coding transcript ADAM10-015.
The most highly ranked genes with DTU were ADAM10, VMP1, MTUS1, GPR126 (ADGRG6), RPS24, GALNT11, ASAH1, SLK, C6orf89, and GDPD5 (Table S4). ADAM10 had the most significant DTU of all genes, expressing two transcripts (ADAM10-001, ADAM10-015) with significantly changing proportions (Figure 5). Of the genes exclusively significant in DTU analysis, ASAH1 was the most highly ranked gene by FDR. Three transcripts (ASAH1-001, ASAH1-002, ASAH1-006) were expressed and had significantly changing proportions (Figure S8). A total of 77 DTU genes were also found in DTE analysis, with 27 of those genes also found in DGE analysis (Figure 2) and 50 found in DTE but not in DGE (Table S5). Four transcripts highlighted in Figure 4 were also featured in the 15 transcripts shown in Figure 3 (ADAM10-015, RPS24-001, TFPI2-002 and VMP1-004).
The VMP1 and GPR126 genes showed the greatest changes in proportion in their transcripts, VMP1-001, VMP1-004, and GPR126-009 while TFPI2-001 and TFPI2-002 were the most highly expressed transcripts with statistically significantly changing proportions (Figure 4). VMP1 exhibited no change in gene expression, yet statistically significant changes were observed in transcript expression. Two transcript isoforms of VMP1 had statistically significant changes in proportion (FDR < 0.05), including a shorter, protein-coding, transcript with 12 exons (VMP1-001), and a longer, non-protein-coding transcript featuring a retained intron (VMP1-004) (Figure S7). GPR126 showed no change in gene expression between 6–10 weeks’ and 11–23 weeks’ gestation despite significant changes at the transcript level. Transcript-level DTU showed GPR126-009 and GRP126-003 had the most statistically significant changes in proportions, while GPR126-009 had the greatest magnitude of change in proportion while GPR126-003 had a smaller magnitude of change (Figure S9). TFPI2 exhibited DTU between a non-coding transcript featuring a retained intron (TFPI2-002) that was DE in transcripts and increased proportions in 11–23 weeks’ gestation placenta, against a protein coding transcript (TFPI2-001) which decreased in proportion (Figure S10). Other transcripts exhibiting both DTE and DTU included MTUS1-005, RPS24-001, PSG6-003, and a processed transcript ADAM10-015, while the transcripts RPS24-003, PSG6-008, PTPRJ-001, PTPRJ-002 were only identified using DTU analysis (Figure S11).
ADAM metalloproteinase domain 10 (ADAM10) gene was the most statistically significant in DTU, with large changes in proportion observed for transcripts ADAM10-001 and ADAM10-015 (Figure 5). ADAM10-001 is a longer protein-coding transcript with a larger 3′ UTR region, ADAM10-015 and ADAM10-008 are long non-coding transcripts, and ADAM10-002 is targeted by NMD. ADAM10 was not considered significant in the DGE analysis, but differential expression was observed for ADAM10-015 and a subtle decrease in expression was detected in ADAM10-002 with DTE analysis. The remaining two transcripts showed no change in expression between gestational windows. While no differential expression was identified for the ADAM10 gene in the DGE analysis, an overall decrease was observed which mirrored the decrease in ADAM10-15. Importantly, whilst the proportion of ADAM10-001 appeared to change, the overall levels were constant indicating this putative change was an artefact primarily due to the decrease in ADAM10-015.
3. Discussion
To our knowledge, this study offers the first profile of transcript expression and usage in placenta across early to mid gestation (6–23 weeks’). Sequencing of a large number of samples has allowed detailed characterization of the normal developing placental transcriptome, while highlighting differential usage of specific transcripts. Here we demonstrate the dynamic and complex transcriptional activity occurring in the human placenta during the transition from a low, but physiological for this time in gestation, oxygen environment (6–10 weeks’) to a more “normal” oxygen (11–23 weeks’) environment through DGE, DTE and DTU analyses. The transition of oxygen tension from 6–10 weeks’ to 11–23 weeks’ gestation was marked by distinct changes in transcript-level dynamics and enriched for pathways including cell migration, transmembrane signalling receptor activity, cellular responses to chemical stimulus and organic substances, and defense response.
Integrating DGE, DTE, and DTU analysis results provided a comprehensive representation of the placental transcriptome and highlighted changes in transcript expression and usage in the absence of any change in gene expression. As a result, DE genes with significant changes in expression and proportion in multiple transcripts were observed. For example, CD36, a gene upregulated in hypoxia that possesses a HIF-1 binding site [33] and is involved in mediation of angiogenesis and inflammatory response [34] had 6 DE transcripts and one transcript with changing proportions. NRP2, which encodes the neuropilin-2 receptor that is repressed by hypoxia and regulates both VEGF and SEMA3F activity to induce tumor angiogenesis [35], also exhibited DGE with two DE transcripts and one transcript with changing proportions. Both CD36 and NRP2 had only one transcript with significant proportion changes each but proportion changes cannot theoretically occur with only one transcript. Hence these changes were found to be the result of subtle non-significant changes in the proportion of transcripts from the same gene in the opposite direction of the significant transcript (Figure S3).
Profiling at an individual transcript resolution afforded an overview of transcript configuration within genes. The FLT1 gene is well known to express a soluble isoform (sFLT-1) that can tightly bind VEGF and suppress angiogenic activity [31] and is associated with preeclampsia [36]. Multiple isoforms of FLT1, including a soluble isoform, have been identified in this study through transcript level profiling. The FLT1-201 isoform exhibits alternative splicing at exon 12, resulting in a shorter transcript, while FLT1-203 deviates from FLT1-001 at an alternatively spliced exon 15a to form a 733aa soluble Flt1 (sFlt1-e15a) isoform identified in a previous study [31] which is significantly upregulated in preeclampsia [37,38]. Statistically significant changes in the expression of non-coding transcripts previously associated with cell invasion, migration, and proliferation, were also identified. MSC-AS1, an anti-sense lncRNA, had higher expression at 11–23 weeks’ gestation and has previously been found to enhance proliferation of gastric cancer cells [39]. Higher expression of C1QTNF1-AS1 was identified at 6–10 weeks’ gestation and overexpression of this lncRNA has been shown to impede proliferation, migration and invasion of human hepatocellular carcinoma cells [40].
It is becoming increasingly apparent that the expression of the majority of all genes is driven by expression of the dominant transcript [41] and so a greater overlap between DGE and DTE genes represented in Figure 2 would have been expected. Comparison of discrepancies between DGE and DTE found variable expression patterns between transcripts within a gene appeared to mask detection of significant changes in overall gene expression. Masking occurred through the collapse of similar transcript-level expression patterns to the gene-level, cancellation of transcripts changing in opposing directions, and the presence of a dominant transcript [17]. The top 15 transcripts with the greatest disparities compared to the gene-level highlighted the consequential masking of transcript variability upon aggregation to an overall gene expression value. Dominant expression masked minor DE transcripts in HBA2, MGAT1, and PSG9, while collapse of subtle changes in expression of multiple transcripts to the gene-level masked significant DTE in GDPD5, AZIN1, and PSG5. A switch in dominant isoform was also observed in VMP1 (Figure S7). These examples suggest that many genes with complex transcript expression patterns cannot be identified upon gene-level aggregation. It has also been previously reported that variable transcripts escape identification in DGE analyses, as the presence of alternative isoform usage leads to an inflated FDR [22].
The interpretation of transcript proportions in DTU analysis presents an opportunity for important insights into the biology but opens the door to possible misinterpretation. Whilst it commonly appears that two isoforms are being switched, it is instead likely to be one isoform being up- or down-regulated as required while the remaining isoforms are unchanged. This is potentially a key insight into the underlying biology that is commonly missed by differential expression analysis at the gene-level [28,42]. Increased transcript diversity per gene is possible through modifications by alternative splicing and alternative promoter usage. These modifications can also target a transcript for nonsense-mediated-decay (NMD) or loss of protein coding capability through intron retention [43]. Specific transcript isoforms have been identified herein that are potential candidates for eliciting functional consequences on the development of the human placenta in early gestation, including both protein-coding and non-coding transcripts. Genes with significant DTE also showed changing transcript proportions which were only detectable through sensitive DTU methods.
High enrichment of cell signaling, cell migration, and immune related pathways were identified from genes statistically significant in DGE, DTE and DTU analyses but only two significantly enriched pathways were oxygen related. However, acute responses to hypoxia occur during early placental development (<10 weeks’ gestation) that are mediated by HIF-1α which regulates developmental processes including trophoblast proliferation in villus explant tissues from first trimester placenta [44]. Despite an absence of enrichment of any hypoxia related processes, genes with the highest significance in DTU (FDR < 7.3 × 10−26) including ADAM10, VMP1, and GPR126 are known to be associated with hypoxia responsive processes such as angiogenesis and autophagy.
ADAM10 encodes disintegrin and metalloproteinase domain 10 and is upstream of the NOTCH and VEGF signaling pathways, of which the VEGF pathway is associated with angiogenesis [45]. ADAM10 has also been shown to mediate the release of the soluble Flt-1 isoform, a known marker of preeclampsia [46] and knockdown of ADAM10 leads to decreased sFlt-1 [47,48]. Even though no change in gene expression was observed, significant DTE and DTU was found in ADAM10 transcripts. Reduced expression and proportion of the non-coding ADAM10-015 transcript occurred simultaneously with an increase in proportion of the protein coding ADAM10-001 transcript which contains the functional disintegrin and peptidase domains [49]. Although the function of ADAM10-015 is unknown, a 10-fold decrease in expression across early gestation may suggest hypoxia responsiveness in that transcript (Figure 5). In fact, increased expression of ADAM10 has previously been shown to be mechanistically linked to hypoxia-induced accumulation of HIF-1α [50]. The ADAM10-002 transcript which was the third most abundant of ADAM10 transcripts is known to undergo NMD, potentially indicating a regulatory role of alternative splicing by targeting transcripts for NMD [51,52].
Changes in transcript expression were observed in GPR126 while gene-level changes were undetected. DTU was observed for GPR126-008, GPR126-009, and GPR126-003 with the greatest changes in proportions occurring in GPR126-009 and GPR126-003, which differ by an 84-nucleotide exon. GPR126-008 and GPR126-009 each possessed a unique exon (Figure S9). Interestingly, a skipping event of exon 23 in GPR126 has been previously shown to be associated with Intrauterine growth restriction (IUGR) and angiogenic-related processes in the human placenta [25]. Interestingly, upon comparison of exon coordinates, it was found that the exon uniquely encoding the GPR126-008 transcript matched the skipped exon associated with IUGR. Knockdown of GPR126 has also been found to inhibit hypoxia-induced angiogenesis in mouse retina [53]. GPR126-009 was more highly expressed in placenta from 11–23 weeks’ compared to 6–10 weeks’ gestation while both GPR126-003 and GPR126-008 significantly decreased in proportions. These suggest potential opposing modes of regulation for each individual transcript in response to hypoxia.
Vacuole membrane protein 1 (VMP1) is a gene known to be involved in autophagy. A steady increase in proportion and expression of the non-coding VMP-004 transcript that features a retained intron was observed, where it replaced the protein-coding VMP1-001 as the dominant transcript around 9–11 weeks’ gestation. Intron retention is mediated through multiple levels of regulation and in mature transcripts it may serve to either alter the resulting protein or cause the transcript to completely lose protein coding potential [54]. HIF-1α induced autophagy has been shown to be reduced with down-regulation of VMP1 in human colon cancer cell lines [55]. A hypoxia signature miRNA, miR-210, has also been shown to down-regulate VMP1 expression and consequently promote cell migration and invasion [56]. Down-regulation of VMP1-004 was observed in early (6–10 weeks’) gestation (Figure S7), coincident with hypoxic conditions in which increased levels of hypoxia-induced autophagy, miR-210 expression, cell migration and invasion have been reported [44,57].
The ASAH1 gene expressed three transcript variants that result in differences in the protein active site upon translation. Uniprot annotation [58] of ASAH1-001 showed that it possesses four sites, Cys143, Arg162, Asn320, and Arg333, necessary for the activation of acid ceramidase [59], while ASAH1-006 contains a retained intron that prevents protein translation. Interestingly, a study in human melanoma cells showed lysosomal acid ceramidase controls the transition between invasive and proliferative phenotypes, with reduced ASAH1 expression associated with invasive behavior [60]. ASAH1 showed a subtle non-significant increase (|logFC| > 0.4 and FDR = 1) in ASAH1-001 and a significant decrease (|logFC| > 0.9 and FDR < 0.05) in ASAH1-006 expression that was filtered out by the effect size cut-off (|logFC| > 1). However, a significant (FDR < 0.05) increase in ASAH1-001 and decrease in ASAH1-006 proportion was detected in DTU. Relatively lower ASAH1-001 and higher ASAH1-006 proportions were found at 6–10 weeks’ gestation, coinciding with hypoxic conditions and trophoblast invasion in the first trimester [1,3].
In conclusion, this study is the first to profile variable human placental expression and usage of transcripts across early to mid gestation by sequencing samples from a large number of pregnancies. Overall, profiling transcriptional dynamics at an individual transcript resolution captured changes between 6–10 weeks’ and 11–23 weeks’ gestation. These appear to reflect the dynamic changes occurring in placental differentiation and growth as it transitions from a physiologically low oxygen environment to a more “normal“ oxygen environment. Analysis of dynamic placental transcriptional change in early to mid gestation in uncomplicated pregnancies is essential before we can identify aberrant transcription that underpins later pregnancy complications. The next step is to determine functional roles of specific gene isoforms in early human placental development in normal and pathological pregnancy.
4. Materials and Methods
4.1. Data Information and Ethics Statement
The RNA-sequencing data used in this study was sourced from previously sequenced human placental chorionic villus samples from 6–23 weeks’ gestation reported in Breen et al. [10]. All RNA-sequencing data is available for download at NCBI Gene Omnibus (GEO) under the Accession number GSE150830. Ethical approval was obtained from the Queen Elizabeth Hospital Human Research Ethics Committee (HREC/16/TQEH/33; 10 May 2016).
4.2. Data Processing
The data were reanalyzed using selective alignment for identification of individual transcript expression profiles. Initial FastQC reports were generated from raw FASTQ files with FastQC [61] and visualized through the ngsReports R package [62], underwent trimming of sequencing adapters by AdapterRemoval [63] and then selective alignment using the Salmon package (v1.1.0) by specifying the—validateMappings argument during alignment [64]. The selective alignment index was generated using the Ensembl GRCh37 build of the reference human transcriptome [65] with the Gencode v19 GRCh37 reference annotation [66]. As sequence fragments from unannotated regions of the transcriptome may falsely align to annotated transcripts bearing sequence similarity, the index was augmented with decoy transcript sequences to be used during selective alignment to reduce false mappings [67]. Quantification of transcript counts was performed with the default Salmon quant method. Transcript counts were divided by bootstrapping estimates of transcript overdispersion to reduce mapping uncertainty arising from physical overlap of transcript regions, as suggested in the edgeR reference manual [68]. Gene counts were generated by aggregating the raw output of transcript counts from Salmon to the gene-level and were used for DGE analysis. Due to unequal male and female placental sample numbers and high variability of transcript expression, transcripts from the X and Y chromosomes were removed prior to performing any further analyses.
4.3. Differential Expression Analysis
Both gene and transcript expression analyses were performed on counts filtered for >2 CPM in ≥27 samples, with 27 samples representing the smallest sample group in the comparison between 6–10 weeks’ (n = 27) and 11–23 weeks’ (n = 57) gestation. A comparison between global distributions of reads from the two sample groups was performed using quantro [69]. Differences between read distributions were detected (p < 0.05), prompting the use of smooth quantile normalization through qsmooth [70]. DGE and DTE analyses were performed using a generalized linear fit model (glmQLFit) implemented in edgeR [68,71], comparing 6–10 weeks’ and 11–23 weeks’ gestation, with fetal sex as a covariate. The gene-wise statistical tests were performed relative to a specified log fold-change threshold of 1.4 using glmTreat. Significance of DGE and DTE was measured at an FDR < 0.05 and |logFC| > 1. Transcripts in the heatmap were clustered using the Ward’s minimum variance method with dissimilarities squared before clustering (“ward.D2”) [72] and visualized via pheatmap [73]. The DGE analysis performed in this study is a reanalysis of RNA-seq data from a study by Breen et al. (2020), while implementing smooth quantile normalization and the same differential expression testing methods described above to remain consistent with the DTE analysis.
4.4. Differential Transcript Usage
Normalized counts were converted to transcripts per million (TPM) scaled by transcript length using tximport [22], then filtered for transcripts contributing at least 10% of the respective gene expression with >2 read counts in ≥27 samples as above. Using DRIMSeq [74], samples were then tested for DTU [23] using a Dirichlet-multinomial model to test at the gene-level and beta-binomial model to test at the transcript-level with significance of DTU measured at an FDR < 0.05, prior to validation of DTU with stageR [75]. To quantify the magnitude of change in transcript proportions for visualization, the median proportions of each transcript within each sample group, 6–10 weeks’ and 11–23 weeks’ gestation, were calculated. The difference in median proportions between each group was then used to represent the magnitude of proportion change.
4.5. Gene Ontology Enrichment
As GO annotations are only provided comprehensively at the gene-level, gene identifiers were used for enrichment testing for DGE result and both DTE and DTU results. GO enrichment was performed using goseq [76] allowing identification of biases in selection of DGE/DTE/DTU genes through the nullp function. Gene length, transcript length, GC content, and median transcript length per gene information were sourced from ensembldb [77] and used to test for bias. The median transcript length per gene accounted for the greatest bias for DTE and DTU results while gene length was the primary bias in the DGE genes. The Wallenius method was used to determine GO enrichment, accounting for biases, and the resulting over-representative p-values were adjusted via the Benjamini-Hochberg method for false discovery [78].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23094506/s1.
Author Contributions
C.T.R. created the concept. K.J.B., C.T.R. and J.B. conceived and developed experimental plans. T.J.-K., D.M. (Dale McAninch) and D.M. (Dylan McCullough) generated the data. Analysis was performed by K.J.B. with analytical input from J.B., S.M.P., S.L. and M.D.S. K.J.B. wrote the manuscript with intellectual input from C.T.R., J.B., S.M.P., Q.W. and T.B.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This research is supported by NIH NICHD R01 HD089685-01 (Maternal molecular profiles reflect placental function and development across gestation) PI Roberts; an Australian Government Research Training Program (RTP) Scholarship awarded to K.J.B.; a National Health and Medical Research Council Investigator Grant (GNT1174971) awarded to C.T.R. and a Matthew Flinders Professorial Fellowship awarded to C.T.R. and funded by Flinders University.
Institutional Review Board Statement
Ethical approval was obtained from the Queen Elizabeth Hospital Human Research Ethics Committee (HREC/16/TQEH/33).
Informed Consent Statement
Written, informed consent was obtained from all patients prior to collection of tissue.
Data Availability Statement
The full workflow for this study is available at (https://github.com/JBogias/EarlyGestationPlacentaProfile). All sequencing data is available for download at NCBI Gene Expression Omnibus (GEO) under the Accession number: GSE150830.
Acknowledgments
We wish to thank the women who kindly donated their placenta tissue. Without their generosity, this research would not have been possible. We would also like to thank the clinical staff who cared for the women.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gude, N.M.; Roberts, C.T.; Kalionis, B.; King, R.G. Growth and function of the normal human placenta. Thromb. Res. 2004, 114, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.T. IFPA Award in Placentology Lecture: Complicated interactions between genes and the environment in placentation, pregnancy outcome and long term health. Placenta 2010, 31, S47–S53. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, R.; Jauniaux, E.; Hustin, J. Maternal circulation in the first-trimester human placenta--myth or reality? Am. J. Obstet. Gynecol. 1997, 176, 695–705. [Google Scholar] [CrossRef]
- Jauniaux, E.; Watson, A.L.; Hempstock, J.; Bao, Y.P.; Skepper, J.N.; Burton, G.J. Onset of maternal arterial blood flow and placental oxidative stress. A possible factor in human early pregnancy failure. Am. J. Pathol. 2000, 157, 2111–2122. [Google Scholar] [CrossRef]
- Brosens, I.; Pijnenborg, R.; Vercruysse, L.; Romero, R. The “Great Obstetrical Syndromes” are associated with disorders of deep placentation. Am. J. Obstet. Gynecol. 2011, 204, 193–201. [Google Scholar] [CrossRef]
- Burton, G.J.; Cindrova-Davies, T.; Yung, H.W.; Jauniaux, E. Oxygen and development of the human placenta. Reproduction 2020, 161, F53–F65. [Google Scholar] [CrossRef]
- Winn, V.D.; Haimov-Kochman, R.; Paquet, A.C.; Yang, Y.J.; Madhusudhan, M.S.; Gormley, M.; Feng, K.-T.V.; Bernlohr, D.A.; McDonagh, S.; Pereira, L.; et al. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology 2007, 148, 1059–1079. [Google Scholar] [CrossRef]
- Sitras, V.; Fenton, C.; Paulssen, R.; Vårtun, Å.; Acharya, G. Differences in gene expression between first and third trimester human placenta: A microarray study. PLoS ONE 2012, 7, e33294. [Google Scholar] [CrossRef]
- Lim, Y.C.; Li, J.; Ni, Y.; Liang, Q.; Zhang, J.; Yeo, G.S.H.; Lyu, J.; Jin, S.; Ding, C. A complex association between DNA methylation and gene expression in human placenta at first and third trimesters. PLoS ONE 2017, 12, e0181155. [Google Scholar] [CrossRef]
- Breen, J.; McAninch, D.; Jankovic-Karasoulos, T.; McCullough, D.; Smith, M.D.; Bogias, K.J.; Wan, Q.; Choudhry, A.; Hin, N.; Pederson, S.M.; et al. Temporal placental genome wide expression profiles reflect three phases of utero-placental blood flow during early to mid human gestation. Indigenous Genomics, Telethon Kids Institute (Adelaide Office): Adelaide, SA, Australia, 2020. manuscript in preparation.
- Gu, Y.; Sun, J.; Groome, L.J.; Wang, Y. Differential miRNA expression profiles between the first and third trimester human placentas. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E836–E843. [Google Scholar] [CrossRef]
- Smith, M.D.; Pillman, K.; Jankovic-Karasoulos, T.; McAninch, D.; Wan, Q.; Bogias, K.J.; McCullough, D.; Bianco-Miotto, T.; Breen, J.; Roberts, C.T. Large-scale transcriptome-wide profiling of microRNAs in human placenta and maternal plasma at early to mid gestation. RNA Biol. 2021, 18, 507–520. [Google Scholar] [CrossRef] [PubMed]
- Buckberry, S.; Bianco-Miotto, T.; Roberts, C.T. Imprinted and X-linked non-coding RNAs as potential regulators of human placental function. Epigenetics 2014, 9, 81–89. [Google Scholar] [CrossRef] [PubMed]
- McAninch, D.; Roberts, C.T.; Bianco-Miotto, T. Mechanistic Insight into Long Noncoding RNAs and the Placenta. Int. J. Mol. Sci. 2017, 18, 1371. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef]
- Yi, L.; Pimentel, H.; Bray, N.L.; Pachter, L. Gene-level differential analysis at transcript-level resolution. Genome Biol. 2018, 19, 53. [Google Scholar] [CrossRef]
- Roberts, H.J.; Hu, S.; Qiu, Q.; Leung, P.C.K.; Caniggia, I.; Gruslin, A.; Tsang, B.; Peng, C. Identification of novel isoforms of activin receptor-like kinase 7 (ALK7) generated by alternative splicing and expression of ALK7 and its ligand, Nodal, in human placenta. Biol. Reprod. 2003, 68, 1719–1726. [Google Scholar] [CrossRef][Green Version]
- Yang, W.; Ahn, H.; Hinrichs, M.; Torry, R.J.; Torry, D.S. Evidence of a novel isoform of placenta growth factor (PlGF-4) expressed in human trophoblast and endothelial cells. J. Reprod. Immunol. 2003, 60, 53–60. [Google Scholar] [CrossRef]
- Monk, D.; Sanches, R.; Arnaud, P.; Apostolidou, S.; Hills, F.A.; Abu-Amero, S.; Murrell, A.; Friess, H.; Reik, W.; Stanier, P.; et al. Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Hum. Mol. Genet. 2006, 15, 1259–1269. [Google Scholar] [CrossRef]
- Ikeda, T.; Sun, L.; Tsuruoka, N.; Ishigaki, Y.; Yoshitomi, Y.; Yoshitake, Y.; Yonekura, H. Hypoxia down-regulates sFlt-1 (sVEGFR-1) expression in human microvascular endothelial cells by a mechanism involving mRNA alternative processing. Biochem. J. 2011, 436, 399–407. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. [version 2; peer review: 2 approved]. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef] [PubMed]
- Soneson, C.; Matthes, K.L.; Nowicka, M.; Law, C.W.; Robinson, M.D. Isoform prefiltering improves performance of count-based methods for analysis of differential transcript usage. Genome Biol. 2016, 17, 12. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Soneson, C.; Patro, R. Swimming downstream: Statistical analysis of differential transcript usage following Salmon quantification. F1000Research 2018, 7, 952. [Google Scholar] [CrossRef]
- Majewska, M.; Lipka, A.; Paukszto, L.; Jastrzebski, J.P.; Szeszko, K.; Gowkielewicz, M.; Lepiarczyk, E.; Jozwik, M.; Majewski, M.K. Placenta transcriptome profiling in intrauterine growth restriction (IUGR). Int. J. Mol. Sci. 2019, 20, 1510. [Google Scholar] [CrossRef] [PubMed]
- Ruano, C.S.M.; Apicella, C.; Jacques, S.; Gascoin, G.; Gaspar, C.; Miralles, F.; Méhats, C.; Vaiman, D. Alternative splicing in normal and pathological human placentas is correlated to genetic variants. Hum. Genet. 2021, 140, 827–848. [Google Scholar] [CrossRef] [PubMed]
- Georgiadou, D.; Boussata, S.; Keijser, R.; Janssen, D.A.M.; Afink, G.B.; van Dijk, M. Knockdown of splicing complex protein PCBP2 reduces extravillous trophoblast differentiation through transcript switching. Front. Cell Dev. Biol. 2021, 9, 671806. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gonzàlez-Porta, M.; Santos, S.; Brazma, A.; Marioni, J.C.; Aebersold, R.; Venkitaraman, A.R.; Wickramasinghe, V.O. Impact of alternative splicing on the human proteome. Cell Rep. 2017, 20, 1229–1241. [Google Scholar] [CrossRef]
- Nucci, M.; Poon, L.C.; Demirdjian, G.; Darbouret, B.; Nicolaides, K.H. Maternal serum placental growth factor isoforms 1 and 2 at 11–13, 20–24 and 30–34 weeks’ gestation in late-onset pre-eclampsia and small for gestational age neonates. Fetal Diagn. Ther. 2014, 35, 249–257. [Google Scholar] [CrossRef]
- Regnault, T.R.H.; Orbus, R.J.; de Vrijer, B.; Davidsen, M.L.; Galan, H.L.; Wilkening, R.B.; Anthony, R.V. Placental expression of VEGF, PlGF and their receptors in a model of placental insufficiency-intrauterine growth restriction (PI-IUGR). Placenta 2002, 23, 132–144. [Google Scholar] [CrossRef]
- Thomas, C.P.; Andrews, J.I.; Liu, K.Z. Intronic polyadenylation signal sequences and alternate splicing generate human soluble Flt1 variants and regulate the abundance of soluble Flt1 in the placenta. FASEB J. 2007, 21, 3885–3895. [Google Scholar] [CrossRef]
- Schumann, S.; Buck, V.U.; Classen-Linke, I.; Wennemuth, G.; Grümmer, R. Claudin-3, claudin-7, and claudin-10 show different distribution patterns during decidualization and trophoblast invasion in mouse and human. Histochem. Cell Biol. 2015, 144, 571–585. [Google Scholar] [CrossRef] [PubMed]
- Mwaikambo, B.R.; Yang, C.; Chemtob, S.; Hardy, P. Hypoxia up-regulates CD36 expression and function via hypoxia-inducible factor-1- and phosphatidylinositol 3-kinase-dependent mechanisms. J. Biol. Chem. 2009, 284, 26695–26707. [Google Scholar] [CrossRef] [PubMed]
- Febbraio, M.; Hajjar, D.P.; Silverstein, R.L. CD36: A class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J. Clin. Investig. 2001, 108, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Coma, S.; Shimizu, A.; Klagsbrun, M. Hypoxia induces tumor and endothelial cell migration in a semaphorin 3F- and VEGF-dependent manner via transcriptional repression of their common receptor neuropilin 2. Cell Adh. Migr. 2011, 5, 266–275. [Google Scholar] [CrossRef]
- Maynard, S.E.; Min, J.-Y.; Merchan, J.; Lim, K.-H.; Li, J.; Mondal, S.; Libermann, T.A.; Morgan, J.P.; Sellke, F.W.; Stillman, I.E.; et al. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J. Clin. Investig. 2003, 111, 649–658. [Google Scholar] [CrossRef]
- Heydarian, M.; McCaffrey, T.; Florea, L.; Yang, Z.; Ross, M.M.; Zhou, W.; Maynard, S.E. Novel splice variants of sFlt1 are upregulated in preeclampsia. Placenta 2009, 30, 250–255. [Google Scholar] [CrossRef]
- Palmer, K.R.; Kaitu’u-Lino, T.J.; Hastie, R.; Hannan, N.J.; Ye, L.; Binder, N.; Cannon, P.; Tuohey, L.; Johns, T.G.; Shub, A.; et al. Placental-Specific sFLT-1 e15a Protein Is Increased in Preeclampsia, Antagonizes Vascular Endothelial Growth Factor Signaling, and Has Antiangiogenic Activity. Hypertension 2015, 66, 1251–1259. [Google Scholar] [CrossRef]
- Jin, X.; Qiao, L.; Fan, H.; Liao, C.; Zheng, J.; Wang, W.; Ma, X.; Yang, M.; Sun, X.; Zhao, W. Long non-coding RNA MSC-AS1 facilitates the proliferation and glycolysis of gastric cancer cells by regulating PFKFB3 expression. Int. J. Med. Sci. 2021, 18, 546–554. [Google Scholar] [CrossRef]
- Han, W.; Yu, G.; Meng, X.; Hong, H.; Zheng, L.; Wu, X.; Zhang, D.; Yan, B.; Ma, Y.; Li, X.; et al. Potential of C1QTNF1-AS1 regulation in human hepatocellular carcinoma. Mol. Cell. Biochem. 2019, 460, 37–51. [Google Scholar] [CrossRef]
- Gonzàlez-Porta, M.; Frankish, A.; Rung, J.; Harrow, J.; Brazma, A. Transcriptome analysis of human tissues and cell lines reveals one dominant transcript per gene. Genome Biol. 2013, 14, R70. [Google Scholar] [CrossRef]
- Vitting-Seerup, K.; Sandelin, A. The landscape of isoform switches in human cancers. Mol. Cancer Res. 2017, 15, 1206–1220. [Google Scholar] [CrossRef] [PubMed]
- Modrek, B.; Lee, C. A genomic view of alternative splicing. Nat. Genet. 2002, 30, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Caniggia, I.; Mostachfi, H.; Winter, J.; Gassmann, M.; Lye, S.J.; Kuliszewski, M.; Post, M. Hypoxia-inducible factor-1 mediates the biological effects of oxygen on human trophoblast differentiation through TGFbeta(3). J. Clin. Investig. 2000, 105, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Caolo, V.; Swennen, G.; Chalaris, A.; Wagenaar, A.; Verbruggen, S.; Rose-John, S.; Molin, D.G.M.; Vooijs, M.; Post, M.J. ADAM10 and ADAM17 have opposite roles during sprouting angiogenesis. Angiogenesis 2015, 18, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Raikwar, N.S.; Liu, K.Z.; Thomas, C.P. N-terminal cleavage and release of the ectodomain of Flt1 is mediated via ADAM10 and ADAM 17 and regulated by VEGFR2 and the Flt1 intracellular domain. PLoS ONE 2014, 9, e112794. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Wang, G.; Zhu, Z.; Huang, Y.; Gu, H.; Ni, X. Increased ADAM10 expression in preeclamptic placentas is associated with decreased expression of hydrogen sulfide production enzymes. Placenta 2015, 36, 947–950. [Google Scholar] [CrossRef]
- Palmer, K.R.; Tong, S.; Kaitu’u-Lino, T.J. Placental-specific sFLT-1: Role in pre-eclamptic pathophysiology and its translational possibilities for clinical prediction and diagnosis. Mol. Hum. Reprod. 2017, 23, 69–78. [Google Scholar] [CrossRef]
- Gutwein, P.; Mechtersheimer, S.; Riedle, S.; Stoeck, A.; Gast, D.; Joumaa, S.; Zentgraf, H.; Fogel, M.; Altevogt, D.P. ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. FASEB J. 2003, 17, 292–294. [Google Scholar] [CrossRef]
- Barsoum, I.B.; Hamilton, T.K.; Li, X.; Cotechini, T.; Miles, E.A.; Siemens, D.R.; Graham, C.H. Hypoxia induces escape from innate immunity in cancer cells via increased expression of ADAM10: Role of nitric oxide. Cancer Res. 2011, 71, 7433–7441. [Google Scholar] [CrossRef]
- Lewis, B.P.; Green, R.E.; Brenner, S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. USA 2003, 100, 189–192. [Google Scholar] [CrossRef]
- de Lima Morais, D.A.; Harrison, P.M. Large-scale evidence for conservation of NMD candidature across mammals. PLoS ONE 2010, 5, e11695. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Wang, Y.; Huang, H.; Yu, W.; Bai, M.; Zhang, L.; Bryan, B.A.; Wang, Y.; Luo, J.; Li, D.; et al. GPR126 protein regulates developmental and pathological angiogenesis through modulation of VEGFR2 receptor signaling. J. Biol. Chem. 2014, 289, 34871–34885. [Google Scholar] [CrossRef] [PubMed]
- Monteuuis, G.; Wong, J.J.L.; Bailey, C.G.; Schmitz, U.; Rasko, J.E.J. The changing paradigm of intron retention: Regulation, ramifications and recipes. Nucleic Acids Res. 2019, 47, 11497–11513. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.E.; Catrinacio, C.; Ropolo, A.; Rivarola, V.A.; Vaccaro, M.I. A novel HIF-1α/VMP1-autophagic pathway induces resistance to photodynamic therapy in colon cancer cells. Photochem. Photobiol. Sci. 2017, 16, 1631–1642. [Google Scholar] [CrossRef] [PubMed]
- Choudhry, H.; Harris, A.L. Advances in Hypoxia-Inducible Factor Biology. Cell Metab. 2018, 27, 281–298. [Google Scholar] [CrossRef] [PubMed]
- Hung, T.-H.; Hsieh, T.-T.; Chen, S.-F.; Li, M.-J.; Yeh, Y.-L. Autophagy in the human placenta throughout gestation. PLoS ONE 2013, 8, e83475. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [CrossRef] [PubMed]
- Gebai, A.; Gorelik, A.; Li, Z.; Illes, K.; Nagar, B. Structural basis for the activation of acid ceramidase. Nat. Commun. 2018, 9, 1621. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, J.; Garandeau, D.; Pandiani, C.; Gaudel, C.; Bille, K.; Nottet, N.; Garcia, V.; Colosetti, P.; Pagnotta, S.; Bahadoran, P.; et al. Lysosomal acid ceramidase ASAH1 controls the transition between invasive and proliferative phenotype in melanoma cells. Oncogene 2019, 38, 1282–1295. [Google Scholar] [CrossRef]
- Andrews, S. Babraham Bioinformatics–FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 September 2020).
- Ward, C.M.; To, T.-H.; Pederson, S.M. ngsReports: A Bioconductor package for managing FastQC reports and other NGS related log files. Bioinformatics 2020, 36, 2587–2588. [Google Scholar] [CrossRef]
- Lindgreen, S. AdapterRemoval: Easy cleaning of next-generation sequencing reads. BMC Res. Notes 2012, 5, 337. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Frankish, A.; Diekhans, M.; Ferreira, A.-M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Malik, L.; Sarkar, H.; Zakeri, M.; Almodaresi, F.; Soneson, C.; Love, M.I.; Kingsford, C.; Patro, R. Alignment and mapping methodology influence transcript abundance estimation. Genome Biol. 2020, 21, 239. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Hicks, S.C.; Irizarry, R.A. Quantro: A data-driven approach to guide the choice of an appropriate normalization method. Genome Biol. 2015, 16, 117. [Google Scholar] [CrossRef]
- Hicks, S.C.; Okrah, K.; Paulson, J.N.; Quackenbush, J.; Irizarry, R.A.; Bravo, H.C. Smooth quantile normalization. Biostatistics 2018, 19, 185–198. [Google Scholar] [CrossRef]
- Lun, A.T.L.; Chen, Y.; Smyth, G.K. It’s DE-licious: A Recipe for Differential Expression Analyses of RNA-seq Experiments Using Quasi-Likelihood Methods in edgeR. Methods Mol. Biol. 2016, 1418, 391–416. [Google Scholar] [PubMed]
- Ward, J.H. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package Version 1.0.12. 2019. Available online: https://cran.r-project.org/package=pheatmap (accessed on 31 March 2022).
- Nowicka, M.; Robinson, M.D. DRIMSeq: A Dirichlet-multinomial framework for multivariate count outcomes in genomics. F1000Research 2016, 5, 1356. [Google Scholar] [CrossRef]
- Van den Berge, K.; Soneson, C.; Robinson, M.D.; Clement, L. stageR: A general stage-wise method for controlling the gene-level false discovery rate in differential expression and differential transcript usage. Genome Biol. 2017, 18, 151. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Rainer, J.; Gatto, L.; Weichenberger, C.X. ensembldb: An R package to create and use Ensembl-based annotation resources. Bioinformatics 2019, 35, 3151–3153. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).