Transcriptomics and RNA-Based Therapeutics as Potential Approaches to Manage SARS-CoV-2 Infection
Abstract
1. Introduction
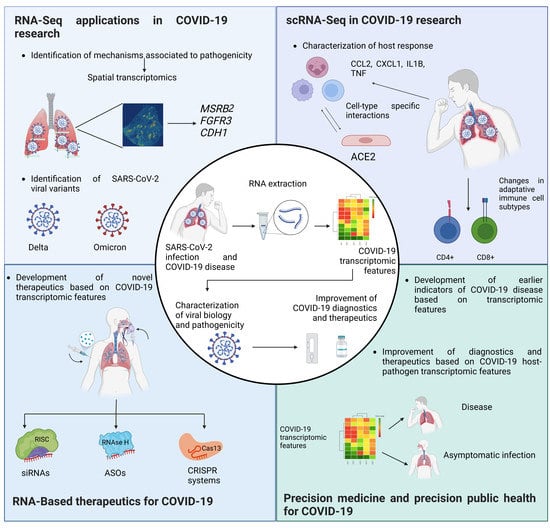
2. The Use of Transcriptomics as a Tool for Understanding SARS-CoV-2 Biology
2.1. Viral Variants and Quasispecies for SARS-CoV-2
2.2. Metabolic Pathways Defined by Transcriptome Analysis in SARS-CoV-2 Infection
2.3. New RNA-Seq Pipelines for SARS-CoV-2 Research
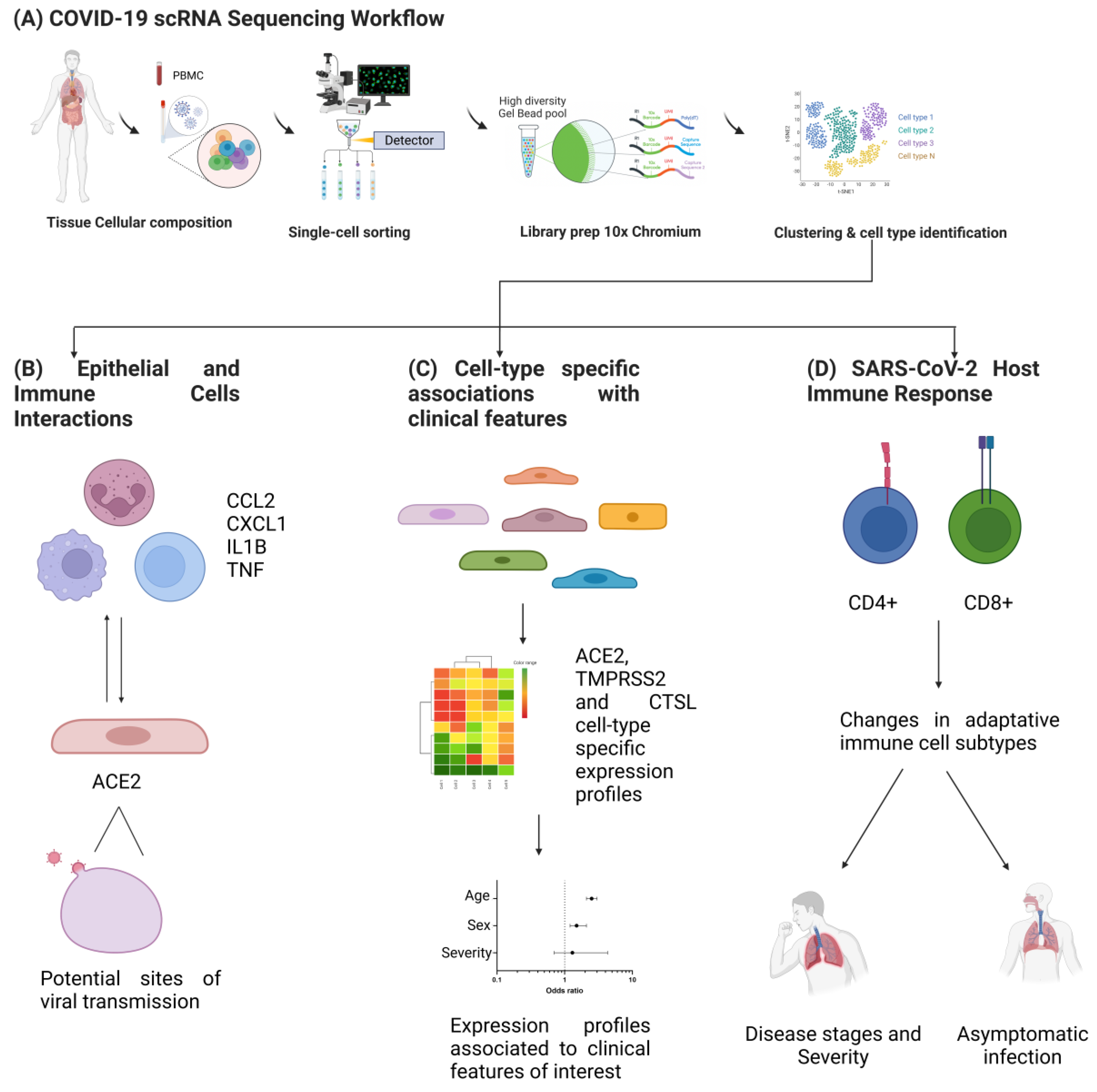
3. Single-Cell Transcriptomics in SARS-CoV-2 Research
3.1. Pipelines for scRNAseq Data Analysis
3.2. Application of scRNAseq in SARS-CoV-2 Research
4. Next-Generation Sequencing Platforms for SARS-CoV-2 Genome Research
Direct RNA Sequencing
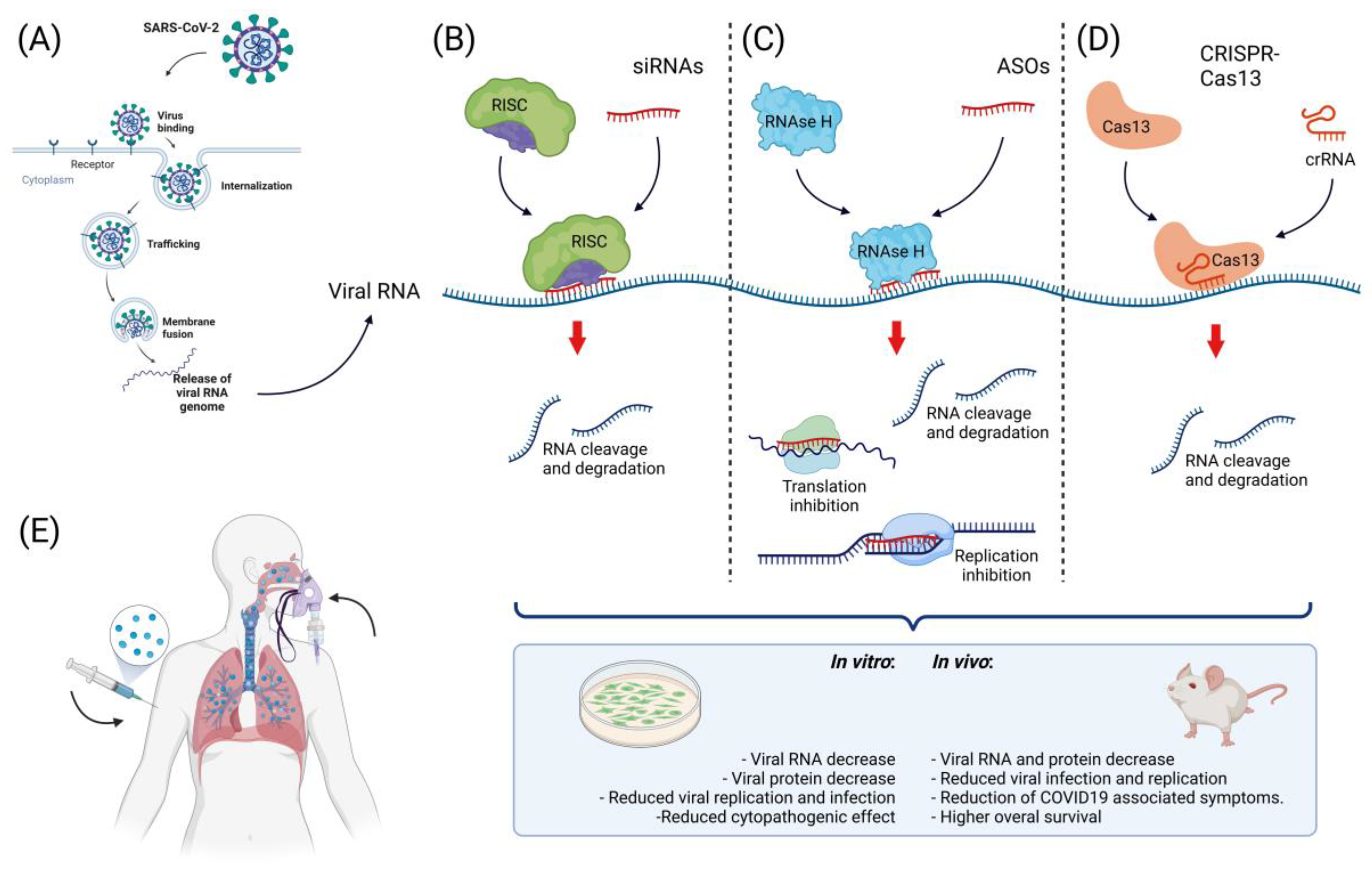
5. The Landscape of Nucleic Acid-Based Therapies in SARS-CoV-2
5.1. Small Interfering RNAs
5.2. MicroRNAs (miRNAs)
5.3. Antisense Oligonucleotides
5.4. CRISPR-Cas
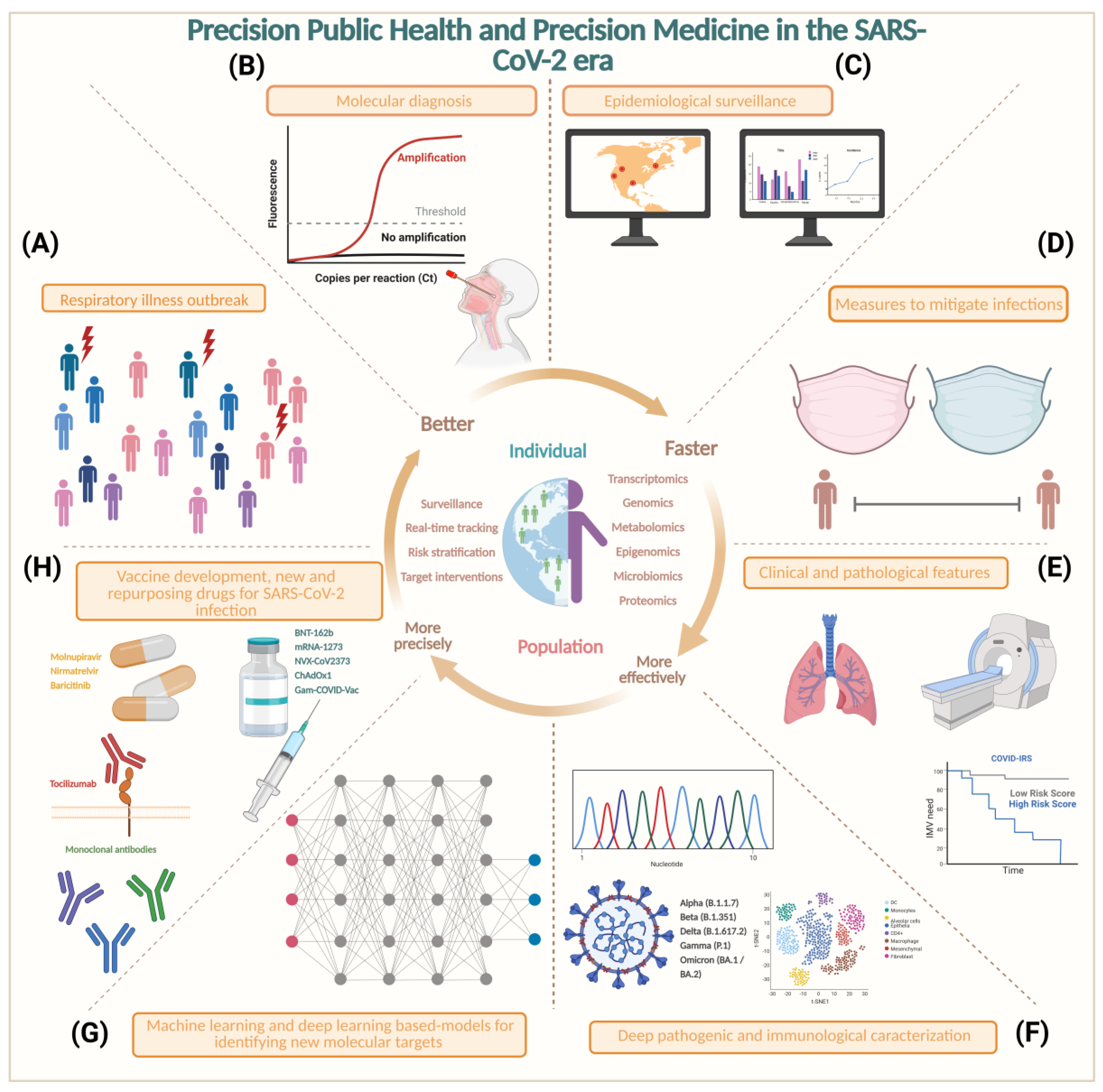
6. Precision Medicine and Precision Public Health in SARS-CoV-2
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.-Y.; Yang, J.-S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liu, K.; Zhang, H.; Zhang, L.; Bian, Y.; Huang, L. CoV-Seq, a New Tool for SARS-CoV-2 Genome Analysis and Visualization: Development and Usability Study. J. Med. Internet Res. 2020, 22, e22299. [Google Scholar] [CrossRef] [PubMed]
- Skowronski, D.M.; De Serres, G. Safety and Efficacy of the BNT162b2 MRNA Covid-19 Vaccine. N. Engl. J. Med. 2021, 384, 1576–1577. [Google Scholar] [CrossRef]
- Carter, L.J.; Garner, L.V.; Smoot, J.W.; Li, Y.; Zhou, Q.; Saveson, C.J.; Sasso, J.M.; Gregg, A.C.; Soares, D.J.; Beskid, T.R.; et al. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent. Sci. 2020, 6, 591–605. [Google Scholar] [CrossRef]
- Agrawal, S. RNA Therapeutics Are Stepping Out of the Maze. Trends Mol. Med. 2020, 26, 1061–1064. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Barh, D.; Tiwari, S.; Weener, M.E.; Azevedo, V.; Góes-Neto, A.; Gromiha, M.M.; Ghosh, P. Multi-Omics-Based Identification of SARS-CoV-2 Infection Biology and Candidate Drugs against COVID-19. Comput. Biol. Med. 2020, 126, 104051. [Google Scholar] [CrossRef]
- Nyholm, L.; Koziol, A.; Marcos, S.; Botnen, A.B.; Aizpurua, O.; Gopalakrishnan, S.; Limborg, M.T.; Gilbert, M.T.P.; Alberdi, A. Holo-Omics: Integrated Host-Microbiota Multi-Omics for Basic and Applied Biological Research. iScience 2020, 23, 101414. [Google Scholar] [CrossRef]
- Maulding, N.D.; Seiler, S.; Pearson, A.; Kreusser, N.; Stuart, J. Dual RNA-Seq Analysis of SARS-CoV-2 Correlates Specific Human Transcriptional Response Pathways Directly to Viral Expression. Sci. Rep. 2022, 12, 1329. [Google Scholar] [CrossRef]
- Finkel, Y.; Mizrahi, O.; Nachshon, A.; Weingarten-Gabbay, S.; Morgenstern, D.; Yahalom-Ronen, Y.; Tamir, H.; Achdout, H.; Stein, D.; Israeli, O.; et al. The Coding Capacity of SARS-CoV-2. Nature 2021, 589, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated Transmissibility and Impact of SARS-CoV-2 Lineage B.1.1.7 in England. Science 2021, 372, e3055. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, A.; McMenamin, J.; Taylor, B.; Robertson, C. SARS-CoV-2 Delta VOC in Scotland: Demographics, Risk of Hospital Admission, and Vaccine Effectiveness. Lancet 2021, 397, 2461–2462. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; St Denis, K.J.; Hoelzemer, A.; Lam, E.C.; Nitido, A.D.; Sheehan, M.L.; Berrios, C.; Ofoman, O.; Chang, C.C.; Hauser, B.M.; et al. MRNA-Based COVID-19 Vaccine Boosters Induce Neutralizing Immunity against SARS-CoV-2 Omicron Variant. Cell 2022, 185, 457–466.e4. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Casner, R.G.; Nair, M.S.; Wang, M.; Yu, J.; Cerutti, G.; Liu, L.; Kwong, P.D.; Huang, Y.; Shapiro, L.; et al. Increased Resistance of SARS-CoV-2 Variant P.1 to Antibody Neutralization. Cell Host Microbe 2021, 29, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Garcia-Knight, M.A.; Khalid, M.M.; Servellita, V.; Wang, C.; Morris, M.K.; Sotomayor-González, A.; Glasner, D.R.; Reyes, K.R.; Gliwa, A.S.; et al. Transmission, Infectivity, and Antibody Neutralization of an Emerging SARS-CoV-2 Variant in California Carrying a L452R Spike Protein Mutation. medRxiv 2021. [Google Scholar] [CrossRef]
- Jangra, S.; Ye, C.; Rathnasinghe, R.; Stadlbauer, D.; Alshammary, H.; Amoako, A.A.; Awawda, M.H.; Beach, K.F.; Bermúdez-González, M.C.; Chernet, R.L.; et al. SARS-CoV-2 Spike E484K Mutation Reduces Antibody Neutralisation. Lancet Microbe 2021, 2, e283–e284. [Google Scholar] [CrossRef]
- Annavajhala, M.K.; Mohri, H.; Wang, P.; Nair, M.; Zucker, J.E.; Sheng, Z.; Gomez-Simmonds, A.; Kelley, A.L.; Tagliavia, M.; Huang, Y.; et al. A Novel and Expanding SARS-CoV-2 Variant, B.1.526, Identified in New York. medRxiv 2021. [Google Scholar] [CrossRef]
- Greaney, A.J.; Loes, A.N.; Crawford, K.H.D.; Starr, T.N.; Malone, K.D.; Chu, H.Y.; Bloom, J.D. Comprehensive Mapping of Mutations in the SARS-CoV-2 Receptor-Binding Domain That Affect Recognition by Polyclonal Human Plasma Antibodies. Cell Host Microbe 2021, 29, 463–476.e6. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; Lam, E.C.; Denis, K.S.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 Variants Escape Neutralization by Vaccine-Induced Humoral Immunity. Cell 2021, 184, 2372–2383.e9. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed]
- Bhoyar, R.C.; Jain, A.; Sehgal, P.; Divakar, M.K.; Sharma, D.; Imran, M.; Jolly, B.; Ranjan, G.; Rophina, M.; Sharma, S.; et al. High Throughput Detection and Genetic Epidemiology of SARS-CoV-2 Using COVIDSeq next-Generation Sequencing. PLoS ONE 2021, 16, e0247115. [Google Scholar] [CrossRef]
- Domingo, E.; Perales, C. Viral Quasispecies. PLoS Genet. 2019, 15, e1008271. [Google Scholar] [CrossRef]
- Nowak, M.A. What Is a Quasispecies? Trends Ecol. Evol. 1992, 7, 118–121. [Google Scholar] [CrossRef]
- Chaudhry, M.Z.; Eschke, K.; Grashoff, M.; Abassi, L.; Kim, Y.; Brunotte, L.; Ludwig, S.; Šafranko, Ž.M.; Kurolt, I.-C.; Markotić, A.; et al. SARS-CoV-2 Quasispecies Mediate Rapid Virus Evolution and Adaptation. J. Virol. 2020, 96, 5. [Google Scholar] [CrossRef]
- Fuentes-Prior, P. Priming of SARS-CoV-2 S Protein by Several Membrane-Bound Serine Proteinases Could Explain Enhanced Viral Infectivity and Systemic COVID-19 Infection. J. Biol. Chem. 2021, 296, 100135. [Google Scholar] [CrossRef] [PubMed]
- Al Khatib, H.A.; Benslimane, F.M.; Elbashir, I.E.; Coyle, P.V.; Al Maslamani, M.A.; Al-Khal, A.; Al Thani, A.A.; Yassine, H.M. Within-Host Diversity of SARS-CoV-2 in COVID-19 Patients With Variable Disease Severities. Front. Cell. Infect. Microbiol. 2020, 10, 575613. [Google Scholar] [CrossRef]
- Li, C.; Chen, J.; Lv, S.; Li, J.; Li, L.; Hu, X. Whole-Transcriptome RNA Sequencing Reveals Significant Differentially Expressed MRNAs, MiRNAs, and LncRNAs and Related Regulating Biological Pathways in the Peripheral Blood of COVID-19 Patients. Mediat. Inflamm. 2021, 2021, e6635925. [Google Scholar] [CrossRef]
- Kumar, N.; Mishra, B.; Mehmood, A.; Athar, M.; Mukhtar, M.S. Integrative Network Biology Framework Elucidates Molecular Mechanisms of SARS-CoV-2 Pathogenesis. iScience 2020, 23, 101526. [Google Scholar] [CrossRef]
- Christensen, E.E.; Jørgensen, M.J.; Nore, K.G.; Dahl, T.B.; Yang, K.; Ranheim, T.; Huse, C.; Lind, A.; Nur, S.; Stiksrud, B.; et al. Critical COVID-19 Is Associated with Distinct Leukocyte Phenotypes and Transcriptome Patterns. J. Intern. Med. 2021, 290, 677–692. [Google Scholar] [CrossRef] [PubMed]
- Salgado-Albarrán, M.; Navarro-Delgado, E.I.; Del Moral-Morales, A.; Alcaraz, N.; Baumbach, J.; González-Barrios, R.; Soto-Reyes, E. Comparative Transcriptome Analysis Reveals Key Epigenetic Targets in SARS-CoV-2 Infection. NPJ Syst. Biol. Appl. 2021, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, P.; Raj, A.S.; Roy, S.; Kumar, N.S.; Kumar, H. Comparative Transcriptome Analysis of SARS-CoV, MERS-CoV, and SARS-CoV-2 to Identify Potential Pathways for Drug Repurposing. Comput. Biol. Med. 2021, 128, 104123. [Google Scholar] [CrossRef]
- Lara-Ureña, N.; García-Domínguez, M. Relevance of BET Family Proteins in SARS-CoV-2 Infection. Biomolecules 2021, 11, 1126. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.M.; Zhang, C.; Wang, Z.; Ni, Z.; Zhang, S.; Yang, S.; Huang, X.; Mo, L.; Li, J.; Lee, B.; et al. A Multi-Omics Investigation of the Composition and Function of Extracellular Vesicles along the Temporal Trajectory of COVID-19. Nat. Metab. 2021, 3, 909–922. [Google Scholar] [CrossRef]
- Longo, S.K.; Guo, M.G.; Ji, A.L.; Khavari, P.A. Integrating Single-Cell and Spatial Transcriptomics to Elucidate Intercellular Tissue Dynamics. Nat. Rev. Genet. 2021, 22, 627–644. [Google Scholar] [CrossRef]
- Margaroli, C.; Benson, P.; Sharma, N.S.; Madison, M.C.; Robison, S.W.; Arora, N.; Ton, K.; Liang, Y.; Zhang, L.; Patel, R.P.; et al. Spatial Mapping of SARS-CoV-2 and H1N1 Lung Injury Identifies Differential Transcriptional Signatures. Cell Rep. Med. 2021, 2, 100242. [Google Scholar] [CrossRef]
- Desai, N.; Neyaz, A.; Szabolcs, A.; Shih, A.R.; Chen, J.H.; Thapar, V.; Nieman, L.T.; Solovyov, A.; Mehta, A.; Lieb, D.J.; et al. Temporal and Spatial Heterogeneity of Host Response to SARS-CoV-2 Pulmonary Infection. Nat. Commun. 2020, 11, 6319. [Google Scholar] [CrossRef]
- Conesa, A.; Beck, S. Making Multi-Omics Data Accessible to Researchers. Sci. Data 2019, 6, 251. [Google Scholar] [CrossRef]
- Sabato, L.D.; Vaccari, G.; Knijn, A.; Ianiro, G.; Bartolo, I.D.; Morabito, S. SARS-CoV-2 RECoVERY: A Multi-Platform Open-Source Bioinformatic Pipeline for the Automatic Construction and Analysis of SARS-CoV-2 Genomes from NGS Sequencing Data. bioRxiv 2021, 425365. [Google Scholar] [CrossRef]
- da Rosa, R.L.; Yang, T.S.; Tureta, E.F.; de Oliveira, L.R.S.; Moraes, A.N.S.; Tatara, J.M.; Costa, R.P.; Borges, J.S.; Alves, C.I.; Berger, M.; et al. SARSCOVIDB—A New Platform for the Analysis of the Molecular Impact of SARS-CoV-2 Viral Infection. ACS Omega 2021, 6, 3238–3243. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.T.; Altschuler, K.; Zhan, S.H.; Chan, Y.A.; Deverman, B.E. COVID-19 CG Enables SARS-CoV-2 Mutation and Lineage Tracking by Locations and Dates of Interest. eLife 2021, 10, e63409. [Google Scholar] [CrossRef] [PubMed]
- Hwang, B.; Lee, J.H.; Bang, D. Single-Cell RNA Sequencing Technologies and Bioinformatics Pipelines. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, S.; Teichmann, S.A. Single Cell Transcriptomics Comes of Age. Nat. Commun. 2020, 11, 4307. [Google Scholar] [CrossRef] [PubMed]
- Svensson, V.; Vento-Tormo, R.; Teichmann, S.A. Exponential Scaling of Single-Cell RNA-Seq in the Past Decade. Nat. Protoc. 2018, 13, 599–604. [Google Scholar] [CrossRef]
- Andrews, T.S.; Kiselev, V.Y.; McCarthy, D.; Hemberg, M. Tutorial: Guidelines for the Computational Analysis of Single-Cell RNA Sequencing Data. Nat. Protoc. 2021, 16, 1–9. [Google Scholar] [CrossRef]
- Gao, C.; Zhang, M.; Chen, L. The Comparison of Two Single-Cell Sequencing Platforms: BD Rhapsody and 10x Genomics Chromium. Curr. Genom. 2020, 21, 602–609. [Google Scholar] [CrossRef]
- See, P.; Lum, J.; Chen, J.; Ginhoux, F. A Single-Cell Sequencing Guide for Immunologists. Front. Immunol. 2018, 9, 2425. [Google Scholar] [CrossRef]
- Zheng, G.X.Y.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively Parallel Digital Transcriptional Profiling of Single Cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef]
- Singh, M.; Bansal, V.; Feschotte, C. A Single-Cell RNA Expression Map of Human Coronavirus Entry Factors. Cell Rep. 2020, 32, 108175. [Google Scholar] [CrossRef]
- Luecken, M.D.; Theis, F.J. Current Best Practices in Single-cell RNA-seq Analysis: A Tutorial. Mol. Syst. Biol. 2019, 15, e8746. [Google Scholar] [CrossRef] [PubMed]
- 10X Genomics Getting Started with Single Cell Gene Expression 2021. Available online: https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation (accessed on 31 July 2022).
- Zhang, J.-Y.; Wang, X.-M.; Xing, X.; Xu, Z.; Zhang, C.; Song, J.-W.; Fan, X.; Xia, P.; Fu, J.-L.; Wang, S.-Y.; et al. Single-Cell Landscape of Immunological Responses in Patients with COVID-19. Nat. Immunol. 2020, 21, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M.; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef] [PubMed]
- Wolock, S.L.; Lopez, R.; Klein, A.M. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst. 2019, 8, 281–291.e9. [Google Scholar] [CrossRef] [PubMed]
- Zeming, K.K.; Thakor, N.V.; Zhang, Y.; Chen, C.-H. Real-Time Modulated Nanoparticle Separation with an Ultra-Large Dynamic Range. Lab Chip 2016, 16, 75–85. [Google Scholar] [CrossRef]
- Young, M.D.; Behjati, S. SoupX Removes Ambient RNA Contamination from Droplet-Based Single-Cell RNA Sequencing Data. GigaScience 2020, 9, giaa151. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate Alignment of Transcriptomes in the Presence of Insertions, Deletions and Gene Fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating Single-Cell Transcriptomic Data across Different Conditions, Technologies, and Species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Berg, D.A.; Zhu, Y.; Shin, J.Y.; Song, J.; Bonaguidi, M.A.; Enikolopov, G.; Nauen, D.W.; Christian, K.M.; Ming, G.; et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades Underlying Adult Neurogenesis. Cell Stem Cell 2015, 17, 360–372. [Google Scholar] [CrossRef]
- Ji, Z.; Ji, H. TSCAN: Pseudo-Time Reconstruction and Evaluation in Single-Cell RNA-Seq Analysis. Nucleic Acids Res. 2016, 44, e117. [Google Scholar] [CrossRef]
- Ren, X.; Wen, W.; Fan, X.; Hou, W.; Su, B.; Cai, P.; Li, J.; Liu, Y.; Tang, F.; Zhang, F.; et al. COVID-19 Immune Features Revealed by a Large-Scale Single-Cell Transcriptome Atlas. Cell 2021, 184, 1895–1913.e19. [Google Scholar] [CrossRef]
- Teichmann, S.; Regev, A. The Network Effect: Studying COVID-19 Pathology with the Human Cell Atlas. Nat. Rev. Mol. Cell Biol. 2020, 21, 415–416. [Google Scholar] [CrossRef]
- Delorey, T.M.; Ziegler, C.G.K.; Heimberg, G.; Normand, R.; Yang, Y.; Segerstolpe, A.; Abbondanza, D.; Fleming, S.J.; Subramanian, A.; Montoro, D.T.; et al. A Single-Cell and Spatial Atlas of Autopsy Tissues Reveals Pathology and Cellular Targets of SARS-CoV-2. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jin, K.; Bardes, E.E.; Mitelpunkt, A.; Wang, J.Y.; Bhatnagar, S.; Sengupta, S.; Krummel, D.P.; Rothenberg, M.E.; Aronow, B.J. An Interactive Single Cell Web Portal Identifies Gene and Cell Networks in COVID-19 Host Responses. iScience 2021, 24, 103115. [Google Scholar] [CrossRef]
- Chua, R.L.; Lukassen, S.; Trump, S.; Hennig, B.P.; Wendisch, D.; Pott, F.; Debnath, O.; Thürmann, L.; Kurth, F.; Völker, M.T.; et al. COVID-19 Severity Correlates with Airway Epithelium–Immune Cell Interactions Identified by Single-Cell Analysis. Nat. Biotechnol. 2020, 38, 970–979. [Google Scholar] [CrossRef]
- Sungnak, W. SARS-CoV-2 Entry Factors Are Highly Expressed in Nasal Epithelial Cells Together with Innate Immune Genes. Nat. Med. 2020, 26, 14. [Google Scholar] [CrossRef] [PubMed]
- The NHLBI LungMap Consortium; The Human Cell Atlas Lung Biological Network; Muus, C.; Luecken, M.D.; Eraslan, G.; Sikkema, L.; Waghray, A.; Heimberg, G.; Kobayashi, Y.; Vaishnav, E.D.; et al. Single-Cell Meta-Analysis of SARS-CoV-2 Entry Genes across Tissues and Demographics. Nat. Med. 2021, 27, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Sen, K.; Datta, S.; Ghosh, A.; Jha, A.; Ahad, A.; Chatterjee, S.; Suranjika, S.; Sengupta, S.; Bhattacharya, G.; Shriwas, O.; et al. Single-Cell Immunogenomic Approach Identified SARS-CoV-2 Protective Immune Signatures in Asymptomatic Direct Contacts of COVID-19 Cases. Front. Immunol. 2021, 12, 733539. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, O. Single-Cell Transcriptomics: Technology and Applications. In Single-Cell Omics; Elsevier: Amsterdam, The Netherlands, 2019; pp. 231–251. ISBN 978-0-12-814919-5. [Google Scholar]
- Zhang, J.; Wang, W.; Huang, J.; Wang, X.; Zeng, Y. How Far Is Single-cell Sequencing from Clinical Application? Clin. Transl. Med. 2020, 10, e117. [Google Scholar] [CrossRef] [PubMed]
- Ozsolak, F.; Milos, P.M. RNA Sequencing: Advances, Challenges and Opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Garalde, D.R.; Snell, E.A.; Jachimowicz, D.; Sipos, B.; Lloyd, J.H.; Bruce, M.; Pantic, N.; Admassu, T.; James, P.; Warland, A.; et al. Highly Parallel Direct RNA Sequencing on an Array of Nanopores. Nat. Methods 2018, 15, 201–206. [Google Scholar] [CrossRef]
- Weirather, J.L.; de Cesare, M.; Wang, Y.; Piazza, P.; Sebastiano, V.; Wang, X.-J.; Buck, D.; Au, K.F. Comprehensive Comparison of Pacific Biosciences and Oxford Nanopore Technologies and Their Applications to Transcriptome Analysis. F1000Research 2017, 6, 100. [Google Scholar] [CrossRef]
- Moor, A.E.; Itzkovitz, S. Spatial Transcriptomics: Paving the Way for Tissue-Level Systems Biology. Curr. Opin. Biotechnol. 2017, 46, 126–133. [Google Scholar] [CrossRef]
- Tang, A.D.; Soulette, C.M.; van Baren, M.J.; Hart, K.; Hrabeta-Robinson, E.; Wu, C.J.; Brooks, A.N. Full-Length Transcript Characterization of SF3B1 Mutation in Chronic Lymphocytic Leukemia Reveals Downregulation of Retained Introns. Nat. Commun. 2020, 11, 1438. [Google Scholar] [CrossRef]
- Wyman, D.; Balderrama-Gutierrez, G.; Reese, F.; Jiang, S.; Rahmanian, S.; Forner, S.; Matheos, D.; Zeng, W.; Williams, B.; Trout, D.; et al. A Technology-Agnostic Long-Read Analysis Pipeline for Transcriptome Discovery and Quantification. bioRxiv 2020. [Google Scholar] [CrossRef]
- World Health Organization COVID-19 Weekly Epidemiological Update 2021. Available online: https://apps.who.int/iris/handle/10665/351137 (accessed on 31 July 2022).
- Centers for Disease Control and Prevention SARS-CoV-2 Variant Classifications and Definitions 2021. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html (accessed on 31 July 2022).
- Shu, Y.; McCauley, J. GISAID: Global Initiative on Sharing All Influenza Data—From Vision to Reality. Euro Surveill. 2017, 22, 30494. [Google Scholar] [CrossRef]
- Pillay, S.; Giandhari, J.; Tegally, H.; Wilkinson, E.; Chimukangara, B.; Lessells, R.; Moosa, Y.; Mattison, S.; Gazy, I.; Fish, M.; et al. Whole Genome Sequencing of SARS-CoV-2: Adapting Illumina Protocols for Quick and Accurate Outbreak Investigation during a Pandemic. Genes 2020, 11, 949. [Google Scholar] [CrossRef] [PubMed]
- Elbe, S.; Buckland-Merrett, G. Data, Disease and Diplomacy: GISAID’s Innovative Contribution to Global Health: Data, Disease and Diplomacy. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Mignardi, M.; Nilsson, M. Fourth-Generation Sequencing in the Cell and the Clinic. Genome Med. 2014, 6, 31. [Google Scholar] [CrossRef]
- Cocquet, J.; Chong, A.; Zhang, G.; Veitia, R.A. Reverse Transcriptase Template Switching and False Alternative Transcripts. Genomics 2006, 88, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Tyson, J.R.; James, P.; Stoddart, D.; Sparks, N.; Wickenhagen, A.; Hall, G.; Choi, J.H.; Lapointe, H.; Kamelian, K.; Smith, A.D.; et al. Improvements to the ARTIC Multiplex PCR Method for SARS-CoV-2 Genome Sequencing Using Nanopore. bioRxiv 2020. [Google Scholar] [CrossRef]
- Campos, J.H.C.; Maricato, J.T.; Braconi, C.T.; Antoneli, F.; Janini, L.M.R.; Briones, M.R.S. Direct RNA Sequencing Reveals SARS-CoV-2 M6A Sites and Possible Differential DRACH Motif Methylation among Variants. Viruses 2021, 13, 2108. [Google Scholar] [CrossRef]
- Vacca, D.; Fiannaca, A.; Tramuto, F.; Cancila, V.; La Paglia, L.; Mazzucco, W.; Gulino, A.; La Rosa, M.; Maida, C.M.; Morello, G.; et al. Direct RNA Nanopore Sequencing of SARS-CoV-2 Extracted from Critical Material from Swabs. Life. 2022, 12, 69. [Google Scholar] [CrossRef]
- Viehweger, A.; Krautwurst, S.; Lamkiewicz, K.; Madhugiri, R.; Ziebuhr, J.; Hölzer, M.; Marz, M. Direct RNA Nanopore Sequencing of Full-Length Coronavirus Genomes Provides Novel Insights into Structural Variants and Enables Modification Analysis. Genome Res. 2019, 29, 1545–1554. [Google Scholar] [CrossRef]
- Athanasopoulou, K.; Boti, M.A.; Adamopoulos, P.G.; Skourou, P.C.; Scorilas, A. Third-Generation Sequencing: The Spearhead towards the Radical Transformation of Modern Genomics. Life 2021, 12, 30. [Google Scholar] [CrossRef]
- Vandelli, A.; Monti, M.; Milanetti, E.; Armaos, A.; Rupert, J.; Zacco, E.; Bechara, E.; Delli Ponti, R.; Tartaglia, G.G. Structural Analysis of SARS-CoV-2 Genome and Predictions of the Human Interactome. Nucleic Acids Res. 2020, 48, 11270–11283. [Google Scholar] [CrossRef] [PubMed]
- Bojkova, D.; Klann, K.; Koch, B.; Widera, M.; Krause, D.; Ciesek, S.; Cinatl, J.; Münch, C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 2020, 586, 469–472. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Genomic Sequencing of SARS-CoV-2 A Guide to Implementation for Maximum Impact on Public Health 8 January 2021. Available online: https://www.who.int/publications/i/item/9789240018440 (accessed on 31 July 2022).
- Husso, T.; Ylä-Herttuala, S.; Turunen, M.P. A New Gene Therapy Approach for Cardiovascular Disease by Non-Coding RNAs Acting in the Nucleus. Mol. Ther. Nucleic Acids 2014, 3, e197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, L.; Zhang, Y.; Fang, D. Alleviation of Neurological Disease by RNA Editing. Methods 2021, 194, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-Coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Liu, Y.; Chemparathy, A.; Pande, T.; La Russa, M.; Qi, L.S. A Comprehensive Analysis and Resource to Use CRISPR-Cas13 for Broad-Spectrum Targeting of RNA Viruses. Cell Rep. Med. 2021, 2, 100245. [Google Scholar] [CrossRef] [PubMed]
- Klim, J.R.; Vance, C.; Scotter, E.L. Antisense Oligonucleotide Therapies for Amyotrophic Lateral Sclerosis: Existing and Emerging Targets. Int. J. Biochem. Cell Biol. 2019, 110, 149–153. [Google Scholar] [CrossRef]
- Abu-Baker, A.; Kharma, N.; Perreault, J.; Grant, A.; Shekarabi, M.; Maios, C.; Dona, M.; Neri, C.; Dion, P.A.; Parker, A.; et al. RNA-Based Therapy Utilizing Oculopharyngeal Muscular Dystrophy Transcript Knockdown and Replacement. Mol. Ther. Nucleic Acids 2019, 15, 12–25. [Google Scholar] [CrossRef]
- Ledford, H. Gene-Silencing Technology Gets First Drug Approval after 20-Year Wait. Nature 2018, 560, 291–292. [Google Scholar] [CrossRef] [PubMed]
- van de Berg, D.; Kis, Z.; Behmer, C.F.; Samnuan, K.; Blakney, A.K.; Kontoravdi, C.; Shattock, R.; Shah, N. Quality by Design Modelling to Support Rapid RNA Vaccine Production against Emerging Infectious Diseases. NPJ Vaccines 2021, 6, 65. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Oehm, P.; Derhovanessian, E.; Jabulowsky, R.A.; Vormehr, M.; Gold, M.; Maurus, D.; Schwarck-Kokarakis, D.; Kuhn, A.N.; Omokoko, T.; et al. An RNA Vaccine Drives Immunity in Checkpoint-Inhibitor-Treated Melanoma. Nature 2020, 585, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Dowdy, S.F. Overcoming Cellular Barriers for RNA Therapeutics. Nat. Biotechnol. 2017, 35, 8. [Google Scholar] [CrossRef] [PubMed]
- Francisco, V.; Rebelo, C.; Rodrigues, A.F.; Blersch, J.; Fernandes, H.; Ferreira, L. A High-Throughput Screening Platform to Identify Nanocarriers for Efficient Delivery of RNA-Based Therapies. Methods 2020, 190, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Juliano, R.L.; Ming, X.; Carver, K.; Laing, B. Cellular Uptake and Intracellular Trafficking of Oligonucleotides: Implications for Oligonucleotide Pharmacology. Nucleic Acid Ther. 2014, 24, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Chow, M.Y.T.; Qiu, Y.; Lam, J.K.W. Inhaled RNA Therapy: From Promise to Reality. Trends Pharmacol. Sci. 2020, 41, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Finkel, R.S.; Mercuri, E.; Darras, B.T.; Connolly, A.M.; Kuntz, N.L.; Kirschner, J.; Chiriboga, C.A.; Saito, K.; Servais, L.; Tizzano, E.; et al. Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N. Engl. J. Med. 2017, 377, 1723–1732. [Google Scholar] [CrossRef]
- Neil, E.E.; Bisaccia, E.K. Nusinersen: A Novel Antisense Oligonucleotide for the Treatment of Spinal Muscular Atrophy. J. Pediatr. Pharmacol. Ther. 2019, 24, 194–203. [Google Scholar] [CrossRef]
- De Wel, B.; Goosens, V.; Sobota, A.; Van Camp, E.; Geukens, E.; Van Kerschaver, G.; Jagut, M.; Claes, K.; Claeys, K.G. Nusinersen Treatment Significantly Improves Hand Grip Strength, Hand Motor Function and MRC Sum Scores in Adult Patients with Spinal Muscular Atrophy Types 3 and 4. J. Neurol. 2021, 268, 923–935. [Google Scholar] [CrossRef]
- Adams, D.; Polydefkis, M.; González-Duarte, A.; Wixner, J.; Kristen, A.V.; Schmidt, H.H.; Berk, J.L.; Losada López, I.A.; Dispenzieri, A.; Quan, D.; et al. Long-Term Safety and Efficacy of Patisiran for Hereditary Transthyretin-Mediated Amyloidosis with Polyneuropathy: 12-Month Results of an Open-Label Extension Study. Lancet Neurol. 2021, 20, 49–59. [Google Scholar] [CrossRef]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.-C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef]
- Coelho, T.; Adams, D.; Silva, A.; Lozeron, P.; Hawkins, P.N.; Mant, T.; Perez, J.; Chiesa, J.; Warrington, S.; Tranter, E.; et al. Safety and Efficacy of RNAi Therapy for Transthyretin Amyloidosis. N. Engl. J. Med. 2013, 369, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Niktab, I.; Haghparast, M.; Beigi, M.-H.; Megraw, T.L.; Kiani, A.; Ghaedi, K. Design of Advanced SiRNA Therapeutics for the Treatment of COVID-19. Meta Gene 2021, 29, 100910. [Google Scholar] [CrossRef] [PubMed]
- Steinbach, J.M.; Weller, C.E.; Booth, C.J.; Saltzman, W.M. Polymer Nanoparticles Encapsulating SiRNA for Treatment of HSV-2 Genital Infection. J. Control. Release 2012, 162, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Zhao, F.; Sun, H.; Wang, Z.; Huang, Y.; Zhu, W.; Xu, F.; Mei, S.; Liu, X.; Zhang, D.; et al. CRISPR-Cas13a Inhibits HIV-1 Infection. Mol. Ther. Nucleic Acids 2020, 21, 147–155. [Google Scholar] [CrossRef]
- Cao, C.; Cai, Z.; Xiao, X.; Rao, J.; Chen, J.; Hu, N.; Yang, M.; Xing, X.; Wang, Y.; Li, M.; et al. The Architecture of the SARS-CoV-2 RNA Genome inside Virion. Nat. Commun. 2021, 12, 3917. [Google Scholar] [CrossRef]
- Liu, C.; Zhou, Q.; Li, Y.; Garner, L.V.; Watkins, S.P.; Carter, L.J.; Smoot, J.; Gregg, A.C.; Daniels, A.D.; Jervey, S.; et al. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. ACS Cent. Sci. 2020, 6, 315–331. [Google Scholar] [CrossRef]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.-J.; Jiang, S. The Spike Protein of SARS-CoV—A Target for Vaccine and Therapeutic Development. Nat. Rev. Microbiol. 2009, 7, 226–236. [Google Scholar] [CrossRef]
- Liu, Y.; Soh, W.T.; Kishikawa, J.; Hirose, M.; Nakayama, E.E.; Li, S.; Sasai, M.; Suzuki, T.; Tada, A.; Arakawa, A.; et al. An Infectivity-Enhancing Site on the SARS-CoV-2 Spike Protein Targeted by Antibodies. Cell 2021, 184, 3452–3466.e18. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell Entry Mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Kim, J.H.; Marks, F.; Clemens, J.D. Looking beyond COVID-19 Vaccine Phase 3 Trials. Nat. Med. 2021, 27, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Jalkanen, P.; Kolehmainen, P.; Häkkinen, H.K.; Huttunen, M.; Tähtinen, P.A.; Lundberg, R.; Maljanen, S.; Reinholm, A.; Tauriainen, S.; Pakkanen, S.H.; et al. COVID-19 MRNA Vaccine Induced Antibody Responses against Three SARS-CoV-2 Variants. Nat. Commun. 2021, 12, 3991. [Google Scholar] [CrossRef] [PubMed]
- McDonald, I.; Murray, S.M.; Reynolds, C.J.; Altmann, D.M.; Boyton, R.J. Comparative Systematic Review and Meta-Analysis of Reactogenicity, Immunogenicity and Efficacy of Vaccines against SARS-CoV-2. NPJ Vaccines 2021, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Nalbandian, A.; Sehgal, K.; Gupta, A.; Madhavan, M.V.; McGroder, C.; Stevens, J.S.; Cook, J.R.; Nordvig, A.S.; Shalev, D.; Sehrawat, T.S.; et al. Post-Acute COVID-19 Syndrome. Nat. Med. 2021, 27, 601–615. [Google Scholar] [CrossRef]
- Mizrahi, B.; Shilo, S.; Rossman, H.; Kalkstein, N.; Marcus, K.; Barer, Y.; Keshet, A.; Shamir-Stein, N.; Shalev, V.; Zohar, A.E.; et al. Longitudinal Symptom Dynamics of COVID-19 Infection. Nat. Commun. 2020, 11, 6208. [Google Scholar] [CrossRef]
- Al-Aly, Z.; Xie, Y.; Bowe, B. High-Dimensional Characterization of Post-Acute Sequelae of COVID-19. Nature 2021, 594, 259–264. [Google Scholar] [CrossRef]
- di Filippo, L.; Frara, S.; Giustina, A. The Emerging Osteo-Metabolic Phenotype of COVID-19: Clinical and Pathophysiological Aspects. Nat. Rev. Endocrinol. 2021, 17, 445–446. [Google Scholar] [CrossRef]
- Legrand, M.; Bell, S.; Forni, L.; Joannidis, M.; Koyner, J.L.; Liu, K.; Cantaluppi, V. Pathophysiology of COVID-19-Associated Acute Kidney Injury. Nat. Rev. Nephrol. 2021, 17, 751–764. [Google Scholar] [CrossRef]
- Nishiga, M.; Wang, D.W.; Han, Y.; Lewis, D.B.; Wu, J.C. COVID-19 and Cardiovascular Disease: From Basic Mechanisms to Clinical Perspectives. Nat. Rev. Cardiol. 2020, 17, 543–558. [Google Scholar] [CrossRef]
- Hao, F.; Tam, W.; Hu, X.; Tan, W.; Jiang, L.; Jiang, X.; Zhang, L.; Zhao, X.; Zou, Y.; Hu, Y.; et al. A Quantitative and Qualitative Study on the Neuropsychiatric Sequelae of Acutely Ill COVID-19 Inpatients in Isolation Facilities. Transl. Psychiatry 2020, 10, 355. [Google Scholar] [CrossRef]
- Mirfazeli, F.S.; Sarabi-Jamab, A.; Jahanbakhshi, A.; Kordi, A.; Javadnia, P.; Shariat, S.V.; Aloosh, O.; Almasi-Dooghaee, M.; Faiz, S.H.R. Neuropsychiatric Manifestations of COVID-19 Can Be Clustered in Three Distinct Symptom Categories. Sci. Rep. 2020, 10, 20957. [Google Scholar] [CrossRef] [PubMed]
- Kashif, A.; Chaudhry, M.; Fayyaz, T.; Abdullah, M.; Malik, A.; Anwer, J.M.A.; Inam, S.H.A.; Fatima, T.; Iqbal, N.; Shoaib, K. Follow-up of COVID-19 Recovered Patients with Mild Disease. Sci. Rep. 2021, 11, 13414. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-Nucleotide RNAs Mediate RNA Interference in Cultured Mammalian Cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Lendeckel, W.; Tuschl, T. RNA Interference Is Mediated by 21- and 22-Nucleotide RNAs. Genes Dev. 2001, 15, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Ameres, S.L.; Martinez, J.; Schroeder, R. Molecular Basis for Target RNA Recognition and Cleavage by Human RISC. Cell 2007, 130, 101–112. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Shi, Y.; Yang, D.H.; Xiong, J.; Jia, J.; Huang, B.; Jin, Y.X. Inhibition of Genes Expression of SARS Coronavirus by Synthetic Small Interfering RNAs. Cell Res. 2005, 15, 193–200. [Google Scholar] [CrossRef]
- Wu, C.-J.; Huang, H.-W.; Liu, C.-Y.; Hong, C.-F.; Chan, Y.-L. Inhibition of SARS-CoV Replication by SiRNA. Antivir. Res. 2005, 65, 45–48. [Google Scholar] [CrossRef]
- Wang, Z.; Ren, L.; Zhao, X.; Hung, T.; Meng, A.; Wang, J.; Chen, Y.-G. Inhibition of Severe Acute Respiratory Syndrome Virus Replication by Small Interfering RNAs in Mammalian Cells. J. Virol. 2004, 78, 7523–7527. [Google Scholar] [CrossRef]
- Li, B.; Tang, Q.; Cheng, D.; Qin, C.; Xie, F.Y.; Wei, Q.; Xu, J.; Liu, Y.; Zheng, B.; Woodle, M.C.; et al. Using SiRNA in Prophylactic and Therapeutic Regimens against SARS Coronavirus in Rhesus Macaque. Nat. Med. 2005, 11, 944–951. [Google Scholar] [CrossRef]
- Gallicano, G.I.; Casey, J.L.; Fu, J.; Mahapatra, S. Molecular Targeting of Vulnerable RNA Sequences in SARS CoV-2: Identifying Clinical Feasibility. Gene Ther. 2020, 29, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Juliano, R.L. The Delivery of Therapeutic Oligonucleotides. Nucleic Acids Res. 2016, 44, 6518–6548. [Google Scholar] [CrossRef]
- Idris, A.; Davis, A.; Supramaniam, A.; Acharya, D.; Kelly, G.; Tayyar, Y.; West, N.; Zhang, P.; McMillan, C.L.D.; Soemardy, C.; et al. A SARS-CoV-2 Targeted SiRNA-Nanoparticle Therapy for COVID-19. Mol. Ther. 2021, 29, 2219–2226. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Y.; Putral, L.N.; Liang, M.; Chang, H.-I.; Davies, N.M.; McMillan, N.A.J. Development of a Novel Method for Formulating Stable SiRNA-Loaded Lipid Particles for in Vivo Use. Pharm. Res. 2009, 26, 512–522. [Google Scholar] [CrossRef]
- McCaskill, J.; Singhania, R.; Burgess, M.; Allavena, R.; Wu, S.; Blumenthal, A.; McMillan, N.A. Efficient Biodistribution and Gene Silencing in the Lung Epithelium via Intravenous Liposomal Delivery of SiRNA. Mol. Ther. Nucleic Acids 2013, 2, e96. [Google Scholar] [CrossRef] [PubMed]
- Khaitov, M.; Nikonova, A.; Shilovskiy, I.; Kozhikhova, K.; Kofiadi, I.; Vishnyakova, L.; Nikolskii, A.; Gattinger, P.; Kovchina, V.; Barvinskaia, E.; et al. Silencing of SARS-CoV-2 with Modified SiRNA-Peptide Dendrimer Formulation. Allergy 2021, 76, 2840–2854. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of MiR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef]
- Lam, J.K.W.; Chow, M.Y.T.; Zhang, Y.; Leung, S.W.S. SiRNA Versus MiRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef]
- Doench, J.G.; Sharp, P.A. Specificity of MicroRNA Target Selection in Translational Repression. Genes Dev. 2004, 18, 504–511. [Google Scholar] [CrossRef]
- Alshaer, W.; Zureigat, H.; Al Karaki, A.; Al-Kadash, A.; Gharaibeh, L.; Hatmal, M.M.; Aljabali, A.A.A.; Awidi, A. SiRNA: Mechanism of Action, Challenges, and Therapeutic Approaches. Eur. J. Pharmacol. 2021, 905, 174178. [Google Scholar] [CrossRef]
- Hennig, T.; Prusty, A.B.; Kaufer, B.B.; Whisnant, A.W.; Lodha, M.; Enders, A.; Thomas, J.; Kasimir, F.; Grothey, A.; Klein, T.; et al. Selective Inhibition of MiRNA Processing by a Herpesvirus-Encoded MiRNA. Nature 2022, 605, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Gahlawat, A.W.; Witte, T.; Haarhuis, L.; Schott, S. A Novel Circulating MiRNA Panel for Non-Invasive Ovarian Cancer Diagnosis and Prognosis. Br. J. Cancer 2022. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Strand, S.H.; Schmidt, L.; Weiss, S.; Borre, M.; Kristensen, H.; Rasmussen, A.K.I.; Daugaard, T.F.; Kristensen, G.; Stroomberg, H.V.; Røder, M.A.; et al. Validation of the Four-MiRNA Biomarker Panel MiCaP for Prediction of Long-Term Prostate Cancer Outcome. Sci. Rep. 2020, 10, 10704. [Google Scholar] [CrossRef]
- Li, Q.; Lowey, B.; Sodroski, C.; Krishnamurthy, S.; Alao, H.; Cha, H.; Chiu, S.; El-Diwany, R.; Ghany, M.G.; Liang, T.J. Cellular MicroRNA Networks Regulate Host Dependency of Hepatitis C Virus Infection. Nat. Commun. 2017, 8, 1789. [Google Scholar] [CrossRef] [PubMed]
- Moffett, H.F.; Cartwright, A.N.R.; Kim, H.-J.; Godec, J.; Pyrdol, J.; Äijö, T.; Martinez, G.J.; Rao, A.; Lu, J.; Golub, T.R.; et al. The MicroRNA MiR-31 Inhibits CD8+ T Cell Function in Chronic Viral Infection. Nat. Immunol. 2017, 18, 791–799. [Google Scholar] [CrossRef]
- Li, C.; Hu, X.; Li, L.; Li, J.-H. Differential MicroRNA Expression in the Peripheral Blood from Human Patients with COVID-19. J. Clin. Lab. Anal. 2020, 34, e23590. [Google Scholar] [CrossRef]
- Samy, A.; Maher, M.A.; Abdelsalam, N.A.; Badr, E. SARS-CoV-2 Potential Drugs, Drug Targets, and Biomarkers: A Viral-Host Interaction Network-Based Analysis. Sci. Rep. 2022, 12, 11934. [Google Scholar] [CrossRef]
- Khan, A.-A.-K.; Sany, R.U.; Islam, S.; Islam, A.B.M.K. Epigenetic Regulator MiRNA Pattern Differences Among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 World-Wide Isolates Delineated the Mystery Behind the Epic Pathogenicity and Distinct Clinical Characteristics of Pandemic COVID-19. Front. Genet. 2020, 11, 765. [Google Scholar] [CrossRef]
- Panda, M.; Kalita, E.; Singh, S.; Kumar, K.; Rao, A.; Prajapati, V.K. MiRNA-SARS-CoV-2 Dialogue and Prospective Anti-COVID-19 Therapies. Life Sci. 2022, 305, 120761. [Google Scholar] [CrossRef]
- Ahmed, J.Q.; Maulud, S.Q.; Dhawan, M.; Priyanka; Choudhary, O.P.; Jalal, P.J.; Ali, R.K.; Tayib, G.A.; Hasan, D.A. MicroRNAs in the Development of Potential Therapeutic Targets against COVID-19: A Narrative Review. J. Infect. Public Health 2022, 15, 788–799. [Google Scholar] [CrossRef]
- Vitravene Study Group. A Randomized Controlled Clinical Trial of Intravitreous Fomivirsen for Treatment of Newly Diagnosed Peripheral Cytomegalovirus Retinitis in Patients with Aids. Am. J. Ophthalmol. 2002, 133, 467–474. [Google Scholar] [CrossRef]
- Janssen, H.L.A.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV Infection by Targeting MicroRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Zhang, Z.; Zhang, W.; Feng, X.; Xu, L. Prodrug-Type Antisense Oligonucleotides with Enhanced Nuclease Stability and Anti-Tumour Effects. Eur. J. Pharm. Sci. 2021, 162, 105832. [Google Scholar] [CrossRef] [PubMed]
- Kole, R. RNA Therapeutics: Beyond RNA Interference and Antisense Oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140. [Google Scholar] [CrossRef] [PubMed]
- DeVos, S.L.; Miller, T.M. Antisense Oligonucleotides: Treating Neurodegeneration at the Level of RNA. Neurotherapeutics 2013, 10, 486–497. [Google Scholar] [CrossRef]
- Le, B.T.; Raguraman, P.; Kosbar, T.R.; Fletcher, S.; Wilton, S.D.; Veedu, R.N. Antisense Oligonucleotides Targeting Angiogenic Factors as Potential Cancer Therapeutics. Mol. Ther. Nucleic Acids 2019, 14, 142–157. [Google Scholar] [CrossRef]
- Berber, B.; Aydin, C.; Kocabas, F.; Guney-Esken, G.; Yilancioglu, K.; Karadag-Alpaslan, M.; Caliseki, M.; Yuce, M.; Demir, S.; Tastan, C. Gene Editing and RNAi Approaches for COVID-19 Diagnostics and Therapeutics. Gene Ther. 2020, 28, 290–305. [Google Scholar] [CrossRef]
- Crooke, S.T. Molecular Mechanisms of Antisense Oligonucleotides. Nucleic Acid Ther. 2017, 27, 70–77. [Google Scholar] [CrossRef]
- Lim, K.H.; Han, Z.; Jeon, H.Y.; Kach, J.; Jing, E.; Weyn-Vanhentenryck, S.; Downs, M.; Corrionero, A.; Oh, R.; Scharner, J.; et al. Antisense Oligonucleotide Modulation of Non-Productive Alternative Splicing Upregulates Gene Expression. Nat. Commun. 2020, 11, 3501. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA Editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Gouni-Berthold, I. The Role of Antisense Oligonucleotide Therapy against Apolipoprotein-CIII in Hypertriglyceridemia. Atheroscler. Suppl. 2017, 30, 19–27. [Google Scholar] [CrossRef]
- Zorzi, F.; Angelucci, E.; Sedda, S.; Pallone, F.; Monteleone, G. Smad7 Antisense Oligonucleotide-Based Therapy for Inflammatory Bowel Diseases. Dig. Liver Dis. 2013, 45, 552–555. [Google Scholar] [CrossRef] [PubMed]
- Dulla, K.; Slijkerman, R.; van Diepen, H.C.; Albert, S.; Dona, M.; Beumer, W.; Turunen, J.J.; Chan, H.L.; Schulkens, I.A.; Vorthoren, L.; et al. Antisense Oligonucleotide-Based Treatment of Retinitis Pigmentosa Caused by USH2A Exon 13 Mutations. Mol. Ther. 2021, 29, 2441–2455. [Google Scholar] [CrossRef] [PubMed]
- Yuen, M.-F.; Gane, E.; Kim, D.J.; Chan, H.; Surujbally, B.; Pavlovic, V.; Triyatni, M.; Grippo, J.; Kim, H.J.; Leerapun, A.; et al. RO7062931 Antisense Oligonucleotide Phase 1 Study Demonstrates Target Engagement in Patients with Chronic Hepatitis B on Established Nucleos(t)Ide Therapy. J. Hepatol. 2020, 73, S51. [Google Scholar] [CrossRef]
- Billioud, G.; Kruse, R.L.; Carrillo, M.; Whitten-Bauer, C.; Gao, D.; Kim, A.; Chen, L.; McCaleb, M.L.; Crosby, J.R.; Hamatake, R.; et al. In Vivo Reduction of Hepatitis B Virus Antigenemia and Viremia by Antisense Oligonucleotides. J. Hepatol. 2016, 64, 781–789. [Google Scholar] [CrossRef]
- Chery, J.; Petri, A.; Wagschal, A.; Lim, S.-Y.; Cunningham, J.; Vasudevan, S.; Kauppinen, S.; Näär, A.M. Development of Locked Nucleic Acid Antisense Oligonucleotides Targeting Ebola Viral Proteins and Host Factor Niemann-Pick C1. Nucleic Acid Ther. 2018, 28, 273–284. [Google Scholar] [CrossRef]
- Lenartowicz, E.; Nogales, A.; Kierzek, E.; Kierzek, R.; Martínez-Sobrido, L.; Turner, D.H. Antisense Oligonucleotides Targeting Influenza A Segment 8 Genomic RNA Inhibit Viral Replication. Nucleic Acid Ther. 2016, 26, 277–285. [Google Scholar] [CrossRef]
- Deas, T.S.; Bennett, C.J.; Jones, S.A.; Tilgner, M.; Ren, P.; Behr, M.J.; Stein, D.A.; Iversen, P.L.; Kramer, L.D.; Bernard, K.A.; et al. In Vitro Resistance Selection and In Vivo Efficacy of Morpholino Oligomers against West Nile Virus. Antimicrob. Agents Chemother. 2007, 51, 2470–2482. [Google Scholar] [CrossRef]
- Sun, L.; Li, P.; Ju, X.; Rao, J.; Huang, W.; Ren, L.; Zhang, S.; Xiong, T.; Xu, K.; Zhou, X.; et al. In Vivo Structural Characterization of the SARS-CoV-2 RNA Genome Identifies Host Proteins Vulnerable to Repurposed Drugs. Cell 2021, 184, 1865–1883.e20. [Google Scholar] [CrossRef]
- Hasan, M.M.; Ragnarsson, L.; Cardoso, F.C.; Lewis, R.J. Transfection Methods for High-Throughput Cellular Assays of Voltage-Gated Calcium and Sodium Channels Involved in Pain. PLoS ONE 2021, 16, e0243645. [Google Scholar] [CrossRef]
- Clark, D.P.; Pazdernik, N.J.; McGehee, M.R. Manipulation of Nucleic Acids. In Molecular Biology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 132–166. ISBN 978-0-12-813288-3. [Google Scholar]
- Ahn, D.-G.; Lee, W.; Choi, J.-K.; Kim, S.-J.; Plant, E.P.; Almazán, F.; Taylor, D.R.; Enjuanes, L.; Oh, J.-W. Interference of Ribosomal Frameshifting by Antisense Peptide Nucleic Acids Suppresses SARS Coronavirus Replication. Antivir. Res. 2011, 91, 1–10. [Google Scholar] [CrossRef]
- Kelly, J.A.; Woodside, M.T.; Dinman, J.D. Programmed −1 Ribosomal Frameshifting in Coronaviruses: A Therapeutic Target. Virology 2021, 554, 75–82. [Google Scholar] [CrossRef]
- Jacobsen, L.B.; Calvin, S.A.; Colvin, K.E.; Wright, M. FuGENE 6 Transfection Reagent: The Gentle Power. Methods 2004, 33, 104–112. [Google Scholar] [CrossRef]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef]
- Workman, R.E.; Pammi, T.; Nguyen, B.T.K.; Graeff, L.W.; Smith, E.; Sebald, S.M.; Stoltzfus, M.J.; Euler, C.W.; Modell, J.W. A Natural Single-Guide RNA Repurposes Cas9 to Autoregulate CRISPR-Cas Expression. Cell 2021, 184, 675–688.e19. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Barrangou, R.; van der Oost, J. (Eds.) CRISPR-Cas Systems; Springer: Berlin/Heidelberg, Germany, 2013; ISBN 978-3-642-34656-9. [Google Scholar]
- Sioud, M. (Ed.) RNA Interference and CRISPR Technologies: Technical Advances and New Therapeutic Opportunities (Methods in Molecular Biology); Springer: New York, NY, USA, 2020; Volume 2115, ISBN 978-1-07-160289-8. [Google Scholar]
- Soares, F.; Chen, B.; Lee, J.B.; Ahmed, M.; Ly, D.; Tin, E.; Kang, H.; Zeng, Y.; Akhtar, N.; Minden, M.D.; et al. CRISPR Screen Identifies Genes That Sensitize AML Cells to Double-Negative T-Cell Therapy. Blood 2021, 137, 2171–2181. [Google Scholar] [CrossRef]
- Zeballos, C.M.A.; Gaj, T. Next-Generation CRISPR Technologies and Their Applications in Gene and Cell Therapy. Trends Biotechnol. 2021, 39, 692–705. [Google Scholar] [CrossRef]
- Liao, H.-K.; Gu, Y.; Diaz, A.; Marlett, J.; Takahashi, Y.; Li, M.; Suzuki, K.; Xu, R.; Hishida, T.; Chang, C.-J.; et al. Use of the CRISPR/Cas9 System as an Intracellular Defense against HIV-1 Infection in Human Cells. Nat. Commun. 2015, 6, 6413. [Google Scholar] [CrossRef]
- Yin, D.; Ling, S.; Wang, D.; Dai, Y.; Jiang, H.; Zhou, X.; Paludan, S.R.; Hong, J.; Cai, Y. Targeting Herpes Simplex Virus with CRISPR–Cas9 Cures Herpetic Stromal Keratitis in Mice. Nat. Biotechnol. 2021, 39, 567–577. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, Classification and Evolution of CRISPR-Cas Systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Mancilla, A.; Wessels, H.-H.; Legut, M.; Kadina, A.; Mabuchi, M.; Walker, J.; Robb, G.B.; Holden, K.; Sanjana, N.E. Chemically Modified Guide RNAs Enhance CRISPR-Cas13 Knockdown in Human Cells. Cell Chem. Biol. 2021, 29, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Smargon, A.A.; Shi, Y.J.; Yeo, G.W. RNA-Targeting CRISPR Systems from Metagenomic Discovery to Transcriptomic Engineering. Nat. Cell Biol. 2020, 22, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Abbott, T.R.; Dhamdhere, G.; Liu, Y.; Lin, X.; Goudy, L.; Zeng, L.; Chemparathy, A.; Chmura, S.; Heaton, N.S.; Debs, R.; et al. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell 2020, 181, 865–876.e12. [Google Scholar] [CrossRef] [PubMed]
- Fareh, M.; Zhao, W.; Hu, W.; Casan, J.M.L.; Kumar, A.; Symons, J.; Zerbato, J.M.; Fong, D.; Voskoboinik, I.; Ekert, P.G.; et al. Reprogrammed CRISPR-Cas13b Suppresses SARS-CoV-2 Replication and Circumvents Its Mutational Escape through Mismatch Tolerance. Nat. Commun. 2021, 12, 4270. [Google Scholar] [CrossRef]
- Dufait, I.; Liechtenstein, T.; Lanna, A.; Bricogne, C.; Laranga, R.; Padella, A.; Breckpot, K.; Escors, D. Retroviral and Lentiviral Vectors for the Induction of Immunological Tolerance. Scientifica 2012, 2012, 694137. [Google Scholar] [CrossRef]
- Behr, M.; Zhou, J.; Xu, B.; Zhang, H. In Vivo Delivery of CRISPR-Cas9 Therapeutics: Progress and Challenges. Acta Pharm. Sin. B 2021, 11, 2150–2171. [Google Scholar] [CrossRef]
- Blanchard, E.L.; Vanover, D.; Bawage, S.S.; Tiwari, P.M.; Rotolo, L.; Beyersdorf, J.; Peck, H.E.; Bruno, N.C.; Hincapie, R.; Michel, F.; et al. Treatment of Influenza and SARS-CoV-2 Infections via MRNA-Encoded Cas13a in Rodents. Nat. Biotechnol. 2021, 39, 717–726. [Google Scholar] [CrossRef]
- Patel, A.K.; Kaczmarek, J.C.; Bose, S.; Kauffman, K.J.; Mir, F.; Heartlein, M.W.; DeRosa, F.; Langer, R.; Anderson, D.G. Inhaled Nanoformulated MRNA Polyplexes for Protein Production in Lung Epithelium. Adv. Mater. 2019, 31, 1805116. [Google Scholar] [CrossRef]
- König, I.R.; Fuchs, O.; Hansen, G.; von Mutius, E.; Kopp, M.V. What Is Precision Medicine? Eur. Respir. J. 2017, 50, 1700391. [Google Scholar] [CrossRef]
- Ashley, E.A. Towards Precision Medicine. Nat. Rev. Genet. 2016, 17, 507–522. [Google Scholar] [CrossRef]
- Zeggini, E.; Gloyn, A.L.; Barton, A.C.; Wain, L.V. Translational Genomics and Precision Medicine: Moving from the Lab to the Clinic. Science 2019, 365, 1409–1413. [Google Scholar] [CrossRef] [PubMed]
- Khoury, M.J.; Holt, K.E. The Impact of Genomics on Precision Public Health: Beyond the Pandemic. Genome Med. 2021, 13, 67. [Google Scholar] [CrossRef]
- Khoury, M.J.; Armstrong, G.L.; Bunnell, R.E.; Cyril, J.; Iademarco, M.F. The Intersection of Genomics and Big Data with Public Health: Opportunities for Precision Public Health. PLoS Med. 2020, 17, e1003373. [Google Scholar] [CrossRef]
- Precision Public Health as a Key Tool in the COVID-19 Response|Genetics and Genomics|JAMA|JAMA Network. Available online: https://jamanetwork-com.pbidi.unam.mx:2443/journals/jama/fullarticle/2769563 (accessed on 21 February 2022).
- From Precision Medicine to Precision Public Health: The Dialogue Continues. Available online: https://blogs.cdc.gov/genomics/2022/01/25/from-precision-medicine-2/ (accessed on 31 July 2022).
- Callaway, E. Why Does the Omicron Sub-Variant Spread Faster than the Original? Nature 2022, 602, 556–557. [Google Scholar] [CrossRef]
- Arnold, C. Is Precision Public Health the Future—Or a Contradiction? Nature 2022, 601, 18–20. [Google Scholar] [CrossRef]
- DeMerle, K.; Angus, D.C.; Seymour, C.W. Precision Medicine for COVID-19: Phenotype Anarchy or Promise Realized? JAMA 2021, 325, 2041. [Google Scholar] [CrossRef]
- Bilkey, G.A.; Burns, B.L.; Coles, E.P.; Mahede, T.; Baynam, G.; Nowak, K.J. Optimizing Precision Medicine for Public Health. Front. Public Health 2019, 7, 42. [Google Scholar] [CrossRef]
- Wang, S.; Yao, X.; Ma, S.; Ping, Y.; Fan, Y.; Sun, S.; He, Z.; Shi, Y.; Sun, L.; Xiao, S.; et al. A Single-Cell Transcriptomic Landscape of the Lungs of Patients with COVID-19. Nat. Cell Biol. 2021, 23, 1314–1328. [Google Scholar] [CrossRef]
- Mallapaty, S. Where Did Omicron Come from? Three Key Theories. Nature 2022, 602, 26–28. [Google Scholar] [CrossRef]
- Emanuel, E.J.; Persad, G.; Upshur, R.; Thome, B.; Parker, M.; Glickman, A.; Zhang, C.; Boyle, C.; Smith, M.; Phillips, J.P. Fair Allocation of Scarce Medical Resources in the Time of Covid-19. N. Engl. J. Med. 2020, 382, 2049–2055. [Google Scholar] [CrossRef]
- Supady, A.; Curtis, J.R.; Abrams, D.; Lorusso, R.; Bein, T.; Boldt, J.; Brown, C.E.; Duerschmied, D.; Metaxa, V.; Brodie, D. Allocating Scarce Intensive Care Resources during the COVID-19 Pandemic: Practical Challenges to Theoretical Frameworks. Lancet Respir. Med. 2021, 9, 430–434. [Google Scholar] [CrossRef]
- Williams, I.; Essue, B.; Nouvet, E.; Sandman, L.; Razavi, S.D.; Noorulhuda, M.; Goold, S.; Danis, M.; Biemba, G.; Abelson, J.; et al. Priority Setting during the COVID-19 Pandemic: Going beyond Vaccines. BMJ Glob. Health 2021, 6, e004686. [Google Scholar] [CrossRef]
- Doshi, P. Will Covid-19 Vaccines Save Lives? Current Trials Aren’t Designed to Tell Us. BMJ 2020, 371, m4037. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Luu, L.D.W.; Li, J.; Cui, X.; Yao, H.; Chen, S.; Fu, J.; Wang, L.; Wang, C.; et al. Single-Cell Transcriptomic Atlas Reveals Distinct Immunological Responses between COVID-19 Vaccine and Natural SARS-CoV-2 Infection. J. Med. Virol. 2022, 94, 5304–5324. [Google Scholar] [CrossRef]
- Waltz, E. How Nasal-Spray Vaccines Could Change the Pandemic. Nature 2022, 609, 240–242. [Google Scholar] [CrossRef]
- CDC Newsroom. Available online: https://www.cdc.gov/media/releases/2021/s1227-isolation-quarantine-guidance.html (accessed on 25 February 2022).
- Mueller, Y.M.; Schrama, T.J.; Ruijten, R.; Schreurs, M.W.J.; Grashof, D.G.B.; van de Werken, H.J.G.; Lasinio, G.J.; Álvarez-Sierra, D.; Kiernan, C.H.; Castro Eiro, M.D.; et al. Stratification of Hospitalized COVID-19 Patients into Clinical Severity Progression Groups by Immuno-Phenotyping and Machine Learning. Nat. Commun. 2022, 13, 915. [Google Scholar] [CrossRef]
- Siemieniuk, R.A.; Bartoszko, J.J.; Ge, L.; Zeraatkar, D.; Izcovich, A.; Kum, E.; Pardo-Hernandez, H.; Qasim, A.; Martinez, J.P.D.; Rochwerg, B.; et al. Drug Treatments for Covid-19: Living Systematic Review and Network Meta-Analysis. BMJ 2020, 370, m2980. [Google Scholar] [CrossRef]
- Kalil, A.C.; Patterson, T.F.; Mehta, A.K.; Tomashek, K.M.; Wolfe, C.R.; Ghazaryan, V.; Marconi, V.C.; Ruiz-Palacios, G.M.; Hsieh, L.; Kline, S.; et al. Baricitinib plus Remdesivir for Hospitalized Adults with Covid-19. N. Engl. J. Med. 2021, 384, 795–807. [Google Scholar] [CrossRef]
- Watson, J.; Whiting, P.F.; Brush, J.E. Interpreting a Covid-19 Test Result. BMJ 2020, 369, m1808. [Google Scholar] [CrossRef]
- Drain, P.K. Rapid Diagnostic Testing for SARS-CoV-2. N. Engl. J. Med. 2022, 386, 264–272. [Google Scholar] [CrossRef]
- Garcia-Gordillo, J.A.; Camiro-Zúñiga, A.; Aguilar-Soto, M.; Cuenca, D.; Cadena-Fernández, A.; Khouri, L.S.; Rayek, J.N.; Mercado, M.; The ARMII Study Group. COVID-IRS: A Novel Predictive Score for Risk of Invasive Mechanical Ventilation in Patients with COVID-19. PLoS ONE 2021, 16, e0248357. [Google Scholar] [CrossRef]
- Kc, G.B.; Bocci, G.; Verma, S.; Hassan, M.M.; Holmes, J.; Yang, J.J.; Sirimulla, S.; Oprea, T.I. A Machine Learning Platform to Estimate Anti-SARS-CoV-2 Activities. Nat. Mach. Intell. 2021, 3, 527–535. [Google Scholar] [CrossRef]
- Tregoning, J.S.; Flight, K.E.; Higham, S.L.; Wang, Z.; Pierce, B.F. Progress of the COVID-19 Vaccine Effort: Viruses, Vaccines and Variants versus Efficacy, Effectiveness and Escape. Nat. Rev. Immunol. 2021, 21, 626–636. [Google Scholar] [CrossRef]
- Bernal, A.J.; da Silva, M.M.G.; Musungaie, D.B.; Kovalchuk, E.; Gonzalez, A.; Reyes, V.D.; Martín-Quirós, A.; Caraco, Y.; Williams-Diaz, A.; Brown, M.L.; et al. Molnupiravir for Oral Treatment of Covid-19 in Nonhospitalized Patients. N. Engl. J. Med. 2022, 386, 509–520. [Google Scholar] [CrossRef]
- Hammond, J.; Leister-Tebbe, H.; Gardner, A.; Abreu, P.; Bao, W.; Wisemandle, W.; Baniecki, M.; Hendrick, V.M.; Damle, B.; Simón-Campos, A.; et al. Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with Covid-19. N. Engl. J. Med. 2022, 386, 1397–1408. [Google Scholar] [CrossRef]
- Dolgin, E. The Tangled History of MRNA Vaccines. Nature 2021, 597, 318–324. [Google Scholar] [CrossRef]
- Ari, Ş.; Arikan, M. Next-Generation Sequencing: Advantages, Disadvantages, and Future. In Plant Omics: Trends and Applications; Hakeem, K.R., Tombuloğlu, H., Tombuloğlu, G., Eds.; Springer: Cham, Switzerland, 2016; pp. 109–135. ISBN 978-3-319-31703-8. [Google Scholar]
- Illumina COVIDSeq Test|SARS-CoV-2 NGS Test (for the COVID-19 Coronavirus). Available online: https://www.illumina.com/products/by-type/ivd-products/covidseq.html (accessed on 18 May 2022).
- Liu, L.; Li, Y.; Li, S.; Hu, N.; He, Y.; Pong, R.; Lin, D.; Lu, L.; Law, M. Comparison of Next-Generation Sequencing Systems. J. Biomed. Biotechnol. 2012, 2012, 251364. [Google Scholar] [CrossRef]
- Sequencing Support—Coverage Calculator. Available online: https://support.illumina.com/downloads/sequencing_coverage_calculator.html (accessed on 18 May 2022).
- Sequencing Platforms|Compare NGS Platform Applications & Specifications. Available online: https://www.illumina.com/systems/sequencing-platforms.html (accessed on 18 May 2022).
- Bcl2fastq Conversion Software. Available online: https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html (accessed on 18 May 2022).
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet.J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Product Comparison. Available online: http://nanoporetech.com/products/comparison (accessed on 18 May 2022).
- Nanopore Sequencing the SARS-CoV-2 Genome: Introduction to Protocol. Available online: http://nanoporetech.com/resource-centre/nanopore-sequencing-sars-cov-2-genome-introduction-protocol (accessed on 18 May 2022).
- Wick, R.R.; Judd, L.M.; Holt, K.E. Performance of Neural Network Basecalling Tools for Oxford Nanopore Sequencing. Genome Biol. 2019, 20, 129. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Loman, N.J.; Quick, J.; Simpson, J.T. A Complete Bacterial Genome Assembled de Novo Using Only Nanopore Sequencing Data. Nat. Methods 2015, 12, 733–735. [Google Scholar] [CrossRef]
- Medaka; Oxford Nanopore Technologies. 2022. Available online: https://github.com/nanoporetech/medaka (accessed on 18 May 2022).
- Chip Calculator. Available online: https://www.ampliseq.com/otherContent/help-content/help_html/GUID-970D8FA8-0057-4ED4-B730-DF97A2F870AD.html (accessed on 18 May 2022).
- SARS-CoV-2 Research Using GeneStudio S5 System—MX. Available online: //www.thermofisher.com/mx/es/home/life-science/sequencing/dna-sequencing/microbial-sequencing/microbial-identification-ion-torrent-next-generation-sequencing/viral-typing/coronavirus-research/genestudio-s5-system.html (accessed on 18 May 2022).
- Ion GeneStudio S5 Specs—MX. Available online: //www.thermofisher.com/mx/es/home/life-science/sequencing/next-generation-sequencing/ion-torrent-next-generation-sequencing-workflow/ion-torrent-next-generation-sequencing-run-sequence/ion-s5-ngs-targeted-sequencing/ion-s5-specifications.html (accessed on 18 May 2022).
- Dohm, J.C.; Peters, P.; Stralis-Pavese, N.; Himmelbauer, H. Benchmarking of Long-Read Correction Methods. NAR Genom. Bioinform. 2020, 2, lqaa037. [Google Scholar] [CrossRef] [PubMed]
- Pacific Biosciences Estimating Library Yield on the PacBio RS II. 2015. Available online: https://www.pacb.com/wp-content/uploads/Pacbio-Guidelines-Project-Submission-SMRT-Sequencing-DNA-v2.1.pdf (accessed on 18 May 2022).
- Coronavirus Sequencing|SARS-CoV-2 (Covid-19) Surveillance. Available online: https://www.pacb.com/research-focus/microbiology/public-health/covid-19-sequencing-tools-and-resources/ (accessed on 18 May 2022).

| Software | Category | Reference * |
|---|---|---|
| TopHat2 v2.1.1 | Read mapping | https://ccb.jhu.edu/software/tophat/index.shtml Cole Trapnell, Lior Pachter and Steven Salzberg at the University of Maryland, UC Berkeley, USA [58] |
| STAR v2.7.10a | Read mapping | https://github.com/alexdobin/STAR Alexander Dobin, Cold Spring Harbor Laboratory, NY, USA [59] |
| HISAT2 v2.2.2 | Read mapping | http://daehwankimlab.github.io/hisat2/ Kim, D., Langmead, B. & Salzberg, S. Baltimore, Maryland, USA [60] |
| Cufflinks v0.17.3 | Expression quantification | http://cole-trapnell-lab.github.io/cufflinks/ Cole Trapnell, et al. University of Maryland, College Park, Maryland, USA [61] |
| RSEM v1.1.17 | Expression quantification | https://github.com/deweylab/RSEM Bo Li and Colin N Dewey. University of Wisconsin-Madison, Madison, WI, USA [62] |
| StringTie v2.1.0 | Expression quantification | https://ccb.jhu.edu/software/stringtie/ MIT License Johns Hopkins University, Baltimore, Maryland, USA [63] |
| Cell Ranger v3.1.0 | Align reads, generate feature-barcode matrices, perform clustering and other secondary analyses | https://github.com/10XGenomics/cellranger © 2022 10x Genomics. California USA [52] |
| Seurat v4.0 | Filter based on RNA, number of detect genes, and number of total UMIs, | https://satijalab.org/seurat/ Hao, et al., Center for Genomics and Systems Biology, New York University, New York, USA [64] |
| Scrublet v0.2.1 | Single-cell remover of doublets | https://github.com/swolock/scrublet Samuel Wolock, MIT [55] |
| SoupX v1.2.2 | estimation and removal of cell free mRNA contamination | https://github.com/constantAmateur/SoupX Matthew Young. Wellcome Trust Sanger Institute, Cellular Genetics, Wellcome Genome Campus, Hinxton, CB10 1SA, UK [57] |
| Waterfall | Dimensionality reduction | https://doi.org/10.1016/j.stem.2015.07.013 Jaehoon Shin et al. Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA [65] |
| TSCAN v1.0 | Dimensionality reduction | https://github.com/zji90/TSCAN Zhicheng Ji, Hongkai Ji. Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA [66] |
| Study | Status | Interventions | Outcome Measures | Enrolled Patients | Locations | Identifier |
|---|---|---|---|---|---|---|
| COVID-2019 Vaccine Immune Response Based on Single Cell Multi-Omics | Recruiting | Biological: recent vaccination | Changes in classification of human peripheral blood mononuclear cells | 50 | China | NCT04871932 |
| Virological and Immunological Monitoring in Patients (Suspected of/Confirmed With) COVID-19 | Active, not recruiting | Procedure: blood draw Procedure: bronchoalveolar lavage Procedure: SARS-CoV-2 swabs | Identification of cytokines and chemokines associated with COVID-19 severity and outcome Identification of cellular subsets that can predict COVID-19 severity and outcome SARS-CoV-2 sequencing | 109 | Belgium | NCT04904692 |
| COVID-19 in Baselland: Investigation and Validation of Serological Diagnostic Assays and Epidemiological Study of SARS-CoV-2 Specific Antibody Responses COVID-19 | Recruiting | Diagnostic test: blood draw Diagnostic test: fingertip tests for POC assays Diagnostic test: saliva collection Diagnostic test: collection of swabs | Qualitative method validation (yes/no) Quantitative method validation (antibody concentrations) Immune cell repertoire sequencing | 550 | Switzerland | NCT04483908 |
| Myeloid Cells in Patients with COVID-19 Pneumonia | Not yet recruiting | Other: blood sampling Other: nasal brushing | Myeloid cell subpopulation phenotype Myeloid cell functions Myeloid cell transcriptomic and proteomic study. | 120 | France | NCT04590261 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arriaga-Canon, C.; Contreras-Espinosa, L.; Rebollar-Vega, R.; Montiel-Manríquez, R.; Cedro-Tanda, A.; García-Gordillo, J.A.; Álvarez-Gómez, R.M.; Jiménez-Trejo, F.; Castro-Hernández, C.; Herrera, L.A. Transcriptomics and RNA-Based Therapeutics as Potential Approaches to Manage SARS-CoV-2 Infection. Int. J. Mol. Sci. 2022, 23, 11058. https://doi.org/10.3390/ijms231911058
Arriaga-Canon C, Contreras-Espinosa L, Rebollar-Vega R, Montiel-Manríquez R, Cedro-Tanda A, García-Gordillo JA, Álvarez-Gómez RM, Jiménez-Trejo F, Castro-Hernández C, Herrera LA. Transcriptomics and RNA-Based Therapeutics as Potential Approaches to Manage SARS-CoV-2 Infection. International Journal of Molecular Sciences. 2022; 23(19):11058. https://doi.org/10.3390/ijms231911058
Chicago/Turabian StyleArriaga-Canon, Cristian, Laura Contreras-Espinosa, Rosa Rebollar-Vega, Rogelio Montiel-Manríquez, Alberto Cedro-Tanda, José Antonio García-Gordillo, Rosa María Álvarez-Gómez, Francisco Jiménez-Trejo, Clementina Castro-Hernández, and Luis A. Herrera. 2022. "Transcriptomics and RNA-Based Therapeutics as Potential Approaches to Manage SARS-CoV-2 Infection" International Journal of Molecular Sciences 23, no. 19: 11058. https://doi.org/10.3390/ijms231911058
APA StyleArriaga-Canon, C., Contreras-Espinosa, L., Rebollar-Vega, R., Montiel-Manríquez, R., Cedro-Tanda, A., García-Gordillo, J. A., Álvarez-Gómez, R. M., Jiménez-Trejo, F., Castro-Hernández, C., & Herrera, L. A. (2022). Transcriptomics and RNA-Based Therapeutics as Potential Approaches to Manage SARS-CoV-2 Infection. International Journal of Molecular Sciences, 23(19), 11058. https://doi.org/10.3390/ijms231911058