Abstract
Increased salinity is one of the major consequences of climatic change affecting global crop production. The early stages in the barley (Hordeum vulgare L.) life cycle are considered the most critical phases due to their contributions to final crop yield. Particularly, the germination and seedling development are sensitive to numerous environmental stresses, especially soil salinity. In this study, we aimed to identify SNP markers linked with germination and seedling development at 150 mM NaCl as a salinity treatment. We performed a genome-wide association study (GWAS) using a panel of 208 intermedium-spike barley (H. vulgare convar. intermedium (Körn.) Mansf.) accessions and their genotype data (i.e., 10,323 SNPs) using the genome reference sequence of “Morex”. The phenotypic results showed that the 150 mM NaCl salinity treatment significantly reduced all recorded germination and seedling-related traits compared to the control treatment. Furthermore, six accessions (HOR 11747, HOR 11718, HOR 11640, HOR 11256, HOR 11275 and HOR 11291) were identified as the most salinity tolerant from the intermedium-spike barley collection. GWAS analysis indicated that a total of 38 highly significantly associated SNP markers under control and/or salinity traits were identified. Of these, two SNP markers on chromosome (chr) 1H, two on chr 3H, and one on chr 4H were significantly linked to seedling fresh and dry weight under salinity stress treatment. In addition, two SNP markers on chr 7H were also significantly associated with seedling fresh and dry weight but under control condition. Under salinity stress, one SNP marker on chr 1H, 5H and 7H were detected for more than one phenotypic trait. We found that in most of the accessions exhibiting the highest salinity tolerance, most of the salinity-related QTLs were presented. These results form the basis for detailed studies, leading to improved salt tolerance breeding programs in barley.
1. Introduction
Abiotic stresses such as salinity, drought, low or high temperatures, floods and frost are global environmental phenomena that negatively affect many plant species and are expected to become more severe and widespread [1]. Soil salinity problems exist in more than 100 countries, and their global area is approximately 1.13 billion hectares. In total, 20% of the worldwide cultivated land and 33% of the irrigated land are salt-affected [2,3,4]. The salinity stress significantly reduces plant growth and negatively affects photosynthesis, consequently resulting in biomass and yield reduction [5]. The seed germination and seedling establishment processes are the most critical stages in the plant life cycle that contribute substantially to final yield. Seed germination and seedling emergence take place in the soil rhizosphere where there is plenty of salt accumulation, and thus, these two stages are most sensitive to soil salinity stress [6].
Seed germination and plant growth and development interfere with salinity through (i) essential nutrients uptake imbalance, (ii) osmotic pressure imbalance, i.e., initiating water shortage, (iii) ion toxicity and (iv) production of reactive oxygen species (ROS) that act at the cellular or at whole plant level to cause physiological and biochemical defects, resulting in reduced germination, suppressed seedling growth and poor harvest [7]. The osmotic adjustment may be triggered first under high-saline conditions [8], and then, plants regulate cellular ion homeostasis by the actions of multiple transporters in response to ion toxicity [9,10], while osmolyte adjustment is a key element under salinity and drought stresses [11]. It is known that several stress-regulated transcription factors are activated in response to both salt and drought stresses, such as WRKY transcription factors [12], Apetala 2 (AP2) [13], Basic leucine zipper (bZIP) [14], and MYB [15].
Barley (Hordeum vulgare L.) is among the most important cereal crops worldwide and has been known to be cultivated for about 10,000 years. Naturally, it shows high levels of drought and salinity tolerance such that it has the advantage of growing in marginal environments that are unsuitable for other cereal crops [6,16,17,18] Therefore, it is usually used as a model crop for understanding mechanisms of germination and seedling and plant development in monocots under salinity stress [19,20]. However, salinity causes a significant reduction in barley growth and seed yield. The level of salinity tolerance of the barley genotype depends on the ability to develop and survive under salinity stress [21,22]. Salt stress on barley seed germination and seedling development has been examined in a number of studies to determine how barley responds to salinity and to develop new salt-tolerant lines to be used in breeding programs [6,23]. An increase in salinity levels in barley cultivation resulted in a decrease in germination rate, shoot length, root length, fresh and dry roots weights, and relative growth rate [24].
It is well known that salinity tolerance is controlled by several loci [25]. To reveal and clarify the genetic basis of this complex agronomic trait, genome-wide association studies (GWAS) have been gradually used. In rice, GWAS was executed to identify loci regulating salinity tolerance. GWAS identified new quantitative trait loci (QTL) on chromosomes (chr) 4, 6, and 7 that control salinity tolerance at the seedling stage [26]. In barley, at the germination stage, Mano and Takeda [25] stated 17 QTL controlling abscisic acid (ABA) responses on chr 1H, chr 2H, chr 3H and chr 5H, where the loci located on chr 5H were closely linked to salinity tolerance. Witzel et al. [27] studied QTL mapping using the Oregon Wolfe Barley population and found several regions on chr 2H, 5H and 7H that were associated with salt stress response at the germination stage. Thus far, little is known about the genes and genetic mechanisms associated with the identified QTL for salinity tolerance at germination [28]. The current developments in GWAS technology allow for the genotyping of thousands of gene loci across hundreds of accessions using high-throughput markers to enhance breeding efficiency [29]. GWAS can locate polymorphisms and underlying genetic loci accounting for phenotypic variations [30,31].
To develop new salt-tolerant barley lines to be used in breeding programs, there needs to be screening of the available barley genetic resources and worldwide collections under salinity stress conditions to select the best lines for this goal. To the best of our knowledge, among QTLs governing response to salinity stress tolerance in barley, only a few have been identified at germination and seedling levels. In this study, we screened a collection of 208 worldwide barley collections, so-called intermedium-spike accessions as described by Youssef et al. [32,33] at germination and seedling stages and used GWAS analysis to identify salinity tolerance loci associated with salt tolerance-related traits at germination and seedling stages in this collection. We tested the collection under control and salinity treatment (150 mM NaCl). The germination and seedling growth parameters of these accessions were assessed, and stress tolerance indices were calculated. The GWAS analysis of the tested traits associated with salinity tolerance at germination and seedling stages was conducted using 10,323 single nucleotide polymorphism (SNP) markers. At the germination and seedling stages, we identified 38 putative QTL and 153 HC predictive genes associated with salt-tolerance traits that can be used in future barley breeding resilience programs.
2. Results
2.1. Trait Variability and Heritability Estimates of Intermedium-Spike Barley Population
The analysis of variance for germination and seedling growth parameters revealed highly significant differences among the intermedium-spike barley accessions under control and 150 mM NaCl conditions (Table 1). In addition, the combined ANOVA for all studied parameters revealed highly significant differences among accessions, between treatments and for their interaction as well. Moreover, the coefficient of variation (CV) values were higher under salinity conditions in the majority of traits studied as compared to the control. Wide ranges (minimum and maximum values) of the studied traits were observed under both treatments (Table 2). These results indicate sufficient variability and different responses to salinity stress that exist in the intermedium-spike barley material.

Table 1.
Mean squares (MS) of separate and combined ANOVA of salinity treatments, coefficient of variation (CV%), coefficient of determination (R2), and broad-sense heritability (Hb) for the investigated traits in 208 accessions (A) under two treatments (T); control and salinity treatment (150 mM NaCl).

Table 2.
Means, summary statistics, and relative salinity performance (R%) due to salinity effects on the studied traits as compared to control.
Moderate to high values of coefficient of determination (R2) and heritability (Hb) estimates were noted under both conditions. Under control, the highest R2 and Hb were observed for germination index (GI) (0.95 and 97.6%), while the lowest ones were recorded for water content percentage (WCP) (0.78 and 85.7%). Under salinity, shoot length (ShL) exhibited the highest value of R2 (0.91) and Hb (95.1%), whereas seedling fresh weight (SFW) had the lowest values of R2 (0.61) and Hb (67.8%).
2.2. Means of the Studied Traits and Response to Salinity
The relative salinity performance (R%) on the investigated parameters ranged between −42.07% for seedling vigor index (SVI) and 24.86% for seedling dry weight (SDW) (Table 2). It could be observed that the majority of the parameters were reduced under salinity conditions, while mean germination time (MGT), root:shoot ratio (RSR) and SDW were increased.
Among the 208 accessions tested, 22 of them including HOR 11747, HOR 11718, and HOR 11640 showed 100% germination under salinity conditions. These were the fastest germination accessions, but they recorded the lowest values of MGT with an average of 2.5 days (Table S1). The accessions HOR 5837, HOR 5017, HOR 11256 and HOR 11275 showed the tallest shoots under 150 mM NaCl conditions, while accessions HOR 11291, HOR 11275, HOR 11256, and HOR 6661 recorded the tallest root length (Table S1). Accessions HOR 11291, BCC 747, HOR 11275, and HOR 11256 had the highest WCP under 150 mM NaCl treatments. By these criteria, these accessions could be characterized as salt-tolerant genotypes. Together, we found HOR 11747, HOR 11718, HOR 11640, HOR 11256, HOR 11275 and HOR 11291 to be the most salinity-tolerant accessions in the intermedium-spike barley collection.
2.3. Phenotypic Correlation among Germination and Seedling Growth Parameters
Pearson correlation coefficients were calculated among germination and seedling growth parameters under control and salinity conditions (150 mM NaCl), which are displayed in Figure 1. Under both treatments, the intermedium-spike barley population (208 accessions) displayed a normal distribution for the majority of the studied traits as shown in Figure 1. Strong, positive and highly significant correlations were observed between germination percentage (GP) and each germination rate index (GRI), coefficient of velocity of germination (CVG), GI and SVI under both treatments. MGT was negatively and highly significantly correlated with GP, GRI, CVG and GI under both treatments. Correlation coefficients among germination growth parameters were much higher under control compared to salinity. Furthermore, ShL showed highly significant and positive correlations with SVI, root length (RL), seedling length (SL), SFW and WCP under both treatments. In addition, it could be observed that correlation coefficients among seedling growth parameters were much higher under salinity stress compared to control.
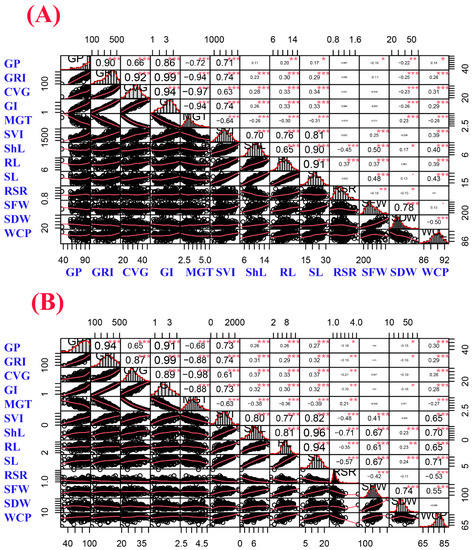
Figure 1.
Correlation matrix of germination and seedling growth parameters under (A) control and (B) salinity conditions (150 mM NaCl). The distribution of each trait is shown on the diagonal. The bivariate scatter plots with a fitted line are displayed on the bottom of the diagonal, and the correlation coefficients plus the significance level as stars are shown on the top of the diagonal. *, **, *** are significant, highly significant, and very highly significant at 0.1, 0.05, 0.01, and 0.001 probability level, respectively. Trait abbreviations are given in Table 1 footnote.
2.4. Salinity Tolerance Indices (STI) and Their Relationships
Eight salinity tolerance indices related to GP, MGT, ShL, RL, SL, SFW, SDW, and WCP were subjected to the analysis of variance and correlation and are displayed in Table 3 and Figure 2. The analysis of variance revealed highly significant differences among the intermedium-spike barley accessions for all tested salinity indices. High coefficients of determination (R2) coupled with high estimates of broad-sense heritability (Hb) were obtained for the salinity indices. They ranged between 0.77 and 85.2% seedling fresh weight tolerance index (SFWTI) and 0.92 and 95.9% mean germination time tolerance index (MGTTI) for R2 and Hb, respectively. The seedling dry weight tolerance index (SDWTI) exhibited the highest mean (1.29) among salinity tolerance indices, while shoot length tolerance index (ShLTI) showed the lowest value (0.65). The minimum and maximum values of the salinity tolerance indices are shown in Table 3. For each line, the mean and summary statistics of the tested indices are displayed in the supplementary Table S2. According to the germination tolerance index (GPTI), the accessions HOR 11275, HOR 11618, HOR 11640 and HOR 11747 exhibited the highest values of GPTI with an average of 1.3. High values of STI indicate that these accessions are tolerant to the stress. For MGTTI, the accessions HOR 11633, HOR 11618, HOR 11718, and HOR 7076 had the highest values of MGTTI and recorded 1.69, 1.66, 1.63 and 1.58, respectively. These accessions completed their germination in a short time and could be considered tolerant genotypes to salinity stress at the germination stage. For the seedling length tolerance index (SLTI), the accessions HOR 7113, HOR 11256, HOR 11291, HOR 5772, and HOR 11275 showed the highest values of the seedling length tolerance index with an average of 1.34 and were considered the tallest seedlings under salinity conditions compared to control. For the water content tolerance index (WCTI), the accessions HOR 11256, HOR 11291 and HOR 12354 had the highest values of WCTI with an average of 0.98, indicating a high water content in their tissues under salinity conditions near to their content under control. These STI results confirmed the superiority of HOR 11747, HOR 11718, HOR 11640, HOR 11256, HOR 11275 and HOR 11291 accessions in salinity tolerance in the intermedium-spike barley collection.

Table 3.
Mean squares (MS) of accessions, coefficient of variation (CV%), coefficient of determination (R2), broad-sense heritability (Hb), and descriptive statistics for the salinity tolerance indices.
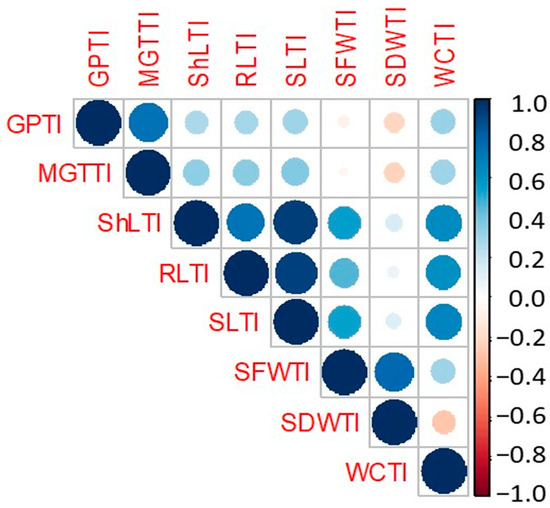
Figure 2.
Pearson correlation matrix among salinity tolerance indices (STI). Trait abbreviations are given in Table 3 footnote.
A Pearson correlation coefficients matrix was calculated among the eight salinity tolerance indices (Figure 2). GPTI was positively and highly significantly correlated with MGTTI, ShLTI, RLTI, and SLTI. In addition, WCTI was associated positively and highly significantly with all salinity tolerance indices except for SDWTI, which was negative.
2.5. QTL Detection
Both phenotypic and genotypic data were subjected to a QTL analysis to detect putative QTL associated with traits related to germination and seedling development under control and salinity stress and for eight salinity tolerance indices (Table S3). One QTL was identified for each of the traits GP, CVG, GI, MGT, SFW and SDW under control conditions and mapped on chr 1H, 2H, 6H and 7H. In addition, all these QTL showed positive additive effects for the major allele. In addition, thirteen QTL were detected for ten investigated traits out of thirteen under salinity conditions and mapped on chromosomes 1H, 2H, 3H, 5H and 7H. Six QTL regions were responsible for salinity tolerance in the intermedium-spike barley collection by increasing their performance mainly for traits CVG, SDW and GI (chr 1H), SVI (chr 5H), ShL (chr 2H) and SL (chr 7H), while the other detected QTL showed negative performance of the accessions for traits GP (Chr 5H), ShL (Chr 5H), SL (Chr 5H), SFW (Chr 1H and Chr 3H) and WCP (Chr 7H). Five QTL were identified for five salinity tolerance indices (ShLTI, RLTI, SLTI, SFWTI, and WCTI) out of eight. The QTL were located on chromosomes 2H and 7H, and the major SNP alleles showed negative estimates (Table 4). We found that the salinity-related QTL associated with marker 3_599466174, located on chr 3H was presented in all six accessions that showed superior salinity tolerance in this collection. The QTL associated with markers 1_546373322, 2_698868743, 6_7696839 and 7_629887676 located on chr 1H, 2H, 6H and 7H, respectively, were found in all six accessions except for HOR 11275.

Table 4.
Putative QTL associated with germination and seedling growth related traits under control and salinity conditions as well as salinity tolerance indices.
2.6. Candidate Gene Prediction
Within the regions flanking each QTL, we searched for the high confidence (HC) genes from the barley reference genome assembly of Morex, version refseq 1 (Table 4 and Table S4) and its corresponding gene annotation [34]. We found 153 HC genes associated with 38 putative QTL. Of these, 22 SNPs were found inside possible HC candidate genes (Table 5), while the other 16 SNPs were located close to the HC genes. Based on their −log10 P values and % R2 values (Table 4), it may be possible that some of these candidate genes are involved in enhancing salinity tolerance at the germination and seedlings stages. For example, the candidate genes coding for the Cytochrome P450 superfamily protein, early nodulin-like protein, Phospholipase A1, Helicase-like protein, MYB domain protein, Homeobox-leucine zipper protein family and kinase family protein were found more than once, indicating a possible role in salinity tolerance during germination and seedling stages in barley. We found that the gene HORVU4Hr1G085320 encoding a NAC domain protein was associated with SNP m4_631432489 and is proposed to regulate the SDW under salinity stress in barley. In addition, we found that duplication genes HORVU1Hr1G090570 and HORVU1Hr1G090580 both encode phospholipase A1-II 6 protein and are associated with marker 1_546373322. The QTL was associated with marker 3_599466174 presented in all six superior salinity tolerant accessions, encoding the NRT1/ PTR FAMILY 5.10 protein (Table 5 and Table S4).

Table 5.
Putative HC candidate genes associated with identified QTL related to control and salinity treatments.
3. Discussion
Salinity has a negative effect on many metabolic processes, causing a reduction in seed germination, seedling growth and plant development due to the effects of salt accumulation in the plant tissues [35,36]. Plant tolerance to salinity is a complex quantitative trait that depends on genetic and physiological factors, and any change induced by salinity is influenced by gene expression [37]. The genetic variation found in wild crop relatives shows a large amount of allelic variation in contrast to commercial elite cultivars [33]. This genetic variation can potentially contribute greatly to improvement of stress tolerance [38]. Among QTLs controlling response to osmotic stress [39,40], ionic stress [27,41] and salinity stress tolerance [28] in barley, only a few have been identified at germination and seedling stages. Therefore, our discovery of novel QTL associated with salinity tolerance traits in the worldwide intermedium-spike barley collection has the potential to improve crop salinity tolerance in barley breeding programs.
The results indicated significant negative effects of the salinity treatment for all seed germination and seedling growth related traits compared to control condition except for the MGT, RSR and SDW that increased under salinity stress. Despite that the accuracy of the results obtained might be due to the tight control of the experiment, these results are in contrast to many research studies that have stated that plant dry weight (DW) was reduced under salinity conditions in different crops including barley [6,42,43,44,45]. Our results were in agreement with what was found by Sayed et al. [46] studying the S42IL introgression library of 50 lines under salinity stress conditions and Angessa et al. [28] in the CM72 x Gairdner doubled haploid barley population. Additionally, Abdul Qados [47] and Allam et al. [48] reported that the fresh and dry matter production was found to increase in studied genotypes from lower to higher salinity levels. In these populations, seeds have been subjected to salt-induced physiological drought stress as a result of the high osmotic pressures generated by the salt environment, causing an observed reduction in germination and seedling growth traits. This process affects the ability of seeds to absorb water from the germination medium, consequently causing prolonged plant development or even preventing plants from absorbing water and consequently seedling and plant growth [49,50]. The intermedium-spike barley collection showed a wide range of salinity tolerance in all of the tested traits (Table 2). Across the population, the maximum reduction due to the salinity treatment was found in SVI by 42.07%, and the minimum reduction was found in WCP by 9.92%, while the maximum increase due to salinity treatment was found in SDW by 24.86%, and the minimum increase was found in MGT by 10.95% compared to the control condition. The salt-tolerance expressed in reducing the loss of water and increasing the seedling dry weight may be due to the increase in osmotic potential in seeds by nutrient uptake, which led to the absorption of more water under salinity stress [51]. The increase in RSR by 11.94% under salinity conditions may also be due to significantly reduced shoot dry matter production, shortened shoots, and elongated roots to obtain water [52]. Notably, where the SDW mean across all accessions under control condition was 30.0 g, 121 accessions showed SDW more than 35.0 g under salinity stress treatment; 77 of these accessions showed WCP >80%. These salt-tolerant accessions may be used in barley breeding programs for the improvement of salinity tolerance to develop environmentally smart cultivars to be used for climatic change scenarios. The STI recorded in this study showed high variation among the accessions. This finding was in agreement with Angessa et al., 2017 [28], and Allel et al., 2019 [53]. We suggested that it is better to use traits with high rates of variation such as STI (Table 3), WCP and SVI than the traits with low rates of variation to select accessions for salinity tolerance breeding programs [24,54]. The STI can be used as an indicator for selecting the salt-tolerance accessions from this collection.
The observed significant differences among the accessions and between the salinity treatments and control indicate various possible responses to salinity stress in this intermedium-spike barley collection. This variation among the accessions was reflected through the high estimates of broad-sense heritability under salinity conditions in the current study. The heritability estimates of the investigated traits were high (between 67.8 in SFW and 95.9 in MGTTI) for all traits under salinity treatment. These findings indicate that salinity tolerance related traits during germination and seedling development are genetically controlled to a high extend. Confirming our findings, high estimates of heritability in the same recorded traits at germination and seedling development under salinity stress were also obtained in other reports [6,42,44,46]. GWAS analysis makes use of historical recombination to identify regions of the genome that are responsive to traits using a high-resolution genome scan [55]. Several QTL have been reported for salt tolerance traits during germination and seedling development [28,46,56]. For example, QTL for salinity tolerance were identified on chromosomes 1H, 2H and 7H in the DH populations TX9425 × Franklin, YYXT × Franklin and in a worldwide collection of 206 barley accessions [57,58]. Some of those QTL were closely linked to significant markers reported in our study.
In this study, the GWAS analysis revealed that 55 significant SNPs, summarized to 38 putative QTL, may regulate salinity tolerance in the intermedium-spike barley collection. For the salinity-related traits as well as salinity tolerance indices, three QTL on chromosome 1H, four QTL on chromosome 2H, one QTL on chromosome 3H, four QTL on chromosome 5H, and three QTL on chromosome 7H were detected. These results confirmed that multiple small-effect genes act in combination to regulate salinity tolerance in barley during germination and seedling development [59]. Of the presented candidate gene families associated with these QTL, the kinase family protein and phospholipase A1 were also identified at germination and seedling stages in rice and barley under salinity stress [44,60]. The presence of phospholipase A1 in HOR 11747, HOR 11718, HOR 11640, HOR 11256 and HOR 11291 might explain their salinity-tolerant superiority since it aids in the biosynthesis of jasmonic acid, which is considered an important stress regulator [61,62]. Plants such as barley, cotton and wheat have been found to use the protein kinase superfamily genes to adapt to drought and salinity stress conditions [24,63,64,65]. In soybean plants with overexpressed protein kinases, the salt tolerance was significantly increased, suggesting that it may play a crucial role in salinity tolerance [66]. In Arabidopsis thaliana, the abscisic acid (ABA)-non-activated protein kinases regulate reactive oxygen species (ROS) homeostasis and trigger gene expression under salinity stress conditions [67]. The NAC domain transcription factor that we identified as a candidate gene associated with marker m4_631432489 enhanced the salinity tolerance in our barley collection, especially in two of the highest tolerant ones, HOR 11718 and HOR 11640. This result is in agreement with Li et al. [68] who found that salt stress influenced the expression level of GmNAC06 in soybean. NAC overexpression caused proline and glycine betaine accumulation in the cells that help to alleviate or avoid the negative effects of ROS. Similarly, it also regulates the Na+/K+ ratios to maintain ionic homeostasis in soybean hairy roots [68]. MYB transcription factors play an important role in abiotic stress responses in different plants such as Arabidopsis [69], peanut [70] and wheat [71]. In our study, we found that MYB domain transcription factors are associated with markers related to salinity tolerance. Through promoting expression of stress-associated genes and controlling osmotic and oxidizing substances, MYB domain transcription factors might help maintain cell homeostasis in response to drought and salt stress [72]. In transgenic Arabidopsis plants, salinity stress prompted lipase expression, enhancing salinity tolerance, which simplifies seed germination, vegetative growth, flowering, and seed set [73]. All of the above-mentioned gene families are found to be associated with stress tolerance in barley, including salinity, as reported in this study. Our results will form the basis for future studies to discover and verify the mechanism by which candidate genes play a role in salinity tolerance during germination and seedling development stages in barley. In this study, various promising barley accessions showed a high degree of salinity tolerance based on their STI, such as HOR 11747, HOR 11718, HOR 11640, HOR 11256, HOR 11275, and HOR 11291. These accessions could be used as the basis for our research plan to create improved salinity-tolerant barley population. Prior to this, we recommend greenhouses and field experiments in order to validate these results using a wider range of plant growth and yield traits under salinity stress conditions.
4. Materials and Methods
4.1. Plant Material and Genotyping
A set of 208 intermedium-spike spring barley accessions (H. vulgare L. convar. intermedium (Körn.) Mansf.) [74] of worldwide origin (Europe, East and West Asia, Africa and Americas) was used in this study. All information about this intermedium-spike barley accession collection and its genetic characterization has been published by Youssef et al. [33].
4.2. Experimental Design and Salinity Stress Treatments
The experiment was conducted at the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben, Germany, in a completely randomized design. The whole set of accessions was surface sterilized with 70% ethanol solution for one minute and rinsed with sterile distilled water several times, then briefly blotted. The seeds were placed on two layers of filter papers (C160; Ahlstrom-Munksjö, GmbH, Dettingen an der Erms, Germany) laying in crystal clear rectangular boxes (V3-92; Licefa GmbH & Co. KG, Bad Salzuflen, Germany). The salinity treatments, with three replicates, were conducted by watering the seeds with 150 mM NaCl (Sodium chloride CELLPURE® ≥ 99.5 %, for cell culture and biochemistry, Carl Roth, GmbH, Karlsruhe, Germany), whereas deionized water was applied as a control treatment, and the seeds were placed in a versatile environmental test chamber (Model No. MLR-352-PE, Panasonic, Kadoma, Japan) for ten days, maintained at 20 ± 2 °C with 50 ± 5% humidity at 12 h light (200 μmol m−2 s−1) and 12 h dark periods per day. The seeds were considered germinated when the radicle reached at least 2 mm in length, and the number of the germinated seeds was counted daily after 24 h of incubation until the end of the experiment.
4.3. Evaluation of Germination and Seedling Growth Parameters
Seeds were counted daily until the 10th day to calculate the following parameters:
- (1)
- Germination percentage (GP in %):
- (2)
- Germination rate index (GRI) was calculated according to Esechie [75] as GRI = (G1 + G2 + … + Gi)/i, where G1 is the germination percentage, calculated daily from day 1 to i = 10. It gives an indication of the percentage of seeds germinating per day during the germination test period.
- (3)
- Coefficient of velocity of germination (CVG) was calculated following Al-Ansari and Ksiksi [76] as follows: CVG = (N1 + N2 + … + Ni) × 100/( N1*T1 + … + Ni*Ti), where N is the number of seeds germinated every day and T is the number of days from seeding corresponding to N. It gives an indication of the speed of germination. CVG values increase when the number of germinated seeds increases and the time required for germination decreases.
- (4)
- Germination index (GI) was calculated according to Benech Arnold et al. [77] as follows: GI = (10 × N1) + (9 × N2) + … + (1 × N10); where N1, N2, …, N10, is the number of seeds germinated on the first, second and subsequent days until the 10th day, and the multipliers (i.e., 10, 9 … etc.) are weights given to the days of the germination. It measures both percentage and speed of germination. High values indicate that seeds germinate early and low values that seeds germinate late.
- (5)
- Mean germination time (MGT) represents the mean time that seeds require to initiate and end germination. It was calculated according to Orchard [78] as follows:where Ni is the number of the newly germinated seeds at time Ti.MGT = Σ(Ti × Ni)/ΣNi
- (6)
- Seedling vigor index (SVI) was calculated according to Abiri et al. [79] with little modification as a multiplication of the final germination percentage by the total seedling length (shoot and root lengths).
- (7)
- Shoot length (ShL in cm) and root length (RL in cm) were measured manually at the tenth day of germination using a scaled ruler for five seedlings from each replicate at the end of the experiment.
- (8)
- Seedling length (SL in cm) was measured as the total of root and shoot lengths.
- (9)
- Root–shoot ratio (RSR) was calculated as a ratio between root length and shoot length.
- (10)
- Seedlings fresh weight (SFW in g) and seedling dry weight (SDW in mg) were recorded by weighing harvested seedlings at day 10 and, respectively, after drying the fresh weight at 80 °C for 72 h to obtain the SDW using an ultra-micro lab balance (Sartorius AC 1215, Germany).
- (11)
- Water content percentage (WCP in %) was calculated based on the following formula:
4.4. Stress Tolerance Indices (STI)
In order to evaluate the growth performance and the variation among genotypes in their tolerance to salinity, stress tolerance indices (STI) were derived for the following eight parameters:
- (1)
- Germination percentage (GPTI, as germination percentage tolerance index);
- (2)
- Mean germination time (MGTTI, as mean germination time tolerance index);
- (3)
- Shoot length (ShLTI, as shoot length tolerance index);
- (4)
- Root length (RLTI, as root length tolerance index);
- (5)
- Seedling length (SLTI, as seedling length tolerance index);
- (6)
- Seedling fresh weight (SFWTI, as seedling fresh weight tolerance index);
- (7)
- Seedling dry weight (SDWTI, as seedling dry weight tolerance index);
- (8)
- Water content % (WCPTI, as water content percentage tolerance index).
The salt tolerance indices (STIs) for these traits were calculated according to the formula of Fernandez [80]:
where Ys and Yp are the traits of interest of the tested genotypes under salinity (stress) and non-stress conditions (control), and is the mean value of the trait under non-stress conditions.
4.5. Statistical Analyses
The separate and combined analyses of variance (ANOVA) of a completely randomized experiment were performed using SAS software v. 9.2 with PROC GLM procedure [81], to test the effect of each treatment and the interaction between the intermedium-spike accessions and salinity treatments. Broad-sense heritability (Hb) estimates were calculated under control and salinity conditions following Padi [82].
where is the genotypic variance, is the phenotypic variance, is the pooled error variance, and r is the number of replicates. Additionally, least square means (Ls-means) were calculated for each genotype using PROC GLM method of SAS software. The phenotypic Pearson correlation matrix analysis among the traits under control and 150 mM NaCl treatments was calculated by R-studio.
4.6. GWAS Analysis
The GWAS was conducted as previously described by Dreissig et al. [83] with the software SAS 9.4 [84] for 192 accessions with valid genotype and phenotype data. In a first step, all SNPs associated with the target trait are selected using a multiple linear regression model (SAS PROC GLMSELECT). Then, 100 repeated subsamples are created with 80% of the accessions, and only those SNPs that improve the prediction of the remaining 20% are selected (according to the minimum average squared error), and all SNPs selected more than once in this step are considered potential cofactors. The potential cofactors were used as input for the final cofactor selection (SAS PROC GLMSELECT based on the Schwarz Bayesian Criterion) in the whole dataset. The selected co-factors are then modeled with SAS PROC REG in the background of a multiple linear regression model where all SNPs are tested for significance. Thus, allele effects, R² and p value are estimated as a function of the cofactors, which enter the model first according to their ranking in the previous step, by applying the model option PARTIALR2 (SEQTESTS). p values were corrected for multiple testing based on Bonferroni [85], SNPs with Bonferroni p < 0.05 were defined as significant.
4.7. GWAS’ Significant QTL Annotation
To determine whether genes surrounding significant loci are enriched for specific GOs, the genes located within a region of ±100 kb next to the significant SNPs were selected as candidates for annotation and pathway analyses. Cluster of Orthologous Groups (COG) analysis of proteins was performed using the NCBI website (http://www.ncbi.nlm.nih.gov/COG/ (accessed on 29 June 2022)).
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231911060/s1.
Author Contributions
H.M.Y. conceived the study. M.A.S., A.M., T.S. (Thomas Schmutzer) and H.M.Y. performed experiments. H.M.Y., M.A.S. and A.M. analyzed data. T.S. (Thorsten Schnurbusch) provided the intermedium-spike barley collection including genotypic data. A.B. provided the lab materials and equipment. H.M.Y., M.A.S., M.H., A.B. and K.P. wrote the paper with input from all co-authors. All authors have read and agreed to the published version of the manuscript.
Funding
German Research Foundation (DFG YO 304/1-1) to H.Y., SusCrop-ERA-NET project BARISTA (BMBF: 031B0811B) to K.P. and H.M.Y. The Swedish Research Council (grant no. VR 2018-05117 to M.H.), the Swedish Research Council for Environment, Agricultural Sciences, and Spatial Planning (grant no. FORMAS 2018-01026 to M.H.), the Erik Philip-Sörensen Foundation (M.H.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Sincere gratitude goes to the Ministry of Higher Education, Arab Republic of Egypt, for the award of a fully funded postdoctoral scholarship to M.A.S. We thank Tarawneh R. and Ibrahim H. for excellent technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef] [PubMed]
- Mohanavelu, A.; Naganna, S.R.; Al-Ansari, N. Irrigation Induced Salinity and Sodicity Hazards on Soil and Groundwater: An Overview of Its Causes, Impacts and Mitigation Strategies. Agriculture 2021, 11, 983. [Google Scholar] [CrossRef]
- Manuel, R.; Machado, A.; Serralheiro, R.P.; Alvino, A.; Freire, M.I.; Ferreira, R. Soil Salinity: Effect on Vegetable Crop Growth. Management Practices to Prevent and Mitigate Soil Salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. Intergovernmental Technical Panel on Soils. Status of the World’s Soil Resources; Food and Agriculture Organization of the United Nations: Rome, Italy, 2015; ISBN 9789251090046. [Google Scholar]
- Kaashyap, M.; Ford, R.; Kudapa, H.; Jain, M.; Edwards, D.; Varshney, R.; Mantri, N. Differential Regulation of Genes Involved in Root Morphogenesis and Cell Wall Modification is Associated with Salinity Tolerance in Chickpea. Sci. Rep. 2018, 8, 4855. [Google Scholar] [CrossRef] [PubMed]
- Mwando, E.; Angessa, T.T.; Han, Y.; Zhou, G.; Li, C. Quantitative Trait Loci Mapping for Vigour and Survival Traits of Barley Seedlings after Germinating under Salinity Stress. Agronomy 2021, 11, 103. [Google Scholar] [CrossRef]
- Xue, W.; Yan, J.; Jiang, Y.; Zhan, Z.; Zhao, G.; Tondelli, A.; Cattivelli, L.; Cheng, J. Genetic dissection of winter barley seedling response to salt and osmotic stress. Mol. Breed. 2019, 39, 137. [Google Scholar] [CrossRef]
- Zhu, M.; Zhou, M.; Shabala, L.; Shabala, S. Linking osmotic adjustment and stomatal characteristics with salinity stress tolerance in contrasting barley accessions. Funct. Plant Biol. 2015, 42, 252–263. [Google Scholar] [CrossRef]
- Arzani, A.; Ashraf, M. Smart Engineering of Genetic Resources for Enhanced Salinity Tolerance in Crop Plants. CRC Crit. Rev. Plant Sci. 2016, 35, 146–189. [Google Scholar] [CrossRef]
- Deinlein, U. Plant salt-tolerance mechanisms. Trends Plant Sci 2014, 19, 371–379. [Google Scholar] [CrossRef]
- Cattivelli, L.; Rizza, F.; Badeck, F.W.; Mazzucotelli, E.; Mastrangelo, A.M.; Francia, E.; Marè, C.; Tondelli, A.; Stanca, A.M. Drought tolerance improvement in crop plants: An integrated view from breeding to genomics. F. Crop. Res. 2008, 105, 1–14. [Google Scholar] [CrossRef]
- Tao, Z.; Kou, Y.; Liu, H.; Li, X.; Xiao, J.; Wang, S. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J. Exp. Bot. 2011, 62, 4863–4874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, M.; Li, L.; Xu, Z.; Chen, X.; Guo, J.; Ma, Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 2009, 60, 3781–3796. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Zhang, D.F.; Fu, J.; Shi, Y.S.; Song, Y.C.; Wang, T.Y.; Li, Y. Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 2012, 235, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Xu, Y.; Ma, Q.; Xu, W.; Wang, T.; Xue, Y.; Chong, K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007, 143, 1739–1751. [Google Scholar] [CrossRef]
- Sayed, M.A.; Nassar, S.M.; Moustafa, E.S.; Said, M.T. Genetic Mapping Reveals Novel Exotic and Elite QTL Alleles for Salinity Tolerance in Barley. Agronomy 2021, 11, 1774. [Google Scholar] [CrossRef]
- Moustafa, E.S.A.; El-Sobky, E.S.E.A.; Farag, H.I.A.; Yasin, M.A.T.; Attia, A.; Rady, M.O.A.; Awad, M.F.; Mansour, E. Sowing date and genotype influence on yield and quality of dual-purpose barley in a salt-affected arid region. Agronomy 2021, 11, 717. [Google Scholar] [CrossRef]
- Ebrahim, F.; Arzani, A.; Rahimmalek, M.; Sun, D.; Peng, J. Salinity tolerance of wild barley Hordeum vulgare ssp. spontaneum. Plant Breed. 2020, 139, 304–316. [Google Scholar] [CrossRef]
- Gharaghanipor, N.; Arzani, A.; Rahimmalek, M.; Ravash, R. Physiological and Transcriptome Indicators of Salt Tolerance in Wild and Cultivated Barley. Front. Plant Sci. 2022, 13, 1039. [Google Scholar] [CrossRef]
- Gorzolka, K.; Kölling, J.; Nattkemper, T.W.; Niehaus, K. Spatio-Temporal Metabolite Profiling of the Barley Germination Process by MALDI MS Imaging. PLoS ONE 2016, 11, e0150208. [Google Scholar] [CrossRef]
- Shelden, M.C.; Dias, D.A.; Jayasinghe, N.S.; Bacic, A.; Roessner, U. Root spatial metabolite profiling of two genotypes of barley (Hordeum vulgare L.) reveals differences in response to short-term salt stress. J. Exp. Bot. 2016, 67, 3731–3745. [Google Scholar] [CrossRef]
- Gupta, S.; Rupasinghe, T.; Callahan, D.L.; Natera, S.H.A.; Smith, P.M.C.; Hill, C.B.; Roessner, U.; Boughton, B.A. Spatio-Temporal Metabolite and Elemental Profiling of Salt Stressed Barley Seeds During Initial Stages of Germination by MALDI-MSI and µ-XRF Spectrometry. Front. Plant Sci. 2019, 10, 1139. [Google Scholar] [CrossRef] [PubMed]
- Askari, H.; Kazemitabar, S.K.; Zarrini, H.N.; Saberi, M.H. Salt tolerance assessment of barley (Hordeum vulgare L.) genotypes at germination stage by tolerance indices. Open Agric. 2016, 1, 37–44. [Google Scholar] [CrossRef]
- Mwando, E.; Han, Y.; Angessa, T.T.; Zhou, G.; Hill, C.B.; Zhang, X.Q.; Li, C. Genome-Wide Association Study of Salinity Tolerance During Germination in Barley (Hordeum vulgare L.). Front. Plant Sci. 2020, 11, 118. [Google Scholar] [CrossRef]
- Mano, Y.; Takeda, K. Mapping quantitative trait loci for salt tolerance at germination and the seedling stage in barley (Hordeum vulgare L.). Euphytica 1997, 94, 263–272. [Google Scholar] [CrossRef]
- Kumar, S.; Dwivedi, S.K.; Singh, S.S.; Jha, S.K.; Elanchezhian, R.; Singh, O.N.; Bhatt, B.P. Identification of drought tolerant rice genotypes by analysing drought tolerance indices and morpho-physiological traits. Sabrao J. Breed. Genet. 2014, 46, 217–230. [Google Scholar]
- Witzel, K.; Weidner, A.; Surabhi, G.K.; Varshney, R.K.; Kunze, G.; Buck-Sorlin, G.H.; BÖrner, A.; Mock, H.P. Comparative analysis of the grain proteome fraction in barley genotypes with contrasting salinity tolerance during germination. Plant. Cell Environ. 2010, 33, 211–222. [Google Scholar] [CrossRef]
- Angessa, T.T.; Zhang, X.Q.; Zhou, G.; Broughton, S.; Zhang, W.; Li, C. Early growth stages salinity stress tolerance in CM72 x Gairdner doubled haploid barley population. PLoS ONE 2017, 12, e0179715. [Google Scholar] [CrossRef]
- Tondelli, A.; Xu, X.; Moragues, M.; Sharma, R.; Schnaithmann, F.; Ingvardsen, C.; Manninen, O.; Comadran, J.; Russell, J.; Waugh, R.; et al. Structural and Temporal Variation in Genetic Diversity of European Spring Two-Row Barley Cultivars and Association Mapping of Quantitative Traits. Plant Genome 2013, 6, plantgenome2013-03. [Google Scholar] [CrossRef]
- Xu, X.; Sharma, R.; Tondelli, A.; Russell, J.; Comadran, J.; Schnaithmann, F.; Pillen, K.; Kilian, B.; Cattivelli, L.; Thomas, W.T.B.; et al. Genome-Wide Association Analysis of Grain Yield-Associated Traits in a Pan-European Barley Cultivar Collection. Plant Genome 2018, 11, 170073. [Google Scholar] [CrossRef]
- Yu, J.; Zhao, W.; Tong, W.; He, Q.; Yoon, M.Y.; Li, F.P.; Choi, B.; Heo, E.B.; Kim, K.W.; Park, Y.J. A Genome-Wide Association Study Reveals Candidate Genes Related to Salt Tolerance in Rice (Oryza sativa) at the Germination Stage. Int. J. Mol. Sci. 2018, 19, 3145. [Google Scholar] [CrossRef]
- Youssef, H.M.; Mascher, M.; Ayoub, M.A.; Stein, N.; Kilian, B.; Schnurbusch, T. Natural diversity of inflorescence architecture traces cryptic domestication genes in barley (Hordeum vulgare L.). Genet. Resour. Crop Evol. 2017, 64, 843–853. [Google Scholar] [CrossRef]
- Youssef, H.M.; Allam, M.; Boussora, F.; Himmelbach, A.; Milner, S.G.; Mascher, M.; Schnurbusch, T. Dissecting the genetic basis of lateral and central spikelet development and grain traits in Intermedium-spike barley (Hordeum vulgare convar. Intermedium). Plants 2020, 9, 1655. [Google Scholar] [CrossRef] [PubMed]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef]
- Bagues, M.; Sarabi, B.; Ghashghaie, J.; Souli, I.; Nagaz, K. The validity of carbon isotope discrimination as a screening criterion for grain yield in two barley landraces under deficit irrigation with saline water in southern Tunisia. Plant Biotechnol. 2018, 35, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Abbas, G.; Saqib, M.; Akhtar, J.; Haq, M.A. ul Interactive effects of salinity and iron deficiency on different rice genotypes. J. Plant Nutr. Soil Sci. 2015, 178, 306–311. [Google Scholar] [CrossRef]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef]
- Soleimani, B.; Sammler, R.; Backhaus, A.; Beschow, H.; Schumann, E.; Mock, H.P.; von Wirén, N.; Seiffert, U.; Pillen, K. Genetic regulation of growth and nutrient content under phosphorus deficiency in the wild barley introgression library S42IL. Plant Breed. 2017, 136, 892–907. [Google Scholar] [CrossRef]
- Bálint, A.F.; Szira, F.; Börner, A.; Galiba, G. Segregation- and association based mapping of loci influencing osmotic tolerance in barley. Acta Biol. Szeged. 2008, 52, 101–102. [Google Scholar]
- Wójcik-Jagła, M.; Rapacz, M.; Tyrka, M.; Kościelniak, J.; Crissy, K.; Zmuda, K. Comparative QTL analysis of early short-time drought tolerance in Polish fodder and malting spring barleys. Theor. Appl. Genet. 2013, 126, 3021–3034. [Google Scholar] [CrossRef]
- Xue, W.; Yan, J.; Zhao, G.; Jiang, Y.; Cheng, J.; Cattivelli, L.; Tondelli, A. A major QTL on chromosome 7HS controls the response of barley seedling to salt stress in the Nure × Tremois population. BMC Genet. 2017, 18, 79. [Google Scholar] [CrossRef]
- Thabet, S.G.; Moursi, Y.S.; Sallam, A.; Karam, M.A.; Alqudah, A.M. Genetic associations uncover candidate SNP markers and genes associated with salt tolerance during seedling developmental phase in barley. Environ. Exp. Bot. 2021, 188, 104499. [Google Scholar] [CrossRef]
- Nayyeripasand, L.; Garoosi, G.A.; Ahmadikhah, A. Genome-Wide Association Study (GWAS) to Identify Salt-Tolerance QTLs Carrying Novel Candidate Genes in Rice During Early Vegetative Stage; 2021; Volume 14, ISBN 0000000151009. [Google Scholar]
- Mwando, E.; Angessa, T.T.; Han, Y.; Li, C. Salinity tolerance in barley during germination—homologs and potential genes. J. Zhejiang Univ. Sci. B 2020, 21, 93–121. [Google Scholar] [CrossRef] [PubMed]
- Dehnavi, A.R.; Zahedi, M.; Ludwiczak, A.; Perez, S.C.; Piernik, A. Effect of salinity on seed germination and seedling development of sorghum (Sorghum bicolor (L.) Moench) genotypes. Agronomy 2020, 10, 859. [Google Scholar] [CrossRef]
- Sayed, M.A.; Tarawneh, R.; Youssef, H.M.; Pillen, K.; Börner, A. Detection and Verification of QTL for Salinity Tolerance at Germination and Seedling Stages Using Wild Barley Introgression Lines. Plants 2021, 10, 2246. [Google Scholar] [CrossRef] [PubMed]
- Abdul Qados, A.M.S. Effect of salt stress on plant growth and metabolism of bean plant Vicia faba (L.). J. Saudi Soc. Agric. Sci. 2011, 10, 7–15. [Google Scholar] [CrossRef]
- Alam, M.A.; Juraimi, A.S.; Rafii, M.Y.; Abdul Hamid, A. Effect of salinity on biomass yield and physiological and stem-root anatomical characteristics of purslane (Portulaca oleracea L.) accessions. Biomed Res. Int. 2015, 2015, 105695. [Google Scholar] [CrossRef]
- Parihar, P.; Singh, S.; Singh, R.; Singh, V.P.; Prasad, S.M. Effect of salinity stress on plants and its tolerance strategies: A review. Environ. Sci. Pollut. Res. 2015, 22, 4056–4075. [Google Scholar] [CrossRef]
- Sayar, R.; Bchini, H.; Mosbahi, M.; Ezzine, M. Effects of salt and drought stresses on germination, emergence and seedling growth of Durum wheat (Triticum durum Desf.). African J. Agric. Res. 2010, 5, 2008–2016. [Google Scholar]
- Zhang, H.; Irving, L.J.; McGill, C.; Matthew, C.; Zhou, D.; Kemp, P. The effects of salinity and osmotic stress on barley germination rate: Sodium as an osmotic regulator. Ann. Bot. 2010, 106, 1027. [Google Scholar] [CrossRef]
- Baǧci, S.A.; Ekiz, H.; Yilmaz, A. Determination of the salt tolerance of some barley genotypes and the characteristics affecting tolerance. Turkish J. Agric. For. 2003, 27, 253–260. [Google Scholar] [CrossRef]
- Allel, D.; Benamar, A.; Badri, M.; Abdelly, C. Evaluation of salinity tolerance indices in North African barley accessions at reproductive stage. Czech J. Genet. Plant Breed. 2019, 55, 61–69. [Google Scholar] [CrossRef]
- Jamshidi, A.; Javanmard, H.R. Evaluation of barley (Hordeum vulgare L.) genotypes for salinity tolerance under field conditions using the stress indices. Ain Shams Eng. J. 2018, 9, 2093–2099. [Google Scholar] [CrossRef]
- Zhao, K.; Tung, C.W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.; Mezey, J.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef] [PubMed]
- Sbei, H.; Sato, K.; Shehzad, T.; Harrabi, M.; Okuno, K. Detection of QTLs for salt tolerance in Asian barley (Hordeum vulgare L.) by association analysis with SNP markers. Breed. Sci. 2015, 64, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Shabala, S.; Ma, Y.; Xu, R.; Zhou, M. Using QTL mapping to investigate the relationships between abiotic stress tolerance (drought and salinity) and agronomic and physiological traits. BMC Genomics 2015, 16, 43. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Zhou, G.; Shabala, S.; Chen, Z.H.; Cai, S.; Li, C.; Zhou, M. Genome-wide association study reveals a new QTL for salinity tolerance in Barley (Hordeum vulgare L.). Front. Plant Sci. 2016, 7, 946. [Google Scholar] [CrossRef]
- Flowers, T.J. Improving crop salt tolerance. J. Exp. Bot. 2004, 55, 307–319. [Google Scholar] [CrossRef]
- Naveed, S.A.; Zhang, F.; Zhang, J.; Zheng, T.Q.; Meng, L.J.; Pang, Y.L.; Xu, J.L.; Li, Z.K. Identification of QTN and candidate genes for Salinity Tolerance at the Germination and Seedling Stages in Rice by Genome-Wide Association Analyses. Sci. Rep. 2018, 8, 6505. [Google Scholar] [CrossRef]
- Browse, J. Jasmonate: An oxylipin signal with many roles in plants. Vitam. Horm. 2005, 72, 431–456. [Google Scholar] [CrossRef]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The defective in anther dehiscience gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2001, 13, 2191–2209. [Google Scholar] [CrossRef]
- Shehzad, M.; Zhou, Z.; Ditta, A.; Cai, X.; Khan, M.; Xu, Y.; Hou, Y.; Peng, R.; Hao, F.; Wang, K.; et al. Genome-Wide Mining and Identification of Protein Kinase Gene Family Impacts Salinity Stress Tolerance in Highly Dense Genetic Map Developed from Interspecific Cross between G. hirsutum L. and G. darwinii G. Watt. Agronomy 2019, 9, 560. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, Q.; Chen, Q.; Yin, X.; Qian, M.; Sun, X.; Yang, Y. Genome-wide survey indicates diverse physiological roles of the barley (Hordeum vulgare L.) calcium-dependent protein kinase genes. Sci. Rep. 2017, 7, 560. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Gómez-Cadenas, A.; Routly, E.L.; Ho, T.H.D.; Simmonds, J.A.; Gulick, P.J. The salt stress-inducible protein kinase gene, Esi47, from the salt-tolerant wheatgrass Lophopyrum elongatum is involved in plant hormone signaling. Plant Physiol. 2001, 125, 1429–1441. [Google Scholar] [CrossRef]
- Qiu, Y.W.; Zhe, F.E.N.G.; Fu, M.M.; Yuan, X.H.; Luo, C.C.; Yu, Y.B.; Feng, Y.Z.; Qi, W.E.I.; Li, F.L. GsMAPK4, a positive regulator of soybean tolerance to salinity stress. J. Integr. Agric. 2019, 18, 372–380. [Google Scholar] [CrossRef]
- Szymańska, K.P.; Polkowska-Kowalczyk, L.; Lichocka, M.; Maszkowska, J.; Dobrowolska, G. SNF1-Related Protein Kinases SnRK2.4 and SnRK2.10 Modulate ROS Homeostasis in Plant Response to Salt Stress. Int. J. Mol. Sci. 2019, 20, 143. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, R.; Jiang, Q.; Sun, X.; Zhang, H.; Hu, Z. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 2021, 105, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.Z.; Jin, S.H.; Jiang, X.Y.; Dong, R.R.; Li, P.; Li, Y.J.; Hou, B.K. Ectopic expression of UGT75D1, a glycosyltransferase preferring indole-3-butyric acid, modulates cotyledon development and stress tolerance in seed germination of Arabidopsis thaliana. Plant Mol. Biol. 2016, 90, 77–93. [Google Scholar] [CrossRef]
- Chen, N.; Yang, Q.; Pan, L.; Chi, X.; Chen, M.; Hu, D.; Yang, Z.; Wang, T.; Wang, M.; Yu, S. Identification of 30 MYB transcription factor genes and analysis of their expression during abiotic stress in peanut (Arachis hypogaea L.). Gene 2014, 533, 332–345. [Google Scholar] [CrossRef]
- Zhao, Y.; Tian, X.; Wang, F.; Zhang, L.; Xin, M.; Hu, Z.; Yao, Y.; Ni, Z.; Sun, Q.; Peng, H. Characterization of wheat MYB genes responsive to high temperatures. BMC Plant Biol. 2017, 17, 208. [Google Scholar] [CrossRef]
- Du, Y.T.; Zhao, M.J.; Wang, C.T.; Gao, Y.; Wang, Y.X.; Liu, Y.W.; Chen, M.; Chen, J.; Zhou, Y.B.; Xu, Z.S.; et al. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef]
- Naranjo, M.Á.; Forment, J.; Roldán, M.; Serrano, R.; Vicente, O. Overexpression of Arabidopsis thaliana LTL1, a salt-induced gene encoding a GDSL-motif lipase, increases salt tolerance in yeast and transgenic plants. Plant. Cell Environ. 2006, 29, 1890–1900. [Google Scholar] [CrossRef] [PubMed]
- Mansfeld, R. Das morphologische System der Saatgerste, Hordeum vulgare L. s. l. Der Züchter 1950, 20, 8–24. [Google Scholar] [CrossRef]
- Esechie, H.A. Interaction of Salinity and Temperature on the Germination of Sorghum. J. Agron. Crop Sci. 1994, 172, 194–199. [Google Scholar] [CrossRef]
- Al-Ansari, F.; Ksiksi, T. A Quantitative Assessment of Germination Parameters: The Case of and. Open Ecol. J. 2016, 9, 13–21. [Google Scholar] [CrossRef]
- Benech Arnold, R.L.; Fenner, M.; Edwards, P.J. Changes in germinability, ABA content and ABA embryonic sensitivity in developing seeds of Sorghum bicolor (L.) Moench. induced by water stress during grain filling. New Phytol. 1991, 118, 339–347. [Google Scholar] [CrossRef]
- Orchard, T.J. Estimating the parameters of plant seedling emergence. Seed Sci. Technol. 1977, 5, 61–69. [Google Scholar]
- Abiri, R.; Shaharuddin, N.A.; Maziah, M.; Norhana, Z.; Yusof, B.; Atabaki, N.; Sahebi, M.; Azizi, P. Quantitative assessment of indica rice germination to hydropriming, hormonal priming and polyethylene glycol priming. Chil. J. Agric. Res. 2016, 76, 392–400. [Google Scholar] [CrossRef]
- Fernandez, G.C.J. Effective selection criteria for assessing plant stress tolerance. In Proceedings of the International Symposium on Adaptation of Vegetables and other Food Crops in Temperature and Water Stress, Shanhua, Taiwan, 13–16 August 1992; pp. 257–270. [Google Scholar] [CrossRef]
- SAS Institute. The SAS System for Windows; Release 9.2; SAS Institute: Cary, NC, USA, 2011; Available online: http://www.sciepub.com/reference/166089 (accessed on 4 July 2021).
- Padi, F.K. Genotype environment interaction and yield stability in a cowpea-based cropping system. Euphytica 2007, 158, 11–25. [Google Scholar] [CrossRef]
- Dreissig, S.; Maurer, A.; Sharma, R.; Milne, L.; Flavell, A.J.; Schmutzer, T.; Pillen, K. Natural variation in meiotic recombination rate shapes introgression patterns in intraspecific hybrids between wild and domesticated barley. New Phytol. 2020, 228, 1852–1863. [Google Scholar] [CrossRef]
- SAS Institute. The SAS System for Windows; Release 9.4; SAS Institute: Cary, NC, USA, 2013. [Google Scholar]
- Bonferroni, C. Teoria Statistica Delle Classi e Calcolo Delle Probabilita, 8th ed.; Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commericiali di Firenze: Firenze, Italy, 1936; pp. 3–62. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).