Abstract
Litchi (Litchi chinensis Sonn.) is an important subtropical and tropical evergreen fruit tree that is seriously affected by chilling stress. In order to identify genes that may be involved in the response to chilling in litchi, we investigate the physiological and biochemical changes under chilling stress and construct 12 RNA-Seq libraries of leaf samples at 0, 4, 8, and 12 days of chilling. The results show that antioxidant enzymes are activated by chilling treatments. Comparing the transcriptome data of the four time points, we screen 2496 chilling-responsive genes (CRGs), from which we identify 63 genes related to the antioxidant system (AO-CRGs) and 54 ABA, 40 IAA, 37 CTK, 27 ETH, 21 BR, 13 GA, 35 JA, 29 SA, and 4 SL signal transduction-related genes. Expression pattern analysis shows that the expression trends of the 28 candidate genes detected by qRT-PCR are similar to those detected by RNA-Seq, indicating the reliability of our RNA-Seq data. Partial Least Squares Structural Equation Modeling (PLS-SEM) analysis of the RNA-Seq data suggests a model for the litchi plants in response to chilling stress that alters the expression of the plant hormone signaling-related genes, the transcription factor-encoding genes LcICE1, LcCBFs, and LcbZIPs, and the antioxidant system-related genes. This study provides candidate genes for the future breeding of litchi cultivars with high chilling resistance, and elucidates possible pathways for litchi in response to chilling using transcriptomic data.
1. Introduction
Chilling is one of the main abiotic stresses affecting the growth and development of plants, geographical distribution, and production [1,2]. It can affect plant growth and development by changing the membrane system, cell osmotic conditions, metabolism, and antioxidant systems [3]. At present, chilling injury is a serious natural disaster in agriculture [4] that leads to a significant reduction in the yield of field crops [5]. Therefore, it is particularly important to investigate the mechanism of chilling injury and the chilling adaptation strategies of plants. These chilling adaptation strategies include chilling resistance underlying physiological and biochemical changes [6], a complex gene network of chilling responses, and post-transcriptional modifications [7,8]. At present, genes functioning in relation to chilling resistance have been identified [9]. Chilling resistance in arabidopsis has been improved by means of genetic engineering [10].
Litchi (Litchi chinensis Sonn.) is native to China and is a fruit tree of the Sapinaceae family [11]. It is an important subtropical and tropical evergreen fruit tree in Southeast Asia. However, chilling injury is one of the main factors that restricts litchi production. Hence, it is important to investigate the mechanism of litchi in response to chilling so as to find ways to increase its resistance. RNA sequencing (RNA-Seq) technology is a powerful tool for transcriptomic studies. This technique has been used to elucidate puffing disorder in citrus [12], to investigate transcriptomic changes in winter wheat [13], to elucidate transcriptomic responses of Lanzhou lily to drought stress [14], and to identify genes involved in low-temperature-induced senescence, as well as ROS-induced flowering in litchi [15,16]. However, up till now, no RNA-Seq data on litchi plants in response to chilling have been published.
In this study, we use RNA-Seq technology to identify differentially expressed genes (DEGs) which might be potentially involved in the chilling resistance of litchi’s underlying phytohormones and antioxidant systems. We assume that hormone-related DEGs might be directly or indirectly related to antioxidant system-related DEGs to regulate litchi’s response to low temperature. Hence, we perform Partial Least Squares Structural Equation Modeling (PLS-SEM) analysis to quantify the relationship among the biological processes based on the transcript levels of the DEGs [17]. The aim of this work is to elucidate pathways for litchi to adapt to chilling stress by phytohormones and antioxidant system-related genes.
2. Results
2.1. Phenotype of Litchi Leaves under Chilling Stress
Figure 1 shows the morphological changes in litchi leaves under chilling stress. It was found that litchi leaves began to curl and the veins turned red from 4 d of treatment.

Figure 1.
Leaf phenotype of ‘Huaizhi’ litchi trees under chilling stress. ‘Huaizhi’ litchi trees were transferred to a growth chamber. Temperature of the chambers dropped from 22 °C/17 °C (day/night temperature, 12 h day and 12 h night) to 2 °C in 4 days, and then remained at 2 °C for another 8 days. The days represent the samples of ‘Huaizhi’ litchi leaves on 0 d, 2 d, 4 d, 6 d, 8 d, 10 d, and 12 d under low-temperature conditions.
2.2. Physiological and Biochemical Changes in Response to Chilling Stress in Litchi Leaves
Chilling stress reduced the Fv/Fm values of litchi leaves (Figure 2A,B). From day 2, Fv/Fm values decreased and at day 6 to 12, the values were around 0.30, while those of the values at normal temperature remained at a high level above 0.75.
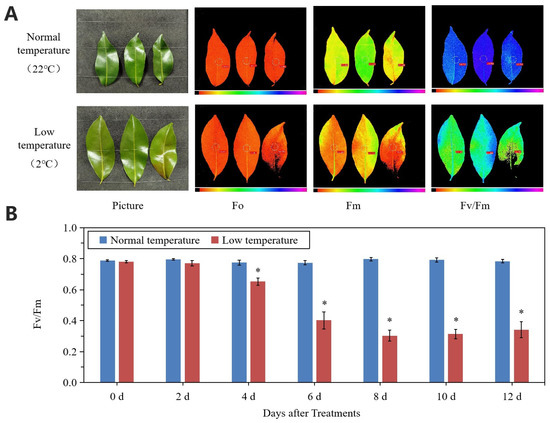
Figure 2.
Changes in leaf chlorophyll fluorescence parameters under normal- and low-temperature conditions. (A) Phenotype and chlorophyll fluorescence imaging for parameters Fo, Fm, and Fv/Fm for leaves of ‘Huaizhi’ at normal temperature as control and low temperature as treatment at the time point of day 12. (B) Fv/Fm values of litchi leaves under low-temperature and normal-temperature conditions. Asterisks (*) indicate significant difference between treatment and control at the same time point (p ≤ 0.05, n = 3, Student’s t-test).
Relative conductivity of litchi leaves increased with the chilling stress. From 4 d to 12 d of treatment, relative conductivity of the treated leaves was significantly higher than that of the controls, whereas the relative water content of the treated leaves was lower than that of the controls (Figure 3A,B). At day 12, the relative conductivity and relative water content of the treated leaves was 64.08% higher and 20.25% lower than that of the controls, respectively. In the treated leaves, activities of SOD, CAT, and POD showed increasing trends, while those of the control leaves were stable and remained at lower levels. The content of H2O2 in the treated leaves increased with the time of treatment and was significantly higher than that in the control leaves (Figure 3C–F).
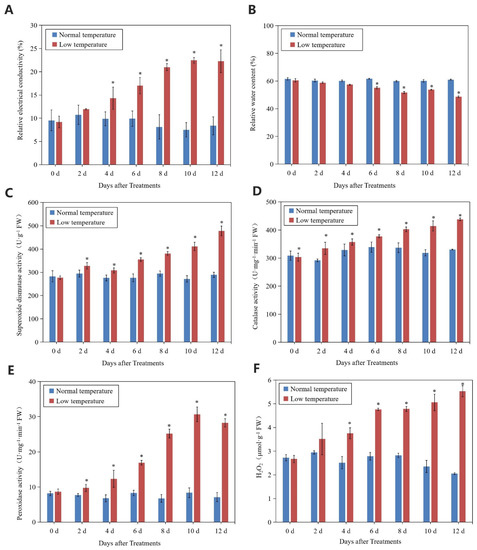
Figure 3.
Changes in relative conductivity (A), relative water content (B), SOD activity (C), CAT activity (D), POD activity (E), and H2O2 content (F) of leaves in ‘Huaizhi’ litchi trees under normal- and low-temperature conditions. ‘Huaizhi’ litchi trees were transferred to a growth chamber. Temperature of the chamber dropped from 22 °C/17 °C (day/night temperature, 12 h day and 12 h night) to 2 °C in 4 days, and then remained at 2 °C for another 8 days. Asterisks (*) indicate significant difference between low temperature as treatment and normal temperature as control at the same time points (p ≤ 0.05, n = 3, Student’s t-test).
2.3. Digital Transcriptomic Analysis
To identify genes in response to chilling stress in litchi, we collected leaves at 0 d, 4 d, 8 d, and 12 d time points of the low-temperature treatment and constructed 12 RNA-Seq libraries. As shown in Table 1, the percentage of Q30 of each sample was more than 93%.

Table 1.
Data quality and alignment analysis.
2.4. Identification of Differentially Expressed Genes
On the basis of FDR ≤0.01 and fold-change ≥2, the DEGs of leaves between the chilling stress (L4d, L8d and L12d) and the control (C0d) were screened (Figure 4). Correlation analysis of the replicates of the four time points (0 d, 4 d, 8 d, and 12 d) showed high repeatability of the sequencing samples (Figure S1). On the whole, down-regulated DEGs were more than the up-regulated ones (Figure 4A–D). At last, we identified 3756, 7489, and 7051 DEGs from the comparison of the samples of 0 d to 4 d, 0 d to 8 d, and 0 d to 12 d, respectively, with an overlap of 2496 (Figure 4D). The 2496 DEGs were defined as chilling responsive genes (CRGs). Then, GO analysis was performed on the DEGs of each sample under chilling stress.
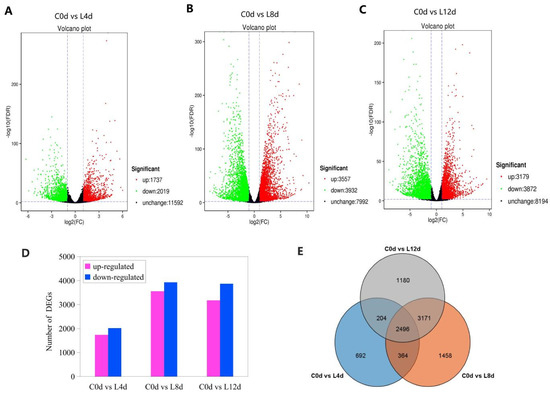
Figure 4.
Differentially expressed genes (A–D) and Venn diagram of the DEGs (E) in response to chilling stress. ‘Huaizhi’ litchi trees were grown in a chamber with 22 °C/17 °C (day/night temperature, 12 h day and 12 h night) as the normal temperature. Temperatures dropped from the normal one to 2 °C for 4 d, and then were maintained at 2 °C for another 8 d. C0d, L4d, L8d, and L12d mean the leaves samples at 0 d, 4 d, 8 d, and 12 d of chilling stress.
2.5. GO and KEGG Enrichment Analysis of the Differentially Expressed Genes
All DEGs were divided into three categories: biological process, cellular components, and molecular function. We found that the cellular process, the metabolic process, and the single-organism process were the most abundant in the category of biological processes (Table S1). In the category of cell elements, cell, cell parts, and membrane parts were the most abundant subcategories. Within the broad category of molecular functions, DEGs were enriched in protein binding, catalytic activity, and transporter activity (Figure S2).
For KEGG pathway analysis, we found that the plant hormone signal transduction pathway was enriched in the DEGs between 0 d and 4 d, 0 d and 8 d, and 0 d and 12 d (Figure 5).
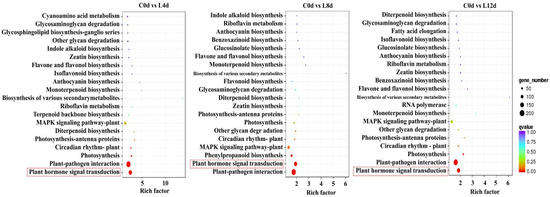
Figure 5.
KEGG enrichment analysis of the DEGs. ‘Huaizhi’ litchi trees were grown in a chamber with 22 °C/17 °C (day/night temperature, 12 h day and 12 h night) as the normal temperature. Temperatures dropped from the normal one to 2 °C for 4 d, and then were maintained at 2 °C for another 8 d. C0d, L4d, L8d, and L12d mean the leaves samples at 0 d, 4 d, 8 d, and 12 d of chilling stress.
2.6. Identification of Plant Hormone Signal-Related Genes Involved in Chilling Stress
From the 2496 CRGs, we screened those related to plant hormone signal transduction pathways (plant hormone signal transduction, ko04075), of which, 54 were related to abscisic acid signaling (ABA-DEGs), 40 were related to auxin signaling (IAA-DEGs), 37 were related to cytokinin signaling (CTK-DEGs), 27 were related to ethylene signaling (ETH-DEGs) (Figure 6), 21 were related to brassinolide signaling (BR-DEGs), 13 were associated with gibberellin signaling (GA-DEGs), 35 were associated with jasmonic acid signaling (JA-DEGs), 29 were associated with salicylic acid (SA-DEGs), and 4 were associated with strigolactone signaling (SL-DEGs) (Figure 7; Table S2).
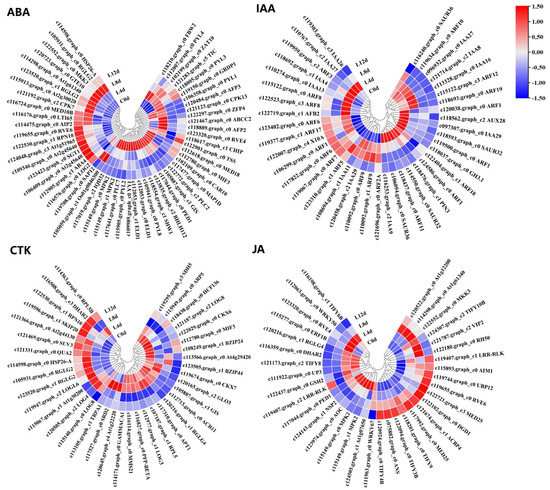
Figure 6.
Heat map diagram showing the expression profiles of the ABA-, IAA-, CTK-, and JA-DEGs under low-temperature conditions. ‘Huaizhi’ litchi trees were grown in a chamber with 22 °C/17 °C (day/night temperature, 12 h day and 12 h night) as the normal temperature. Temperatures dropped from the normal one to 2 °C for 4 d, and then were maintained at 2 °C for another 8 d. C0d, L4d, L8d, and L12d mean the leaves samples at 0 d, 4 d, 8 d, and 12 d of treatment. Fragments per kilobase of transcript per million mapped reads (FPKM) values were normalized to Z-score.
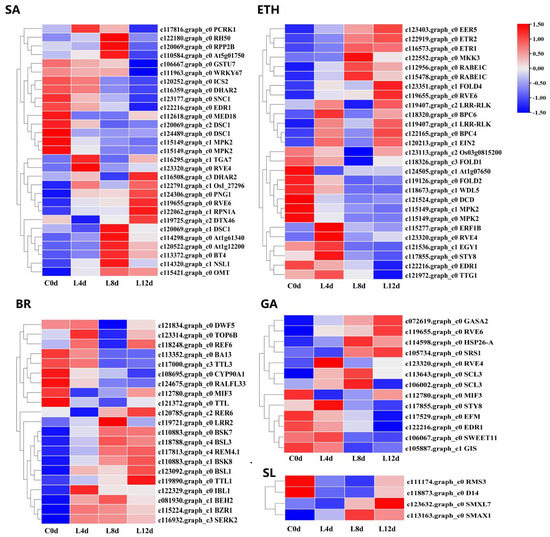
Figure 7.
Heat map diagram showing the expression profiles of the SA-, ETH-, BR-, GA-, and SL-DEGs under low-temperature conditions. ‘Huaizhi’ litchi trees were grown in a chamber with 22 °C/17 °C (day/night temperature, 12 h day and 12 h night) as the normal temperature. Temperatures dropped from the normal one to 2 °C for 4 d, and then were maintained at 2 °C for another 8 d. C0d, L4d, L8d, and L12d mean the leaves samples at 0 d, 4 d, 8 d, and 12 d of treatment. Fragments per kilobase of transcript per million mapped reads (FPKM) values were normalized to Z-score.
2.7. Identification of Antioxidant Enzyme Encoding Genes Involved in Chilling Stress
We focused on the KEGG pathways including ascorbate and aldarate metabolism, glutathione metabolism, peroxisome, phenylpropanoid biosynthesis, and pyruvate metabolism (Figure S3). These pathways contained genes related to the antioxidant system. As a result, we identified 63 DEGs (Figure 8) defined as antioxidant system-related CRGs (AO-CRGs) (Table S3).
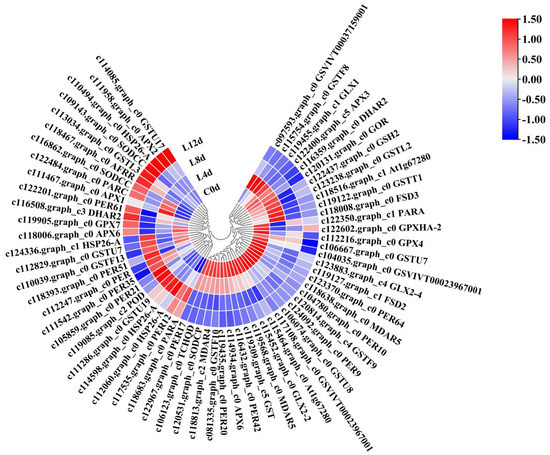
Figure 8.
Heatmap of the antioxidant system-related DEGs in response to low-temperature in litchi. ‘Huaizhi’ litchi trees were grown in a chamber with 22 °C/17 °C (day/night temperature, 12 h day and 12 h night) as the normal temperature. Temperatures dropped from the normal one to 2 °C for 4 d, and then were maintained at 2 °C for another 8 d. C0d, L4d, L8d, and L12d mean leaves samples at 0 d, 4 d, 8 d, and 12 d of treatment. Fragments per kilobase of transcript per million mapped reads (FPKM) values were normalized to Z-score.
2.8. Probable Pathway Involved in Hormonal Control in Response to Chilling Stress
From the CRGs, we also identified transcription factor encoding genes, including LcICE1, LcCBFs, and LcbZIPs. The expression patterns of these CRGs showed that they were all induced by the chilling stress (Figure S4). We also obtained the above nine types of hormone-related DEGs. We suspected that each type of hormone-related DEGs might be directly related to AO-CRGs to regulate litchi response to low temperature, or might be indirectly related to transcription factor-related CRGs (LcICE1, LcCBFs, and LcbZIPs) to affect AO-CRGs, and to regulate the responses of chilling stress in litchi. Hence, we constructed a model using PLS-SEM.
Through PLS modeling data analysis, the influence of collinearity between variables could be effectively eliminated, and the relationship between observed variables and latent variables and between latent variables could be well tested. In the present study, most of the latent variables were significant, and the latent variables were adequately explained by the external model (Table S4). Meanwhile, the extracted mean variance (AVE, measure of convergent validity) was higher than 0.5 (Table S5), indicating that the model was reliable [18]. As shown in Figure 9, each model had two pathways, the direct pathway and the indirect pathway. In the indirect pathway, hormone-related DEGs had positive or negative effects on the three major transcription factors (TFs). In the ETH model, there were high factor loadings due to the high absolute value, such as −0.985, −0.912, and −0.755, higher than the second-order loads in the indirect path (−0.175, 0.231, and −0.042). This suggests that ABA-related DEGs were directly related to AO-CRGs. In addition, SL-DEGs, IAA, CTK, JA, BR, SA, ETH, and GA-DEGs were also directly related to AO-CRGs.
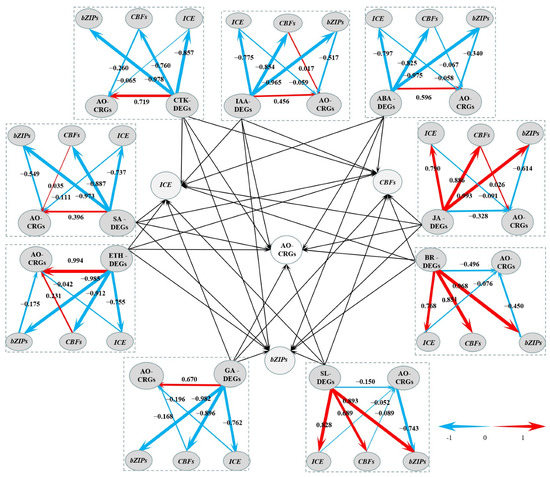
Figure 9.
A graph of the Partial Least Squares Structural Equation Modeling (PLS-SEM). The center part of the graph is based on the other nine parts around it. Levels of the path coefficients are reflected by the width of the red or blue arrows which indicate positive and negative effects, respectively. Values on the arrow indicate the intensity of the factor loading for the pathway. Pathways from hormone-related DEGs to transcription factor-encoding DEGs are first orders, while those from transcription factor encoding DEGs to antioxidant-related CRGs are second orders.
2.9. Confirmation of the RNA-Seq Data by qRT-PCR Analysis
In order to confirm the RNA-Seq data, we randomly selected 28 DEGs for qRT-PCR verification. As shown in Figure 10, the relative expression levels of LcDREB1D, LcAFP2, LcICE1, LcCAX2, LcNAC017, and LcTRX2 increased then decreased during the chilling treatment. The relative expression levels of LcBZIP53, LcBZIP16, LcCOR27, LcMYB3R5, LcHOS1, LcCBF5, LcAFP3, LcSCE1, LcNAC048, LcNAC078, LcMYBC1, and LcNAC082 showed increasing trends.
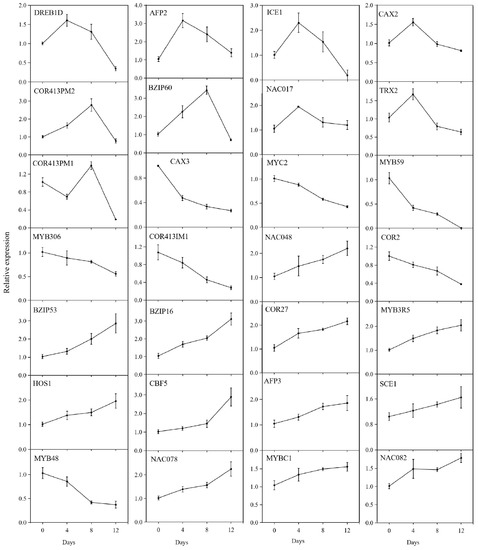
Figure 10.
Expression of the 28 differentially expressed genes determined by quantitative real-time PCR. ‘Huaizhi’ litchi trees were grown in a chamber with 22 °C/17 °C (day/night temperature, 12 h day and 12 h night) as the normal temperature. Temperatures dropped from the normal one to 2 °C for 4 d, and then were maintained at 2 °C for another 8 d. C0d, L4d, L8d, and L12d mean the leaves samples at 0 d, 4 d, 8 d, and 12 d of treatment. Relative transcription was calculated by qRT-PCR using the 2−ΔΔCT method with actin as a reference.
A linear regression analysis of the fold-change in the gene expression ratios between RNA-Seq and qRT-PCR showed a significantly positive correlation between the obtained data from qRT-PCR and RNA-Seq (Figure 11), confirming the reliability of our RNA-Seq data.
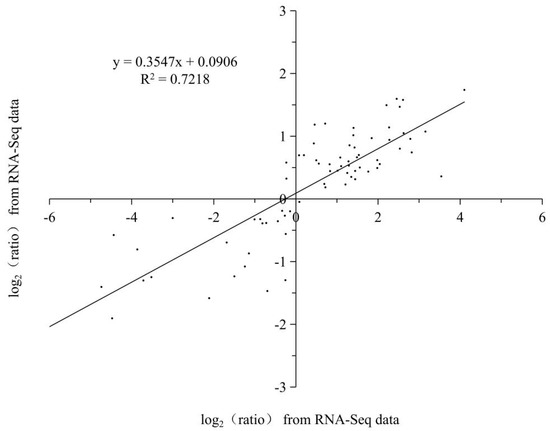
Figure 11.
Coefficient analysis of fold change data between qRT-PCR and RNA-Seq. All the data are from the qRT-PCR analysis and RNA-Seq of the 0 d (C0d), 4 d (L4d), 8 d (L8d), and 12 d (L12d). Scatterplots were generated by the log2 expression ratios from RNA-seq (x-axis) and qRT-PCR (y-axis).
2.10. Model for the Litchi Response to Chilling
We performed PLS-SEM analysis using RNA-Seq data and built a model to quantify their relationship. According to the PLS model, we proposed a model for litchi in response to chilling. As shown in Figure 12, when the litchi trees were subjected to chilling stress, the expression of plant hormone signal-related genes may change, directly affecting the expression pattern of the antioxidant system-related genes, or may regulate the expression of the LcICE1, LcCBFs, and LcbZIPs, and indirectly affecting the antioxidant system-related genes. As a result, LcICE1, LcCBFs, and LcbZIPs may affect the antioxidant system-related genes. The antioxidant system of the litchi trees may be activated, helping the trees to react and adapt to chilling stress.
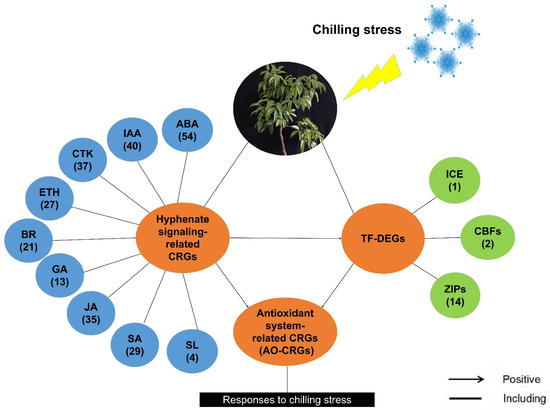
Figure 12.
A proposed model for litchi in response to chilling according to the Partial Least Squares Structural Equation Modeling analyses. Details of the co-expression network of three identified cold-responsive genes and associated hormones are shown. The numbers in the brackets represent the number of the identified chilling responsive genes (CRGs) under the specific items.
3. Discussion
It has been shown that the stability of the cell membrane system indicated by the related conductivity may be affected under chilling stress [19,20]. The membrane system is the site of temperature sensing when plants are exposed to low temperatures [21]. The chlorophyll fluorescence parameter Fv/Fm reflects the maximum quantum efficiency of photosystem II (PSII) photochemistry and has been widely used for stress detection in plants [22]. In the present study, the Fv/Fm value showed a decreasing trend while relative conductivity showed an increasing trend, suggesting that litchi trees were affected by the chilling stress.
The antioxidant system is a key protection mechanism for plants under stressful conditions [23]. Activities of SOD, POD, and CAT, which belong to the antioxidant system, were determined. The results showed that they increased continuously with the time of the chilling treatment, indicating that the antioxidant system of litchi was activated under chilling conditions. However, we still detected an increasing accumulation of H2O2. It is well known that H2O2 can be removed by antioxidant enzymes [24]. It might be that the balance of the synthesis and scavenging of H2O2 was broken by chilling stress [25].
To study the transcriptomic changes in litchi under chilling stress, we sequenced the leaf samples at four time points of the low-temperature treatment and obtained 2496 CRGs. Interestingly, the KEGG enrichment analysis showed that the plant hormone signal transduction pathway was significantly enriched. Plant hormones are trace organic compounds that are induced by plant cells to receive environmental signals and act as signaling molecules [26]. At low concentrations, they can regulate the formation of flowers, stems, and leaves, control the development and ripening of fruits, and regulate the physiological responses of plants to stresses [27]. Our results suggest that plant hormone signal transduction might be an important strategy for litchi to adapt to chilling stress.
The plant hormone abscisic acid (ABA) is known as a signaling substance and is induced by abiotic stresses [28,29]. Abscisic acid receptor kinase 1 (CARK1) phosphorylates RCAR/PYR/PYLs and enhances the inhibition of PP2Cs, leading to the activation of abscisic acid signaling under stress [30]. Aux/IAA proteins are a large family of auxin co-receptors and transcriptional repressors that play central roles in auxin signaling [31]. GH3 encodes an IAA-amino acid synthase which can prevent IAA accumulation and regulate plant growth at low temperatures through an auxin-dependent pathway. In this study, we also identified ABA and IAA signaling transduction component encoding genes in litchi, such as LcABA4, LcIAA9, and LcGH3.1. They were differentially expressed under chilling conditions.
The cytokinin signaling pathway is based on a binary signaling system which is achieved through the continuous transmission of phosphate groups between major components [32]. The loss of function or the mutation of LONELY GUY (LOG), which encodes a cytokinin-activating enzyme, delays root growth in plants under extremely low-temperature conditions [33]. The fine-tuning of ethylene content in tissues affects plant growth and development [34]. Liu et al. [35] found that ERF9 could be induced by ethylene under low-temperature conditions [36]. The transmission of the brassinosteroid signal finally makes the transcription factor family BES1/BZR1 accumulate in a non-phosphorylated state, which activates downstream transcriptional regulation in the nucleus. It then regulates the growth and development of plants. In contrast, the AP2/ERF transcription factor TINY inhibits brassinolide-regulated growth [37]. From the CRGs, we identified four LOG encoding genes, LcLOG3, LcLOG4, LcLOGL6, and LcLOG8, in litchi. We also identified LcERF1B, and LcBZR1D. Expression patterns of these genes showed that they were chilling responsive. Other phytohormone signaling-related genes such the GA, SA, and SL signal-related CRGs in litchi were shown to be chilling-responsive, indicating that phytohormone signals play important roles in the response to chilling in litchi.
As a transmission signal, plant hormones can be directly transmitted to the nucleus to activate the expression of genes to cause corresponding physiological and biochemical changes in plants. Transcription factors can bind to specific DNA sequences to control the expression of particular genes. ICE1 [38,39], CBFs [35,40], and bZIPs [41,42,43,44] are central transcription factors downstream of plant hormone signaling transduction, and act as executors for the hormonal controlled plant growth and development. We hypothesize that they may play an important role in the response of chilling stress in litchi, and might act as mediators in phytohormone signaling and the antioxidant system to regulate the response of litchi to chilling stress. In our previous study, we used PLS-SEM for the first time to elucidate a pathway for high-temperature-induced floral abortion in litchi’s underlying gene expression [17]. In accordance with our previous study, this study further shows that PLS-SEM is valuable in revealing relationships between biological processes based on RNA-Seq data. Future studies should focus on the ICE1, CBFs, and bZIPs underlying the regulation of chilling responses of plant hormones in litchi.
4. Materials and Methods
4.1. Experimental Procedure
Seven-year-old ‘Huaizhi’ litchi trees were planted in 30 L pots containing loam, mushroom cinder, and coconut chaff (3:1:1 (v/v/v)). The trees were cultivated in the experimental orchard of South China Agricultural University, Guangzhou, China (lat. 23°90′40″ N, long. 113°210′18″ E). The trees with mature leaves in the terminal shoots were selected. The trees were transferred to a growth chamber at 22 °C/17 °C (day/night temperature, 12 h day and 12 h night) with a relative humidity of 75–85% and a light intensity of 120 µmol·m−2 s−1. The program for the chilling treatment is shown in Table 2, according to our previous studies [45]. The temperature was maintained at 2 °C from day 4 until the end of the experiment. The fourth and fifth mature compound leaves of the terminal shoots were collected every 2 days. Samples were frozen in liquid nitrogen immediately and stored at −80 °C.

Table 2.
The low-temperature program in the chamber.
4.2. Determination of Maximal Photochemical Efficiency
Leaves were dark-adapted for 20 min, and then the initial fluorescence yield (Fo), maximal fluorescence yield (Fm), and maximal photochemical efficiency (Fv/Fm) were measured by a chlorophyll fluorescence imaging system (Model IMAGING-PAM, WALZ company, Effeltrich, Germany).
4.3. Determination of Relative Conductivity and Relative Water Content
The relative conductivity analysis was preformed according to the method of Redmann et al. [46]. First, 0.1 g of fresh leaves was incubated in 20 mL of distilled water for 12 h, measured with a conductivity meter (Model DDS-307, Shanghai Leici Instrument Inc., Shanghai, China), heated in boiling water for 30 min, cooled to room temperature, and measured for conductivity again. The relative conductivity of the sample was calculated according to the ratio of the conductivity before and after boiling.
Fresh leaves (0.1 g) were kept in distilled water for 24 h and weighed. The leaves were then dried and weighed. Relative water content was calculated as described by Weatherley [47].
4.4. Determination of Activities of Superoxide Dismutase, Catalase, and Peroxidase
Next, 1.0 g leaves were taken and 50 mM (pH 7.8) phosphate buffer in 10 mL was added, then centrifuged at 10,000× g for 20 min at 4 °C. The supernatants were used for the determination of enzyme activity.
Superoxide dismutase (SOD) activity was measured according to the nitrogen blue tetrazolium (NBT) method [48]. The reaction solution containing 50 mM PBS, 130 mM methionine, 75 µM NBT, 50 µM EDTA-Na2, and crude enzyme solution was illuminated at 80 µmol·m−2 s−1 for 25 min. The absorbance was monitored at 560 nm wavelength by a spectrophotometer (Fluoroskan Ascen FL, Thermo, Waltham, MA, USA).
Catalase (CAT) activity was determined by UV absorption method [49,50]. The 3 mL reaction solution contained 15 mM H2O2, 50 mM phosphate buffer, and crude catalase extracts. Catalase activity was determined according to the decrease in the absorbance of H2O2 at 240 nm.
Peroxidase (POD) activity was measured using the guaiacol method described by Gillikin and Graham [51]. The 3 mL reaction solution contained 25 mM guaiacol, 10 mM hydrogen peroxide, 50 mM (pH 7.8) phosphate, and crude extracts. Changes in the absorbance of the reaction solution at a wavelength of 470 nm were determined by a spectrophotometer (Fluoroskan Ascen FL, Thermo).
4.5. Determination of Hydrogen Peroxide Content
The H2O2 content was determined using a commercial kit according to the manufacturer’s instructions (Jiancheng, Nanjing, China). The absorbance measurements were performed using a spectrophotometer (Fluoroskan Ascen FL, Thermo).
4.6. RNA Isolation, cDNA Library Construction, and Analysis
Total RNA was extracted using the RNAprep Pure Plant Kit (polysaccharide and polyphenolic-rich) (Tiangen Biotech, Beijing, China). First-strand cDNA was synthesized using a Reverse Transcriptase M-MLV (RNase H-) system (Takara, Dalian, China) from 1 μg extracted RNA. Sequencing libraries were generated using NEBNext®Ultra™ RNA Library Prep Kit for Illumina® (NEB, Ipswich, MA, USA) following the manufacturers’ instructions. Briefly, Oligo-dT beads (Qiagen, Valencia, CA, USA) were used to enrich mRNA. After fragmentation, the fragmented mRNA was used as a template to synthesize single-stranded cDNA with hexamer random primers. Second-strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. In order to select cDNA fragments of preferentially 240 bp in length, the library fragments were purified with an AMPure XP system (Beckman Coulter, Beverly Hills, CA, USA). Then, PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. At last, PCR products were purified (AMPure XP system), and library quality was assessed on the Agilent Bioanalyzer 2100 system.
4.7. RNA-Seq Data Analysis
Adapters were cut from raw reads, and the low-quality sequences were removed. rRNA sequences were filtered against ribosomal RNA using Bowtie2 (version 2.1.0., Bowtie2. unpublished work. Available online: http://litchidb.genomics.cn, accessed on 12 June 2021). The read counts and fragments per kilobase of the transcript per million mapped reads (FPKM) of each gene were calculated using eXpress (v1.5.1, eXpress. Available online: https://pachterlab.github.io/eXpress/index.html, accessed on 12 June 2021). Q20, Q30, GC-content and sequence duplication level of the clean data were calculated. All the downstream analyses were based on clean data with high quality. The differential expression analysis of two groups was performed using the DESeq R package (1.10.1). Significant DEGs were restricted with false discovery rate (FDR) ≤0.01 and the absolute value of fold-change ≥2 [52]. Gene ontology (GO) enrichment analysis of the DEGs was implemented by the topGO R package-based Kolmogorov–Smirnov test. The enrichment of the GO functional classifications was plotted using the R Bioconductor package GOstats [53]. KEGG enrichment analysis was performed using a website of https://www.genome.jp/kegg/, accessed on 16 June 2021. The dataset is available in the NCBI Short Read Archive (SRA) under the accession number PRJNA845931.
4.8. Quantitative RT-PCR Analysis
Quantitative real-time polymerase chain reaction (qRT-PCR) primers F1/R1 (Table S6) were designed by Primer 6.0 (Premier Biosoft, Palo Alto, CA, USA) and synthesized by Sangon Co., Ltd. (Shanghai, China). The qRT-PCR was performed on a CFX96 real-time PCR machine (Bio-Rad, Hercules, CA, USA) with 2×RealStar Green Power Mixture (GenStar BioSolutions, Beijing, China). qPCR was performed at 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s in 96-well optical reaction plates. Each qRT-PCR analysis was performed in three biological and three technical replicates. The transcript quantification of the genes was performed in relation to Actin, and was calculated using the 2−∆∆CT method [54].
4.9. Statistical Analysis
The IBM SPSS statistical software 23.0 was used to determine significant differences between the treatment and controls. The expression of genes was presented as a heat map diagram using the pheatmap package (http://www.r-project.org/, accessed on 24 June 2022). To explore the relationships between DEGs and the responses of litchi to low temperature, a hypothetical model according to Chen et al. [55] was specified and analyzed with PLS-SEM with the support of the SmartPLS 2.0 M3 software [56]. Standardized path coefficient values were generated with the PLS algorithm by the Path Weighting Scheme using a bootstrapping method to obtain the significance of path coefficients. The sign changes were individual changes. The samples during the calculation of the bootstrapping method were 5000, and the ABA-, IAA-, CTK-, JA-, SA-, ETH-, BR-, GA-, SL-related DEGs models were 54, 40, 37, 35, 29, 27, 21, 13, 4, respectively.
5. Conclusions
Twelve RNA-Seq libraries of the leaf samples were constructed at 0, 4, 8, and 12 d of chilling treatment. We screened 2496 CRGs. From this, 260 plant hormone signal transduction-related CRGs were identified. PLS-SEM analysis of the RNA-Seq data suggested a model for the litchi plants in response to chilling stress by altering the expression of plant hormone signal-related genes, the transcription factor encoding genes LcICE1, LcCBFs, and LcbZIPs, and the antioxidant system-related genes. Our study provides candidate genes for the future breeding of litchi cultivars with high chilling resistance, and elucidates possible pathways for litchi in response to chilling by PLS-SEM analysis using transcriptomic data.
Supplementary Materials
The following supporting information can be downloaded at: www.mdpi.com/article/10.3390/ijms23158424/s1.
Author Contributions
B.Z. designed the research and wrote and revised the manuscript; X.Z. performed the experiments and wrote the manuscript; H.L. and L.H. analyzed the data. All authors have read and agreed to the published version of the manuscript.
Funding
The research was supported by the National Key Research and Development Program of China (2018YFD1000302), the National Natural Science Foundation of China (grant nos. 31772249), the National Litchi and Longan Research System (project no. CARS-33).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available at NCBI SRA accession PRJNA845931.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pokorná, J.; Schwarzerová, K.; Zelenkova, S.; Petrášek, J.; Janotová, I.; Čapková, V.; Opatrný, Z. Sites of actin filament initiation and reorganization in cold-treated tobacco cells. Plant Cell Environ. 2004, 27, 641–653. [Google Scholar] [CrossRef]
- Ensminger, I.; Busch, F.; Huner, N.P. Photostasis and cold acclimation: Sensing low temperature through photosynthesis. Physiol. Plant 2006, 126, 28–44. [Google Scholar] [CrossRef]
- Kaplan, F.; Guy, C.L. β-amylase induction and the protective role of maltose during temperature shock. Plant Physiol. 2004, 135, 1674–1684. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef]
- Wu, F.Z.; Wang, H.X.; Xu, G.H.; Zhang, Z.C. Research progress on the physiological and molecular mechanisms of woody plants under low temperature stress. Sci. Silvae Sin. 2015, 51, 116–128. [Google Scholar]
- Solanke, A.U.; Sharma, A.K. Signal transduction during cold stress in plants. Physiol. Mol. Biol. Plants 2008, 14, 69–79. [Google Scholar] [CrossRef]
- Lee, B.H.; Henderson, D.A.; Zhu, J.K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 2005, 17, 3155–3175. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.K.; Sunkar, R. Gene regulation during cold stress acclimation in plans. Methods Mol. Biol. 2010, 639, 39–55. [Google Scholar]
- Wang, W.; Wang, X.; Huang, M.; Cai, J.; Zhou, Q.; Dai, T.; Jiang, D. Alleviation of field low-temperature stress in winter wheat by exogenous application of salicylic acid. J. Plant Growth Regul. 2021, 40, 811–823. [Google Scholar] [CrossRef]
- Guo, J.R.; Sun, B.X.; He, H.R.; Zhang, Y.F.; Tian, H.Y.; Wang, B.S. Current understanding of bHLH transcription factors in plant abiotic stress tolerance. Int. J. Mol. Sci. 2021, 22, 4921. [Google Scholar] [CrossRef]
- Liu, W.W.; Kim, H.J.; Chen, H.B.; Lu, X.Y.; Zhou, B.Y. Identification of MV-generated ROS responsive EST clones in floral buds of Litchi chinensis Sonn. Plant Cell Rep. 2013, 32, 1361–1372. [Google Scholar] [CrossRef]
- Ibánez, A.M.; Martinelli, F.; Reagan, R.L.; Uratsu, S.L.; Vo, A.; Tinoco, M.A.; Phu, M.L.; Chen, Y.; Rocke, D.M.; Dandekar, A.M. Transcriptome and metabolome analysis of citrus fruit to elucidate puffing disorder. Plant Sci. 2014, 217, 87–98. [Google Scholar] [CrossRef]
- Wang, F.; Ge, S.F.; Xu, X.X.; Xing, Y.; Du, X.; Zhang, X.; Lü, M.X.; Liu, J.Q.; Zhu, Z.L.; Jiang, Y.M. Multiomics analysis reveals new insights into the apple fruit quality decline under high nitrogen conditions. J. Agric. Food Chem. 2021, 69, 5559–5572. [Google Scholar] [CrossRef]
- Li, W.M.; Wang, Y.J.; Ren, H.; Guo, Z.H.; Li, N.; Zhao, C.Z.; Xie, Z.K. Physiological and transcriptomic responses of Lanzhou Lily (Lilium davidii, var. unicolor) to cold stress. PLoS ONE 2020, 15, e0227921. [Google Scholar]
- Wang, C.C.; Liu, H.; Yu, S.; Chen, H.B.; Hu, F.C.; Zhan, H.L.; Pan, X.F.; Lao, Y.H.; Zhong, S.L.; Zhou, B.Y. Identification of genes involved in low temperature-induced senescence of panicle leaf in Litchi chinensis. Genes 2019, 10, 111. [Google Scholar] [CrossRef]
- Lu, X.Y.; Yu, S.; Lü, P.T.; Chen, H.B.; Zhong, S.L.; Zhou, B.Y. Genome-Wide transcriptomic analysis reveals a regulatory network of oxidative stress-induced flowering signals produced in Litchi leaves. Genes 2020, 11, 324. [Google Scholar] [CrossRef]
- Liu, H.; Wang, C.C.; Chen, H.B.; Zhou, B.Y. Genome-wide transcriptome analysis reveals the molecular mechanism of high temperature-induced floral abortion in Litchi chinensis. BMC Genom. 2019, 20, 127. [Google Scholar] [CrossRef]
- Hair, J.F.; Ringle, C.M.; Sarstedt, M. PLS-SEM: Indeed a silver bullet. J. Market Theory. Prac. 2011, 19, 139–152. [Google Scholar] [CrossRef]
- Uemura, M.; Joseph, R.A.; Steponkus, P.L. Cold acclimation of Arabidopsis thaliana (effect on plasma membrane lipid composition and freeze-induced lesions). Plant Physiol. 1995, 109, 15–30. [Google Scholar] [CrossRef]
- Welti, R.; Li, W.Q.; Li, M.Y.; Sang, Y.M.; Biesiada, H.; Zhou, H.E.; Rajashekar, C.B.; Williams, T.D.; Wang, X.M. Profiling membrane lipids in plant stress responses: Role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 2002, 277, 31994–32002. [Google Scholar] [CrossRef]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef]
- Sharma, D.K.; Andersen, S.B.; Ottosen, C.O.; Rosenqvist, E. Wheat cultivars selected for high Fv/Fm under heat stress maintain high photosynthesis, total chlorophyll, stomatal conductance, transpiration and dry matter. Physiol. Plant 2015, 153, 284–298. [Google Scholar] [CrossRef]
- Liang, X.L.; Chen, Q.; Lu, H.; Wu, C.L.; Lu, F.P.; Tang, J.H. Increased activities of peroxidase and polyphenol oxidase enhance cassava resistance to Tetranychus urticae. Exp. App. Acarol. 2017, 71, 195–209. [Google Scholar] [CrossRef]
- Wang, W.B.; Kim, Y.H.; Lee, H.S.; Deng, X.P.; Kwak, S.S. Differential antioxidation activities in two alfalfa cultivars under chilling stress. Plant Biotechnol. Rep. 2009, 3, 301–307. [Google Scholar] [CrossRef]
- Yan, L.; Shah, T.; Cheng, Y.; Lü, y.; Zhang, X.K.; Zou, X.L. Physiological and molecular responses to cold stress in rapeseed (Brassica napus L.). J. Integr. Agr. 2019, 18, 2742–2752. [Google Scholar] [CrossRef]
- Kumar, P.P. Plant hormones and their intricate signaling networks: Unraveling the nexus. Plant Cell Rep. 2013, 32, 731–732. [Google Scholar] [CrossRef][Green Version]
- Janda, M.; Planchais, S.; Djafi, N.; Martinec, J.; Burketova, L.; Valentova, O.; Zachowski, A.; Ruelland, E. Phosphoglycerolipids are master players in plant hormone signal transduction. Plant Cell Rep. 2013, 32, 839–851. [Google Scholar] [CrossRef]
- Mauro, M.F.D.; Iglesias, M.J.; Arce, D.P.; Valle, E.M.; Arnold, R.B.; Tsuda, K.; Yamazaki, K.; Casalongué, C.A.; Godoy, A.V. MBF1s regulate ABA-dependent germination of Arabidopsis seeds. Plant Signal Behav. 2012, 7, 188–192. [Google Scholar] [CrossRef]
- Hewage, K.A.H.; Yang, J.F.; Wang, D.; Hao, G.F.; Yang, G.F.; Zhu, J.K. Chemical manipulation of abscisic acid signaling: A new approach to abiotic and biotic stress management in agriculture. Adv. Sci. 2020, 7, 2001265. [Google Scholar] [CrossRef]
- Li, X.Y.; Kong, X.G.; Huang, Q.; Zhang, Q.; Ge, H.; Zhang, L.; Li, G.M.; Peng, L.; Liu, Z.B.; Wang, J.M.; et al. CARK1 phosphorylates subfamily III members of ABA receptors. J. Exp. Bot. 2019, 70, 519–528. [Google Scholar] [CrossRef]
- Salehin, M.; Li, B.; Tang, M.; Katz, E.; Song, L.; Ecker, J.R.; Kliebenstein, D.J.; Estelle, M. Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Wang, Y.T.; Feng, C.; Wei, Y.; Peng, X.; Guo, X.; Guo, X.W.; Zhai, Z.F.; Li, J.; Shen, X.S.; et al. Overexpression of MsGH3.5 inhibits shoot and root development through the auxin and cytokinin pathways in apple plants. Plant J. 2020, 103, 166–183. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, H.; Kojima, M.; Kuroha, T.; Ishida, T.; Sugimoto, K.; Kiba, T.; Sakakibara, H. Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 2012, 69, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Xin, T.X.; Zhang, Z.; Li, S.; Zhang, S.; Li, Q.; Zhang, Z.H.; Huang, S.W.; Yang, X.Y.V. Genetic regulation of ethylene dosage for cucumber fruit elongation. Plant Cell 2019, 31, 1063–1076. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Shinozaki, K.Y.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Hu, J.; Dahro, B.; Ming, R.; Zhang, Y.; Wang, Y.; Alhag, A.; Li, C.L.; Liu, J.H. ERF108 from Poncirus trifoliata (L.) Raf. functions in cold tolerance by modulating raffinose synthesis through transcriptional regulation of PtrRafS. Plant J. 2021, 108, 705–724. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.L.; Nolan, T.; Jiang, H.; Tang, B.Y.; Zhang, M.C.; Li, Z.H.; Yin, Y.H. The AP2/ERF transcription factor TINY modulates brassinosteroid-regulated plant growth and drought responses in Arabidopsis. Plant Cell 2019, 31, 1788–1806. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998, 16, 433–442. [Google Scholar] [CrossRef]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef]
- Shinozaki, K.; Shinozaki, K.Y. Molecular responses to drought and cold stress. Curr. Opin. Biotechnol. 1996, 7, 161–167. [Google Scholar] [CrossRef]
- Winfield, M.O.; Lu, C.; Wilson, I.D.; Coghill, J.A.; Edwards, K.J. Plant responses to cold: Transcriptome analysis of wheat. Plant Biotechnol. J. 2010, 8, 749–771. [Google Scholar] [CrossRef]
- Kurbidaeva, A.S.; Novokreshchenova, M.G. Genetic control of plant resistance to cold. Russ. J. Genet. 2011, 47, 646–661. [Google Scholar] [CrossRef]
- Li, P.; Zheng, T.; Li, L.; Wang, J.; Cheng, T.; Zhang, Q. Genome-wide investigation of the bZIP transcription factor gene family in Prunus mume: Classification, evolution, expression profile and low-temperature stress responses. Hortic. Plant J. 2022, 8, 230–242. [Google Scholar] [CrossRef]
- Zhou, B.Y.; Li, Y.B.; Wu, C.B.; Chen, H.B.; Yang, R.T. Effect of PP333 on endogenous hormone and water condition of litchi in low temperature. J. Fruit Sci. 2003, 20, 93–95. [Google Scholar]
- Redmann, R.E.; Haraldson, J.; Gusta, L.V. Leakage of UV-absorbing substances as a measure of salt injury in leaf tissue of woody species. Physiol. Plant 1986, 67, 87–91. [Google Scholar] [CrossRef]
- Weatherley, P.E. Studies in the water relations of the cotton plant. I. The field measurement of water deficits in leaves. New Phytol. 1950, 49, 81–97. [Google Scholar] [CrossRef]
- Fridovich, I. Superoxide dismutases. Annu. Rev. Biochem. 1975, 44, 147–159. [Google Scholar] [CrossRef]
- Malay, C.; Greyling, M.M.; Gressel, J. Correlation between Cu/Zn superoxide dismutase and glutathione reductase, and environmental and xenobiotic stress tolerance in maize inbreds. Plant Sci. 1990, 69, 157–166. [Google Scholar]
- Zhou, B.Y.; Guo, Z.F.; Liu, Z.L. Effects of abscisic acid on antioxidant systems of Stylosanthes guianensis (Aublet) Sw. under chilling stress. Crop Sci. 2005, 45, 599–605. [Google Scholar] [CrossRef]
- Gillikin, J.W.; Graham, J.S. Purification and developmental analysis of the major anionic peroxidase from the seed coat of Glycine max (L.). Plant Physiol. 1991, 96, 214–220. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11M, R106. [Google Scholar] [CrossRef]
- Falcon, S.; Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics 2007, 23, 257–258. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, W.L.; Li, J.; Zhu, H.H.; Xu, P.Y.; Chen, J.Z.; Yao, Q. Arbuscular mycorrhizal fungus enhances lateral root formation in Poncirus trifoliata (L.) as revealed by RNA-Seq analysis. Front. Plant Sci. 2017, 8, 20–39. [Google Scholar] [CrossRef]
- Kumar, K. Partial Least Square (PLS) Analysis. Resonance 2021, 26, 429–442. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).