Abstract
Non-specific lipid transfer proteins (nsLTPs) are small cysteine-rich basic proteins which play essential roles in plant growth, development and abiotic/biotic stress response. However, there is limited information about the nsLTP gene (BnLTP) family in rapeseed (Brassica napus). In this study, 283 BnLTP genes were identified in rapeseed, which were distributed randomly in 19 chromosomes of rapeseed. Phylogenetic analysis showed that BnLTP proteins were divided into seven groups. Exon/intron structure and MEME motifs both remained highly conserved in each BnLTP group. Segmental duplication and hybridization of rapeseed’s two sub-genomes mainly contributed to the expansion of the BnLTP gene family. Various potential cis-elements that respond to plant growth, development, biotic/abiotic stresses, and phytohormone signals existed in BnLTP gene promoters. Transcriptome analysis showed that BnLTP genes were expressed in various tissues/organs with different levels and were also involved in the response to heat, drought, NaCl, cold, IAA and ABA stresses, as well as the treatment of fungal pathogens (Sclerotinia sclerotiorum and Leptosphaeria maculans). The qRT-PCR assay validated the results of RNA-seq expression analysis of two top Sclerotinia-responsive BnLTP genes, BnLTP129 and BnLTP161. Moreover, batches of BnLTPs might be regulated by BnTT1 and BnbZIP67 to play roles in the development, metabolism or adaptability of the seed coat and embryo in rapeseed. This work provides an important basis for further functional study of the BnLTP genes in rapeseed quality improvement and stress resistance.
1. Introduction
Rapeseed (Brassica napus), which provides abundant vegetable oil and substantially potential biodiesel feedstocks, is one of the most important oilseed crops worldwide [1]. The growth of the world’s population and the improvement of living standards have increased the global demand for rapeseed oil [2,3]. In addition, the yield and oil quality of rapeseed often suffer from extensive losses, owing to pathogen and biotic/abiotic stresses during rapeseed production worldwide [4,5,6,7]. These situations have increased the demand for the study of the genetic basis of high seed oil accumulation and high stress resistance of rapeseed.
With the development of next-generation sequencing technology, rapeseed breeders have conducted numerous research studies on the molecular mechanism of seed oil biosynthesis and stress resistance in rapeseed using genome-wide association and multiple-omics comparative analyses [8,9,10,11,12] in which various types of candidate genes, including plant non-specific lipid transfer protein (nsLTP) genes, were obtained. The nsLTPs that are plant-specific can bind or transfer many classes of hydrophobic molecules in vitro, e.g., fatty acids, fatty acyl-CoA, phospholipids, glycolipids and cutin monomers [13,14,15]. The nsLTPs have been shown to play crucial roles in various processes of plant growth and development, including seed development and quality, seed oil accumulation, seed germination, post-meiotic anther development, cutin formation and cell wall extension [16,17,18,19,20,21,22,23,24,25]. The nsLTPs also play critical roles in plant responses to abiotic stresses, e.g., cold, heat, salt and drought, as well as biotic stresses, e.g., bacterial and fungal pathogens [21,23,26,27,28,29,30,31]. Therefore, a systematic identification of rapeseed nsLTPs is of extreme importance for the creation of transgenic or gene-stacking rapeseed materials with enhanced seed oil quality and resistance to stresses.
Seeds are an important place for fatty acid synthesis and metabolism, so it is meaningful to have a proper understanding of nsLTPs involved in seed development, metabolism and adaptation. TT1 and bZIP67 are two representative transcription factors (TF) regulating seed coat and seed embryo developmental traits, respectively [32,33]. Therefore, in this study, differentially expressed genes (DEGs) of nsLTP family were identified from the DEG database of transgenic rapeseeds overexpressing BnTT1 and BnbZIP67, and it is speculated that they may be involved in the development and metabolism of seed coat and seed embryo.
The nsLTPs are small, basic proteins that exist in higher plants, which harbor an N-terminal signal peptide and contain an eight-cysteine motif (8CM, C–Xn–C–Xn–CC–Xn–CXC–Xn–C–Xn–C) [34]. The nsLTPs function in the cell secretory pathway [16]. The nsLTPs are located in the plasma membrane [35,36,37], extracellular space [38], cell wall [39] and cytoplasm [40]. The nsLTPs have been reported to play important roles in various developmental and physiological processes [21,40,41]. In recent years, the number of plant genomes sequenced has exploded, which facilitates genome-wide analysis of the plant nsLTP gene family in a variety of plant species. The model dicot Arabidopsis has been reported to contain 79 nsLTP genes [42]. Studies showed that there are 77 in rice [42], 63 in maize [42], 58 in sorghum [42], 461 in wheat [43], 70 in barley [44], 63 in B. rapa [45], 89 in cabbage [46], 83 in potato [47], 64 in tomato [48], 51 in Gossypium arboretum [49], 47 in G. raimondii [49], 91 in G. hirsutum [49], 52 in sesame [50] and 64 in Arachis duranensis [51]. To date, there has been no systemic whole-genome analysis of the nsLTP gene (BnLTPs) family in B. napus under various abiotic/biotic stresses, as well as in seed development and germination, and seed coat color formation, although the reference genomes of different cultivars or ecotypes of B. napus have been published. Herein, we systemically identified BnLTPs in B. napus, and then performed the detailed analyses containing phylogenetic relationship, gene structure, MEME motifs, chromosome location, gene duplication and synteny, SSR loci and cis-element in promoters. We also detected their expression patterns in various organs and tissues and in response to biotic/abiotic stresses and phytohormone treatments. This study will lay a solid foundation for further functional research of BnLTP genes as well as their application in improvement of crop quality and stress resistance in B. napus.
2. Results
2.1. Identification of BnLTP Genes
A total of 283 nsLTP genes were identified in the B. napus genome (Table S1). Gene sequences of BnLTP genes varied from 273 bp to 8989 bp in length, and BnLTP mRNAs ranged from 92 bp to 1809 bp in length. BnLTP proteins varied from 90 aa to 400 aa in length. BnLTP proteins all contained conserved 8CM domains (Figure 1; Table S1). Of the 283 BnLTP proteins, 76 harbored C-terminal GPI anchor signal, whereas it is absent in the remaining 207 BnLTP proteins. The theoretical MWs of BnLTP proteins ranged from 9658.62 Da to 43,916.10 Da. The pI values of BnLTP proteins varied from 3.92 to 11.00. There existed 18–34 aa N-terminal signal peptides in all BnLTP proteins, and thus they all could be substrates for the cell’s secretory pathways. In addition, for further evolutionary relationship analysis, a total of 127, 151 and 106 nsLTP genes were identified in B. rapa, B. oleracea and A. thaliana, respectively (Tables S2–S4).
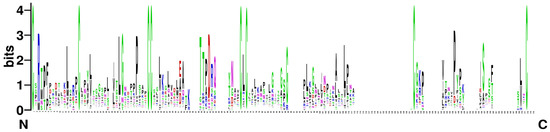
Figure 1.
Sequence logos for the eight-cysteine motif (8CM) of BnLTP proteins. The height of each amino acid residue represents the degree of conservation. The numbers on the x-axis represent the positions in the 8CM. The y-axis shows the information content measured in bits.
2.2. Phylogenetic Relationship Analysis
In total, 667 8CM sequences of nsLTP proteins from B. napus, B. rapa, B. oleracea and A. thaliana (Table S5), were utilized to construct the phylogenetic tree. Evolutionary analysis showed that these 667 nsLTP proteins from four species were separated into seven groups: I, II, III, IV, V, VI and VII (Figure 2). As shown in Table 1, these seven groups all existed in three Brassica species and A. thaliana. In total, there were 170 nsLTP genes from four species in group I, 118 in group II, 68 in group III, 46 in group IV, 98 in group V, 80 in group VI, and 87 in group VII.
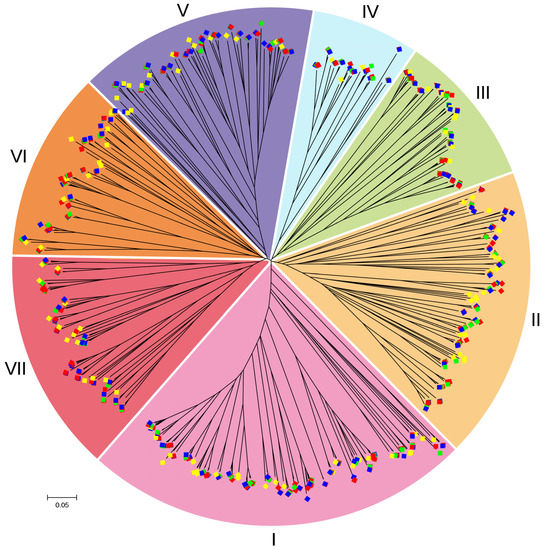
Figure 2.
Phylogenetic relationships of nsLTP proteins among B. napus, B. rapa, B. oleracea and A. thaliana. This tree was generated using the protein sequences of their 8CM domains. BnLTP, BrLTP, BoLTP and AtLTP are shown with the red (Bn), green (Br), blue (Bo) and yellow (At) squares, respectively.

Table 1.
The total number of nsLTP genes in each group of B. napus, B. rapa, B. oleracea and A. thaliana.
2.3. Analysis of Gene Structure and Conserved Motifs
To detect exon-intron organizations of BnLTP genes, their gene structures were illustrated using the GFF3 file from a B. napus genome on the TBtools program (Figure S1). Of 283 BnLTP genes, 143 contained no intron (Table S1). Of the remaining 140 BnLTP genes containing introns, 64 contained 1 intron, 62 had 2 introns, 5 possessed 3 introns, 4 harbored 4 introns, 2 included 5 introns, 2 contained 7 introns, and 1 had 7 introns. To identify conserved motifs in BnLTP protein sequences, MEME tools were used to perform the prediction and analysis. As shown in Figure S1, there existed 20 conserved MEME motifs in 283 BnLTP proteins.
2.4. Chromosome Location
The physical chromosome locations of BnLTP genes were visualized using the TBtools. In total, 239 of 283 BnLTP genes were mapped to 19 chromosomes of B. napus, while the remaining 44 BnLTP genes were mapped to random chromosomes (23 on the A-subgenome, 18 on the C-subgenome, and 3 on chrUnn_random, Figure S2). Notably, these BnLTP genes were unevenly distributed to the chromosomes. Chromosomes A09, C02 and C03 all contained 19 BnLTP genes, chromosomes C01 and C04 both contained 11, chromosomes A01 and A06 both contained 10, chromosomes A05 and C06 both contained 7 and chromosomesA04 and A07 both contained 6. Chromosomes A03, A02, C09, A08, C08 and C07 contained 26, 18, 16, 15, 13, 12 and 9, respectively. In total, there existed 38 clusters containing 92 BnLTPs genes in 15 chromosomes and 4 random chromosomes (Table S6). Chromosome A09 harboring 13 BnLTP genes contained the most clusters (6). Most clusters (27) contained 2 genes, 6 clusters harbored 3 genes and 5 clusters had 4 genes.
2.5. Gene Duplication and Synteny Analysis
To reveal the mechanism underlying the expansion of the nsLTP gene family in B. napus, we detected the types of BnLTP gene duplication. Of 283 BnLTP genes, 191 (67.4%) and 31 (10.9%) genes were derived from segmental duplication and tandem duplication (Table S7). It appears that segmental duplication played important roles in the expansion of the BnLTP gene family. Based on the genome information from B. napus, we analyzed the synteny relationship of BnLTP genes (Figure 3). For the 283 BnLTP genes, 204 syntenic gene pairs were found in B. napus genome in which 40 (19.6%), 23 (11.2%) and 141 (69.1%) gene pairs were obtained among BnA-BnA subgenomes, BnC-BnC subgenomes and BnA-BnC subgenomes, respectively. Overall, the expansion of the BnLTP gene family might have resulted from segmental duplication and hybridization of its two sub-genomes.
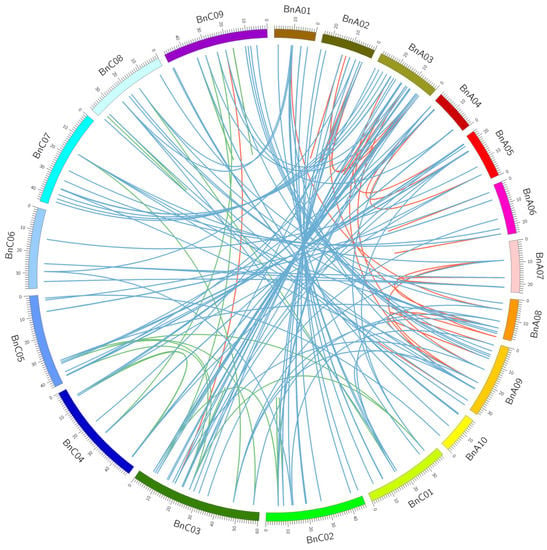
Figure 3.
Genome-wide synteny analysis for nsLTP genes in B. napus. BnA01–10 and BnC01–09 represented chromosomes in A and C sub-genomes in B. napus, respectively. Red, green and blue lines linked the syntenic gene pairs within BnA-BnA, BnC-BnC and BnA-BnC subgenomes of B. napus, respectively.
2.6. Analysis of cis-Regulatory Elements
To identify cis-elements of the promoter regions of BnLTP genes, “Search for CARE” in PlantCARE database using 1.5 kb region upstream of ATG start codon of each BnLTP gene (Table S8) as a query was performed. The results showed that four types of cis-elements were identified in the promoters of BnLTP genes (Table S9). In the first type, plant growth- and development-related cis-elements included sequences associated with differentiation of the palisade mesophyll cells (HD-Zip 1), cell cycle regulation (MSA-like), zein metabolism regulation (O2-site), seed-specific regulation (RY-element), meristem expression (CAT-box), root specific (motif I), circadian control (circadian), endosperm expression (GCN4_motif), endosperm-specific negative expression (AACA_motif) and flavonoid biosynthetic genes regulation (MBSI). In the second type, phytohormone responsive cis-elements included sequences associated with auxin-responsive (TGA-element, AuxRR-core), gibberellin-responsiveness (TATC-box, GARE-motif, P-box), abscisic acid responsiveness (ABRE), MeJA-responsiveness (CGTCA-motif, TGACG-motif) and salicylic acid responsiveness (SARE, TCA-element). In the third type, abiotic stress-related cis-elements included sequences associated with light responsiveness (G-Box, G-box, ACE, 3-AF1 binding site, AAAC-motif, GT1-motif, Sp1, MRE), anoxic specific inducibility (GC-motif), anaerobic induction (ARE), drought-inducibility (MBS), dehydration, low-temp, salt stresses (DRE) and low-temperature responsiveness (LTR). In the fourth type, biotic-stress-related cis-elements included sequences associated with defense and stress responsiveness (TC-rich repeats) and wound responsiveness (WUN-motif).
2.7. Analysis of SSR Loci
MISA-web was used to carry out the analysis of SSR loci. In total, 37 SSR loci were located in BnLTP genes/promoters (Table S10), which were divided into 5 types: mono-(3), di-(23), tri-(4), tetra-(4) and complex-(4) nucleotide repeats. These SSR motifs were mainly harbored in promoters (30), and only a few were found in introns (4), 5′-UTRs (2) and 3′-UTR (1).
2.8. Expression of nsLTPs in Different B. napus Tissues
To examine the expression patterns of BnLTP genes, we analyzed the transcription levels of BnLTP genes in 42 various organs/tissues from different development stages, including roots, stems, leaves, buds, anthocauli, sepals, petals, pistils, stamens, anthers, filaments, seeds, funiculus and silique pods using RNA-seq data (Tables S11 and S12). Of the 283 BnLTP genes, 29 were not expressed at any tissues, whereas the remaining genes were expressed in various tissues with the different levels (Figure 4 and Figure S3), which included organ-specific and constitutive expression patterns. For example, seed-specific genes contained BnLTP017, BnLTP117 and BnLTP210. Of 38 gene clusters, 18 had similar expression patterns among 42 different tissues, and those of the remaining ones were different (Figure S4). Of 74 duplicated gene groups, 50 harbored similar expression patterns across 42 various organs, and 20 had different expression patterns (Figure S5). These results showed that members of some duplicated gene groups (56.8%) or gene clusters (52.6%) with the different expression patterns might have undergone functional divergence.
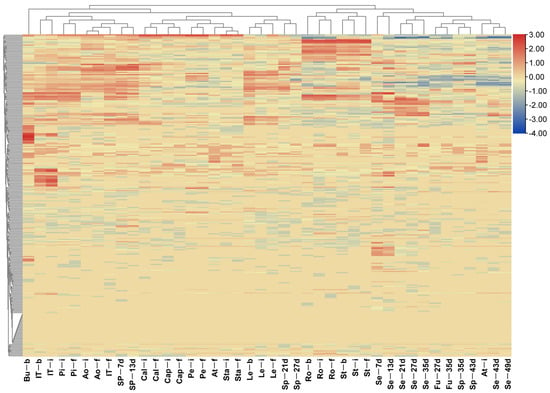
Figure 4.
Expression patterns of BnLTP genes in 42 various tissues and organs. The abbreviations combinations of 42 samples of B. napus cultivar ZS11 are listed in supplementary Table S11. The corresponding heatmap containing BnLTP gene names is shown in supplementary Figure S3.
Totally, transcriptome data of 185 BnLTP genes were obtained for eight LMD-obtained tissues from the early globular seed (Figure S6; Table S13). A total of 50 were expressed in the embryo proper, 90 in the micropylar endosperm, 49 in the peripheral endosperm, 47 in the chalazal endosperm, 36 in the chalazal proliferating tissue, 56 in the chalazal seed coat, 66 in the inner seed coat and 64 in the outer seed coat (FPKM values > 1). In addition, each tissue contained a unique set of BnLTP transcripts. For example, the embryo-specific gene set contained 9 BnLTP genes, and that of micropylar endosperm harbored 34. This result showed that some BnLTP genes might play roles in tissue-specific development processes of the globular seed.
In total, RNA-seq data of 143 BnLTP genes were acquired for three LMD-acquired tissue types of the funiculus (Figure S7; Table S14). A total of 82, 83 and 58 BnLTP genes were expressed in epidermis, cortex and vasculature (FPKM values > 1), respectively, of which 11, 27 and 10 were tissue-specific in corresponding tissues. This result showed the possible crucial roles of BnLTP genes in tissue-specific development processes of the B. napus funiculus.
2.9. Expression of nsLTPs in Response to Biotic Stresses
Plant nsLTPs have been listed as pathogenesis-related protein-14 family [52], and they have been reported to be involved in plant disease resistance reactions [53,54]. Blackleg disease (caused by L. maculans) and white stem rot (caused by S. sclerotiorum) are the most severe diseases of B. napus. We thus detected the expression response of BnLTPs to these two fungal diseases.
In response to S. sclerotiorum, except for BnLTP016, BnLTP031, BnLTP252 and BnLTP265, the transcriptome data of the remaining 279 BnLTP genes were obtained (Figure 5; Table S15), of which 152 were expressed (FPKM values > 1). A total of 72 BnLTP genes were S. sclerotiorum-responsive genes (p < 0.05) of which BnLTP033, BnLTP129, BnLTP161 and BnLTP264, might have more critical roles in resistance to white stem rot. In response to L. maculans, except for BnLTP016, RNA-seq data of the remaining 282 BnLTP genes were obtained (Figure 6; Table S16) of which 174 were expressed (FPKM values > 1). A total of 96 BnLTP genes were L. maculans-responsive genes (p < 0.05) of which BnLTP161 and BnLTP015 might play more important roles in plant defense to blackleg disease.
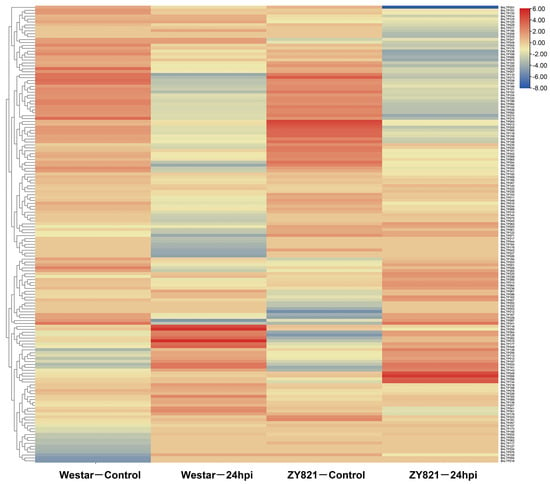
Figure 5.
Expression patterns of BnLTP family genes in the leaves of susceptible (Westar) and tolerant (ZY821) genotypes of rapeseed infected with S. sclerotiorum at 24 h post-inoculation (24 hpi).
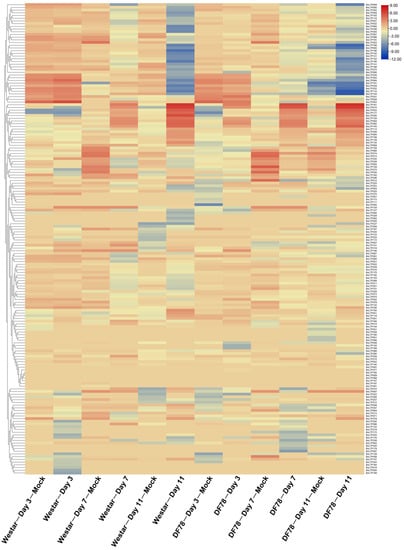
Figure 6.
Expression patterns of BnLTP family genes in the cotyledons of rapeseed resistant (DF78) and susceptible (Westar) lines at 0, 3, 7 and 11 days post-L. maculans inoculation.
2.10. Expression of nsLTPs in Response to Abiotic Stresses
RNA-seq data of 283 BnLTP genes for heat, drought, NaCl, cold and dehydration stresses were all obtained (Figure 7; Tables S17 and S18), and there were 65, 74, 76, 80 and 67 expressed in various samples from the treatments (FPKM values > 1), respectively. We found that the numbers of BnLTP genes (p < 0.05) responding to heat, drought, NaCl, cold and dehydration were 12, 26, 61, 41 and 32, respectively. The responsive mechanisms were different between different low-temperature conditions. Then, we also examined the expression levels of BnLTP genes in two early-maturing B. napus varieties with different cold tolerance under cold accumulations at chilling (CA) and freezing (FA) temperature and cold shocks at the same temperature (CB and FB) condition. A total of 94, 94, 84 and 90 BnLTP genes were expressed under CA, FA, CB and FB treatments, respectively (Figure S8; Table S19); 24, 26, 39 and 48 were CA-, FA-, CB- and FB-responsive genes, respectively.
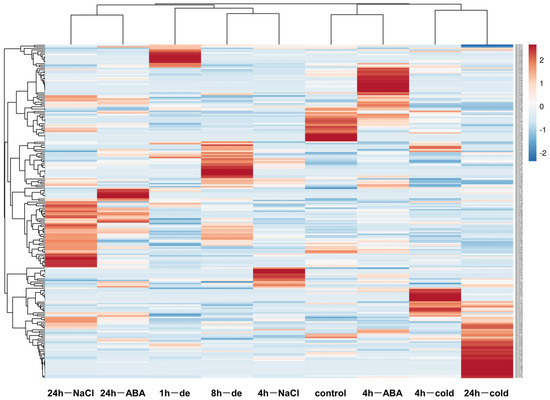
Figure 7.
The heatmap of the expression levels of BnLTP family genes in 3 week-old plants of rapeseed under multiple abiotic stresses (NGDC project ID CRA001775), including dehydration (1 h and 8 h), ABA (25 μM; 4 h and 24 h), NaCl (200 mM; 4 h and 24 h), and cold (4 °C, 4 h and 24 h).
Auxin-responsive elements (TGA-element and AuxRR-core) and abscisic acid-responsive elements (ABRE) were found in the promoters of BnLTP genes, and thus we analyzed their expression patterns under IAA and ABA treatments. Of the 283 BnLTP genes, transcriptome data of 149 and 176 were obtained for IAA and ABA treatment, respectively (Figure 7; Tables S17 and S20). We found that 13 BnLTP genes were IAA-responsive genes, whereas 22 were ABA-responsive genes (p < 0.05).
2.11. Expression of nsLTPs in B. napus Seed Germination
To explore the roles of BnLTP genes in seed germination trait, we examined their expression patterns during different stages of seed germination of three selected accessions with representative germination rates (high, medium or low; C129, C033 and C032, respectively). During 4 time points, 2, 12, 36 and 72 h after imbibition in these 3 accessions, the numbers of expressed BnLTP genes (FPKM values > 1) increased gradually, ranging from 26 to 135 and those of responsive BnLTP genes also increased gradually ranging from 1 to 46 (Figure 8 and Figure S9; Table S21). This result showed that some BnLTP genes might be important in different seed germination processes.
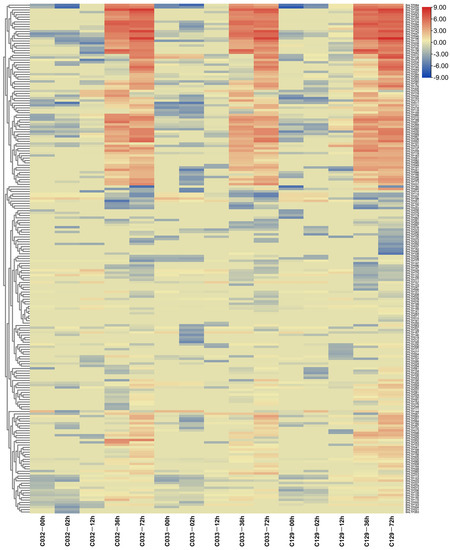
Figure 8.
The heatmap of the expression levels of BnLTP genes at different germination stages of B. napus accessions seeds with representative germination rates (high, medium, or low; C129, C033 and C032, respectively) at 0, 12, 24, 48 and 72 h after imbibition.
2.12. Expression of nsLTPs in B. napus Brown Seeds and Yellow Mutation
Of the 55,637 unigenes identified in 26 d brown seeds and yellow mutation, 64 BnLTP genes were detected (Table S22). Only three BnLTP genes, BnLTP089, BnLTP110 and BnLTP257 were found to be DEGs between the materials, which were all up-regulated in yellow mutation. This result showed that they might play important roles in regulating or co-regulating the seed coat color of B. napus.
2.13. Expression of nsLTPs in B. napus Overexpressing BnTT1 and BnbZIP67
In total, there existed 40, 30 and 32 BnLTP family DEGs in the mid-stage seeds of transgenic rapeseeds overexpressing pBAN::BnTT1 (BOE), pNapA::BnTT1 (NOE) and pNapA::BnbZIP67 (B67O), respectively (Figure 9a; Tables S23–S25). Some 15 BnLTP DEGs were up-regulated in BOE, 11 in NOE, and 16 in B67O, and meanwhile 25 BnLTP DEGs were down-regulated in BOE, 19 in NOE, and 16 in B67O. In addition, we found that two BnLTP DEGs both were up-regulated in BOE and NOE, and nine were all down-regulated in BOE and NOE, while five were up-regulated in NOE, and they were down-regulated in BOE (Figure 9b). These BnLTP genes might be regulated by BnTT1 or BnbZIP67 and play roles in seed development, secondary metabolism, oil accumulation and stress resistance during seed growth and maturation.
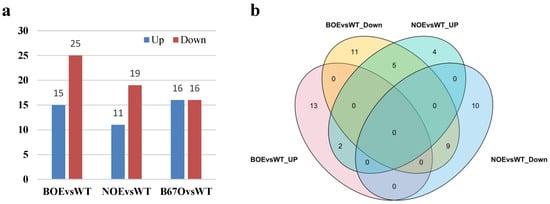
Figure 9.
The numbers of BnLTP family DEGs in transgenic rapeseeds (Westar cultivar) overexpressing pBAN::BnTT1 (BOE, 20 DAP seeds, seed coat-specific promoter), pNapA::BnTT1 (NOE, 20 DAP seeds, seed embryo-specific promoter) and pNapA::BnbZIP67 (B67O, 25 DAP seeds, seed embryo-specific promoter) (a); Venn diagram of the numbers of BnLTP family DEGs in BOE and NOE (b).
2.14. Validation of Selected Sclerotinia-Responsive Genes
To validate the results of RNA-seq expression analysis, two top Sclerotinia-responsive BnLTP genes, BnLTP129 and BnLTP161, were selected to perform the qRT-PCR analysis. Meanwhile, to confirm the availability and feasibility of the plant phenotype imager in the detection of Sclerotinia disease, the chlorophyll fluorescence images of seedling leaves under the condition of Sclerotinia infection were analyzed. At 30 h and 40 h post-infection (hpi), the expression levels of BnLTP129 and BnLTP161 and NPQ values increased compared to that of 20 h post-infection (hpi), while ΦPSII and Fv/Fm values decreased (Figure 10). Under Sclerotinia infection, BnLTP129 and BnLTP161 expression levels both were up-regulated, and the fluorescence parameters varied (Figure 10). This result validated the accuracy of RNA-seq expression data and also showed the application potential of the plant phenotype imager in the detection of Sclerotinia disease in B. napus.
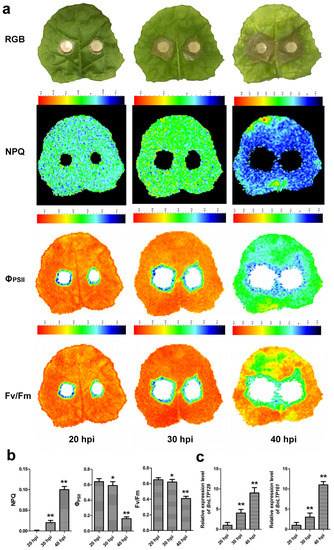
Figure 10.
The impact of S. sclerotiorum infection on photosynthesis of rapeseed leaves (a,b); and the qRT-PCR expression level of two top Sclerotinia-responsive BnLTP genes, BnLTP129 and BnLTP161 (c). Standard images of the RGB, Fv/Fm, ΦPSII and NPQ from S. sclerotiorum-infected rapeseed leaves at 20, 30 and 40 hpi. A false color scale is used for each parameter. The values represent the average ± SD of 3 biological replicates. Fv/Fm, maximum quantum yield of PSII; ΦPSII, effective quantum yield of PSII; NPQ, non-photochemical quenching. * p < 0.05 and ** p < 0.01 compared with samples at 20 hpi.
3. Discussion
The nsLTP proteins belong to the prolamin superfamily with conserved cysteine residues, low molecular mass and a high content of α-helices, which are only found in plants [34,55,56]. The nsLTPs have a hydrophobic cavity that can bind and stabilize various lipid molecules outside membranes [16,17,23,27,57]. An increasing number of studies have shown that plant nsLTP genes play an important role in the response to biotic and abiotic stresses and in plant growth and development [11,18,21,23,25,28,29,31]. To date, with the availability and feasibility of plant reference genomes, the nsLTP gene family has been identified at the genome-wide level in various plant species, including Arabidopsis, Gossypium, sesame, cotton, wheat, potato, tomato, tobacco, barley, maize, cabbage and Chinese cabbage [42,43,44,45,46,47,48,49,50,58,59,60,61]. In this study, 283, 127, 151 and 106 nsLTP genes were identified in B. napus, B. rapa, B. oleracea and A. thaliana genomes, respectively. It appears that the total number of nsLTP genes in allotetraploid B. napus was almost the sum of those in its two diploid parent species, B. oleracea and B. rapa, and three Brassica species contained a higher number of nsLTP genes than that of A. thaliana. This result showed that whole genome triplication of ancestor Brassiceae and allopolyploidization between its two progenitors might have contributed to the expansion of the BnLTP gene family in B. napus. In addition, gene duplication and synteny analysis also further revealed that BnLTP gene expansion was mainly derived from segmental duplication and hybridization of its two sub-genomes, which was also consistent with the previous research results of genome-wide analysis of other gene families in B. napus [62,63].
Numerous studies have showed that some plant nsLTPs were antimicrobial peptides and belonged to the pathogenesis-related protein (PR-14) family members, which possessed extensive antibacterial and antifungal activities [28,29,64]. It has been reported that nsLTP genes played an important role in plant defense to fungal and bacterial pathogens [23,28,29,65]. To date, diverse nsLTPs have been applied in generating plant disease resistance using biotechnological means. For example, overexpression and knockdown of StLTP10 in potato indicated that StLTP10 positively regulated plant resistance to Phytophthora infestans [66]. Cotton GhnsLTPsA10 positively regulated Verticillium wilt and Fusarium wilt resistance in transgenic Arabidopsis and VIGS cotton [11]. Overexpression of AtLTP4.4 in transgenic wheat significantly inhibited the growth of F. graminearum in Bobwhite and RB07 lines in the greenhouse and reduced the size of fungal lesions in in vitro leaf assays [67]. Transgenic Arabidopsis expressing wheat TdLTP4 gene enhanced fungal resistance against Alternaria solani and Botrytis cinerea [68]. Transgenic wheat lines overexpressing TaLTP5 exhibited significantly enhanced resistance to both common root rot (Cochliobolus sativus) and Fusarium head blight (Fusarium graminearum) compared to the untransformed control [69]. Overexpression of the pepper CALTPI gene in Arabidopsis enhanced the resistance against infection by Pseudomonas syringae pv. tomato and Botrytis cinerea [70]. Overexpression of OsLTP1 gene in B. napus enhanced its resistance to S. sclerotiorum [71]. Overexpression of an nsLTPs-like antimicrobial protein gene (LJAMP2) from motherwort (Leonurus japonicus) enhanced resistance to S. sclerotiorum in B. napus, whereas its overexpression in Populus tomentosa also enhanced resistance to fungal pathogens [65,72]. In this study, in response to two fungal pathogens, we identified 72 S. sclerotiorum-responsive BnLTP genes and 96 L. maculans-responsive BnLTP genes. For white stem rot resistance, BnLTP033, BnLTP129, BnLTP161 and BnLTP264, had more critical roles. For blackleg disease resistance, BnLTP161 and BnLTP015 played more roles. Therefore, these BnLTPs could be candidates used to generate the wide broad plant disease resistance.
Many studies have showed that members of the plant nsLTP family played important roles in response to abiotic stress as well as plant hormone signals [68,73,74], which could help plants adapt to changes of environment conditions. For example, a lipid transfer protein variant with a mutant eight-cysteine motif causes photoperiod- and thermo-sensitive dwarfsm in rice [25]. The transgenic plants expressing the pepper CALTPI gene indicated high levels of tolerance to NaCl and drought stresses at various vegetative growth stages [70]. Overexpression of wheat TdLTP4 in Arabidopsis contributed to plant growth under various stress conditions including NaCl, ABA, JA and H2O2 treatments [68]. Previous studies showed that OsLTPL159 was involved in cold tolerance at the early seedling stage in rice, which could be a candidate allele used to improve rice cold tolerance [30]. Overexpression of LTP3 in Arabidopsis conferred enhanced freezing tolerance, whereas loss-of-function and overexpression of LTP3 in Arabidopsis both showed its positive regulatory roles in drought tolerance [75]. Drought-Induced LTP (OsDIL) was mainly responsive to abiotic stresses, including drought, cold, NaCl, and stress-related plant hormone ABA [73]. Transgenic rice plants overexpressing OsDIL were more tolerant to drought stress during vegetative development than wild type. The overexpression of NtLTP4 in Nicotiana tabacum increased the resistance to salt and drought stresses [38]. Here, of the 283 BnLTP genes, 12 were heat-responsive genes, 26 were drought-responsive genes, 61 were NaCl-responsive genes, 41 were cold-responsive genes, 32 were dehydration-responsive genes, 13 were IAA-responsive genes and 22 were ABA-responsive genes. It could be speculated that these BnLTP members played critical roles in response to various abiotic stressors, which also might be applied in molecular breeding of high abiotic stress resistance in B. napus.
Numerous studies have showed that plant nsLTPs played diverse roles in various biological processes, including seed development and maturation as well as seed germinations [21,22,23,50]. Overexpression of SiLTPI.23 in Arabidopsis significantly increased seed oil contents by 17–29% compared with the wild type control [50]. OsLTPL36 was reported to play an essential role in seed development and germination in rice [22]. In A. thaliana, the GPI-anchored lipid transfer proteins acted important roles in the development of seed coats and pollen [76]. In this study, we showed that three BnLTP genes, BnLTP089, BnLTP110 and BnLTP257, were DEGs between 26 d brown seeds and yellow mutants. In yellow mutants, their expression levels were all up-regulated, indicating potential roles of these three BnLTP genes in regulating seed coat color. In addition, based on RNA-seq data of 42 various organs/tissues, we found that several BnLTP genes, including BnLTP059 and BnLTP085, displayed seed-specific expression genes, revealing their possible roles in regulating seed development. Each of eight LMD-obtained tissues from the early globular seed possessed a unique set of BnLTP transcripts, which also might show their important roles in tissue-specific development processes of the globular seed. In the seed germination process of three selected accessions, numbers of expressed and responsive BnLTP genes both gradually increased. Meanwhile, overall, at four time points, high germination rate accession material had more expressed and responsive BnLTP genes. This result showed that the expression of these BnLTP genes might result in high germination rate.
In addition, there existed 30–40 BnLTP DEGs in BOE, NOE and B67O, each of which contained the up-regulated and down-regulated BnLTP genes. TT1 and bZIP67 are two critical TFs that regulate seed coat and seed embryo development and metabolism, respectively [32,33]. These BnLTP DEGs might play important roles in the development, metabolism or adaptability of the seed coat and embryo in rapeseed by responding to BnTT1 and BnbZIP67. Meanwhile, it found that the BnLTP family DEGs might have undergone obvious divergence in regulating physiological functions such as seed development, metabolism and adaptation.
Many studies reported that nsLTPs were defined as small basic proteins [16,17,21,23], but some of the nsLTPs identified in this study, were 400 aa in length, which were atypical nsLTP proteins. This study is not the first attempt to classify nsLTPs. Other researchers attempting a systematic analysis of nsLTPs also found these atypical proteins. For example, some of the nsLTPs in A. duranensis were 244 aa in length [51], and some in cabbage were 258 aa in length [46]. Therefore, nsLTPs need to be redefined after the functions of these atypical nsLTPs were validated using biochemistry and molecular biology experiments, and we also believe that the nsLTP community will eventually resolve this conflict in that time. Additionally, it has been shown that some nsLTPs can bind or transfer lipids [13,14,15], and thus this function of BnLTPs will be studied in our future research program.
4. Materials and Methods
4.1. Sequence Retrieval and Structural Analysis
To identify BnLTP genes, the HMM files of nsLTP domain (PF14368 and PF00234) were downloaded from Pfam database (http://Pfam.sanger.ac.uk/; accessed on 12 March 2020), and then the hmmsearch program (e-value 10e−5; http://hmmer.org; accessed on 12 March 2020) was used to identify BnLTP proteins against B. napus genome v5.0 (http://www.genoscope.cns.fr/brassicanapus/; accessed on 12 December 2019). All candidates were detected using SUPERFAMILY 2 database (https://beta.supfam.org/; accessed on 12 March 2020), sequences examined as non-nsLTP proteins, including proteinase/alpha-amylase inhibitor and seed storage family, 2S albumin, were deleted. The 8CM motifs were manually checked in the remaining sequences, and N-terminal signal peptide was also detected using SignalP-5.0 (http://www.cbs.dtu.dk/services/SignalP/; accessed on 13 March 2020). As results, putative BnLTPs lacking 8CM motifs or N-terminal signal peptide were also excluded. Subsequently, PredGPI (http://gpcr2.biocomp.unibo.it/predgpi/; accessed on 25 March 2020) and TargetP-2.0 (http://www.cbs.dtu.dk/services/TargetP/; accessed on 13 March 2020) were used to predict the presence of C-terminal GPI anchor signal and subcellar location of BnLTP proteins, respectively. To further perform the comparative phylogenetic analysis, AtLTP, BrLTP and BoLTP genes were also identified against Arabidopsis genome v11 (https://www.arabidopsis.org/; accessed on 12 May 2020), B. rapa genome v3.0 (http://brassicadb.cn/; accessed on 29 April 2020), and B. oleracea genome (http://plants.ensembl.org/; accessed on 12 May 2020) using the above method, respectively. The nsLTPs in these four species were named according to their chromosome position. The ProtParam tool (https://web.expasy.org/protparam/; accessed on 15 April 2020) were used to compute the theoretical isoelectric point (pI) and molecular weight (MW) of BnLTPs.
4.2. Phylogenetic Relationship Analysis
The nsLTP protein sequences from B. napus, B. rapa, B. oleracea and A. thaliana were multi-aligned using the MAFFT7 program with default parameters [77], and then the neighbor-joining (NJ) phylogenetic tree was generated using MEGA7 software [78] with the 1000 bootstrap replicates and p-distance model. Sequence logo of 8CM motifs of BnLTP family proteins was analyzed using WebLogo (http://weblogo.threeplusone.com/; accessed on 16 June 2020).
4.3. Analysis of Gene Structure and Conserved Motifs
Gene exon–intron structure information of BnLTP genes was retrieved from the GFF3 file in the B. napus genome v5.0, and conserved motif files of BnLTP proteins were generated on MEME suite 5.1.0 (http://meme-suite.org/tools/meme; accessed on 18 June 2020) with default parameters. Finally, the combined image was visualized with TBtools [79].
4.4. Chromosome Location, Gene Duplication and Synteny Analysis
The chromosome location information of BnLTP genes was obtained from the GFF3 file in B. napus genome v5.0, and the image was visualized with the TBtools [79]. BnLTP gene clusters were identified if two or more BnLTP genes were clustered together on the chromosome, and meanwhile they were separated by no more than three genes. Gene duplication patterns were classified with MCScanX with default parameters. Duplicate gene pairs were determined if the identity of their coding regions was >85%, and they were clustered together in an evolutionary guide tree generated using Clustal Omega tool (https://www.ebi.ac.uk/Tools/msa/clustalo/; accessed on 9 June 2020), which were also annotated as duplicated gene groups. The syntenic relationships of duplicate gene pairs were visualized using Circos (http://circos.ca/software/; accessed on 29 July 2021).
4.5. Promoter cis-Element Analysis
The 1.5 kb regions upstream of BnLTP coding sequences were obtained from the B. napus genome v5.0, and then cis-regulatory elements were predicted on PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; accessed on 18 January 2021).
4.6. Analysis of SSR Loci
SSR loci in BnLTP genes or their promoters were checked on MISA-web [80], with default parameters.
4.7. Expression Analyses Based on Transcriptome Data
Transcriptome data of B. napus in 42 different tissues/organs (project ID PRJNA358784), 9 laser-microdissected tissues from the early globular seed (GSE120360) and 3 laser-microdissected tissues from the funiculus (GSE71859), and in response to two fungal pathogens (Sclerotinia sclerotiorum, 24 hpi, GSE81545; Leptosphaeria maculans, 0, 3, 7 and 11 days post inoculation, GSE777230), heat (40 °C, 3 h) and drought (withdrawing water, 3 days) (GSE156029), cold (4 °C, 12h) and freezing (−4 °C, 12h) (GSE129220), and IAA treatment (GSE105889; 1 μM IAA, 3 h) as well as in brown seeds and yellow mutation (GSE69137) and seed germination progress (GSE137230; 0, 12, 24, 48, and 72 h after imbibition) were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/; accessed on 17 July 2021). Transcriptome data of B. napus under dehydration (1 h and 8 h), salt (200 mM NaCl; 4 h and 24 h), ABA (25 uM ABA; 4 h and 24 h) and cold (4 °C, 4 h and 24 h) treatments were downloaded in NGDC (https://ngdc.cncb.ac.cn/; accessed on 23 July 2021; project ID CRA001775). For two or three biological duplicates of each sample, all analyses were conducted using the average FPKM or TPM values of transcripts. BnLTP genes were considered as differentially expressed genes or stress-responsive genes if |log2(foldchange)| of transcript values of gene expression were > 2 and the FPKM/TPM values > 1. Expression analyses were carried out using DESeq2 R package, and heatmaps were visualized using ClustVis web tool (https://biit.cs.ut.ee/clustvis/; accessed on 23 July 2021) and TBtools [79]. Our group has obtained transgenic rapeseeds overexpressing BnTT1 and BnbZIP67 and performed RNA-seq analysis. In this study, their DEGs database was used to analyze the expression patterns of BnLTP family genes.
4.8. RNA Extraction and qRT-PCR Analysis
Total RNA was isolated from seedling leaves of a Sclerotinia-resistant rapeseed cultivar ZS10 at 20 h, 30 h and 40 h after S. sclerotiorum (1980) inoculation, respectively, using an RNAsimple Total RNA Kit (DP419, Tiangen, Beijing, China). First-strand total cDNA was synthesized with 1 μg of total RNA using the PrimeScript Reagent Kit with gDNA Eraser (Takara, Dalian, China). The qRT-PCR assay was performed using FastStart Universal SYBR Green Master (Roche, Mannheim, Germany) on BIO-RAD CFX96 Real-time PCR System (Bio-Rad, Irvine, CA, USA). Three replicates were carried out for each reaction. Two top Sclerotinia-responsive BnLTP genes, BnLTP129 and BnLTP161, Table S26 were selected to perform the qRT-PCR experiment, and the B. napus ACT7 gene was used as internal control [81].
4.9. Chlorophyll Fluorescence Imaging
The plant phenotype imager (device no. 20A00005; a chlorophyll fluorescence imaging system FluorCam7.0; Photon Systems Instruments, Brno, Czech Republic) was used to analyze the fluorescence parameters (Fv/Fm, NPQ and ΦPSII) and chlorophyll fluorescence images of the rapeseed seedling leaves under 20 h, 30 h and 40 h post Sclerotinia infection [82].
5. Conclusions
In the present study, we performed comprehensive whole-genome mining and analysis of BnLTP family genes in rapeseed, including phylogenetic relationship, exon/intron structure, MEME motifs, synteny analysis, SSR loci, promoter cis-elements and expression patterns. Our work showed critical roles of the BnLTP family genes in plant growth and development, as well as in response to biotic/abiotic stresses and phytohormone treatments. This study will lay a solid foundation for further functional research of BnLTP genes in quality improvement and stress resistance.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23158372/s1.
Author Contributions
Conceptualization, Y.X. and Y.C.; data curation, Y.X. and L.L.; formal analysis, Y.X. and C.Z.; funding acquisition, Y.C.; investigation, Y.X., C.Z., X.L., L.L. and R.S.; methodology, X.L. and H.J.; project administration, Y.C.; resources, Y.X., A.T.I., H.J. and R.S.; software, Y.X., A.T.I. and R.S.; validation, C.Z.; writing—original draft, Y.X. and Y.C.; writing—review and editing, Y.C. and Y.X. undertook the use and maintenance of plant phenotype imager. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31871549; 32001441), Fundamental Research Funds for the Central Universities (XDJK2020C038), Young Eagles Program of Chongqing Municipal Commission of Education (CY220219), Chongqing Municipal and Southwest University Training Program of Innovation and Entrepreneurship for Undergraduates (S202110635162, X202110635094, X202210635398 and X202210635185).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
References
- Woodfield, H.K.; Sturtevant, D.; Borisjuk, L.; Munz, E.; Guschina, I.A.; Chapman, K.; Harwood, J.L. Spatial and temporal mapping of key lipid species in Brassica napus seeds. Plant Physiol. 2017, 173, 1998–2009. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Zeng, X.; Xiong, Q.; Wei, D.; Liao, J.; Xu, Y.; Chen, G.; Zhou, Y.; Dong, H.; Wan, H.; et al. Combining quantitative trait locus and co-expression analysis allowed identification of new candidates for oil accumulation in rapeseed. J. Exp. Bot. 2021, 72, 1649–1660. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Yu, P.; Tian, X.; Guo, L.; Tu, J.; Shen, J.; Yi, B.; Fu, T.; Wen, J.; Liu, K.; et al. DELLA proteins BnaA6.RGA and BnaC7.RGA negatively regulate fatty acid biosynthesis by interacting with BnaLEC1s in Brassica napus. Plant Biotechnol. J. 2021, 19, 2011–2026. [Google Scholar] [CrossRef]
- Fitt, B.D.L.; Brun, H.; Barbetti, M.J.; Rimmer, S.R. World-wide importance of phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus). Eur. J. Plant Pathol. 2006, 114, 3–15. [Google Scholar] [CrossRef]
- del Rio, L.E.; Bradley, C.A.; Henson, R.A.; Endres, G.J.; Hanson, B.K.; McKay, K.; Halvorson, M.; Porter, P.M.; le Gare, D.G.; Lamey, H.A. Impact of Sclerotinia stem rot on yield of canola. Plant Dis. 2007, 91, 191–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, S.; Dunker, S.; Kleinhenz, B.; Rohrig, M.; von Tiedemann, A. Crop loss-related forecasting model for Sclerotinia stem rot in winter oilseed rape. Phytopathology 2007, 97, 1186–1194. [Google Scholar] [CrossRef] [Green Version]
- Raza, A. Eco-physiological and biochemical responses of rapeseed (Brassica napus L.) to abiotic stresses: Consequences and mitigation strategies. J. Plant Growth Regul. 2021, 40, 1368–1388. [Google Scholar] [CrossRef]
- Liu, S.; Fan, C.; Li, J.; Cai, G.; Yang, Q.; Wu, J.; Yi, X.; Zhang, C.; Zhou, Y. A genome-wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor. Appl. Genet. 2016, 129, 1203–1215. [Google Scholar] [CrossRef]
- Wei, L.; Jian, H.; Lu, K.; Filardo, F.; Yin, N.; Liu, L.; Qu, C.; Li, W.; Du, H.; Li, J. Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol. J. 2016, 14, 1368–1380. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Z.; Zhang, C.; Tang, F.; Yang, B.; Zhang, L.; Liu, J.; Huo, Q.; Wang, S.; Li, S.; Wei, L.; et al. Identification of candidate genes controlling oil content by combination of genome-wide association and transcriptome analysis in the oilseed crop Brassica napus. Biotechnol. Biofuels 2019, 12, 216. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, Y.; Sun, Z.; Liu, Z.; Zhang, D.; Yang, J.; Wang, G.; Wu, J.; Ke, H.; Meng, C.; et al. Tissue-specific expression of GhnsLTPs identified via GWAS sophisticatedly coordinates disease and insect resistance by regulating metabolic flux redirection in cotton. Plant J. 2021, 107, 831–846. [Google Scholar] [CrossRef]
- Tang, S.; Zhao, H.; Lu, S.P.; Yu, L.Q.; Zhang, G.F.; Zhang, Y.T.; Yang, Q.Y.; Zhou, Y.M.; Wang, X.M.; Ma, W.; et al. Genome- and transcriptome-wide association studies provide insights into the genetic basis of natural variation of seed oil content in Brassica napus. Mol. Plant 2021, 14, 470–487. [Google Scholar] [CrossRef]
- Yamada, M. Lipid transfer proteins in plants and microorganisms. Plant Cell Physiol. 1992, 33, 1–6. [Google Scholar]
- Kader, J.C. Lipid-transfer proteins: A puzzling family of plant proteins. Trends Plant Sci. 1997, 2, 66–70. [Google Scholar] [CrossRef]
- Han, G.W.; Lee, J.Y.; Song, H.K.; Chang, C.S.; Min, K.; Moon, J.; Shin, D.H.; Kopka, M.L.; Sawaya, M.R.; Yuan, H.S.; et al. Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography. J. Mol. Biol. 2001, 308, 263–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kader, J.C. Lipid-transfer proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 627–654. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, A.d.O.; Gomes, V.M. Role of plant lipid transfer proteins in plant cell physiology—A concise review. Peptides 2007, 28, 1144–1153. [Google Scholar] [CrossRef]
- Chae, K.; Gonong, B.J.; Kim, S.-C.; Kieslich, C.A.; Morikis, D.; Balasubramanian, S.; Lord, E.M. A multifaceted study of stigma/style cysteine-rich adhesin (SCA)-like Arabidopsis lipid transfer proteins (LTPs) suggests diversified roles for these LTPs in plant growth and reproduction. J. Exp. Bot. 2010, 61, 4277–4290. [Google Scholar] [CrossRef]
- Zhang, D.; Liang, W.; Yin, C.; Zong, J.; Gu, F.; Zhang, D. OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 2010, 154, 149–162. [Google Scholar] [CrossRef] [Green Version]
- Edstam, M.M.; Blomqvist, K.; Eklof, A.; Wennergren, U.; Edqvist, J. Coexpression patterns indicate that GPI-anchored non-specific lipid transfer proteins are involved in accumulation of cuticular wax, suberin and sporopollenin. Plant Mol. Biol. 2013, 83, 625–649. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, X.; Lu, C.; Zeng, X.; Li, Y.; Fu, D.; Wu, G. Non-specific lipid transfer proteins in plants: Presenting new advances and an integrated functional analysis. J. Exp. Bot. 2015, 66, 5663–5681. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhou, W.; Lu, Z.; Ouyang, Y.; Su, C.O.; Yao, J. A lipid transfer protein, OsLTPL36, is essential for seed development and seed quality in rice. Plant Sci. 2015, 239, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Salminen, T.A.; Blomqvist, K.; Edqvist, J. Lipid transfer proteins: Classification, nomenclature, structure, and function. Planta 2016, 244, 971–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacq, A.; Pernot, C.; Martinez, Y.; Domergue, F.; Payre, B.; Jamet, E.; Burlat, V.; Pacquit, V.B. The Arabidopsis lipid transfer protein 2 (AtLTP2) is involved in cuticle-cell wall interface integrity and in etiolated hypocotyl permeability. Front. Plant Sci. 2017, 8, 263. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Li, R.; Xu, Y.; Mao, R.; Chen, S.; Chen, L.; Chen, L.; Liu, Y.-G.; Chen, Y. A lipid transfer protein variant with a mutant eight-cysteine motif causes photoperiod- and thermo-sensitive dwarfism in rice. J. Exp. Bot. 2020, 71, 1294–1305. [Google Scholar] [CrossRef]
- Cameron, K.D.; Teece, M.A.; Smart, L.B. Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 2006, 140, 176–183. [Google Scholar] [CrossRef] [Green Version]
- Yeats, T.H.; Rose, J.K.C. The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs). Protein Sci. 2008, 17, 191–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkina, E.I.; Melnikova, D.N.; Bogdanov, I.V.; Ovchinnikova, T.V. Lipid transfer proteins as components of the plant innate immune system: Structure, functions, and applications. Acta Nat. 2016, 8, 47–61. [Google Scholar] [CrossRef]
- Das, K.; Datta, K.; Karmakar, S.; Datta, S.K. Antimicrobial peptides—Small but mighty weapons for plants to fight phytopathogens. Protein Pept. Lett. 2019, 26, 720–742. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, S.; Qin, J.; Sun, C.; Liu, F. The lipid transfer protein OsLTPL159 is involved in cold tolerance at the early seedling stage in rice. Plant Biotechnol. J. 2020, 18, 756–769. [Google Scholar] [CrossRef]
- Balmant, K.M.; Ii, S.R.L.; Duong, B.V.; Zhu, F.; Zhu, N.; Nicklay, J.; Chen, S. Guard cell redox proteomics reveals a role of lipid transfer protein in plant defense. J. Proteom. 2021, 242, 104247. [Google Scholar] [CrossRef] [PubMed]
- Appelhagen, I.; Lu, G.H.; Huep, G.; Schmelzer, E.; Weisshaar, B.; Sagasser, M. TRANSPARENT TESTA1 interacts with R2R3-MYB factors and affects early and late steps of flavonoid biosynthesis in the endothelium of Arabidopsis thaliana seeds. Plant J. 2011, 67, 406–419. [Google Scholar] [CrossRef] [PubMed]
- Mendes, A.; Kelly, A.A.; van Erp, H.; Shaw, E.; Powers, S.J.; Kurup, S.; Eastmond, P.J. bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating fatty acid desaturase 3. Plant Cell 2013, 25, 3104–3116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edstam, M.M.; Viitanen, L.; Salminen, T.A.; Edqvist, J. Evolutionary history of the non-specific lipid transfer proteins. Mol. Plant 2011, 4, 947–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeBono, A.; Yeats, T.H.; Rose, J.K.C.; Bird, D.; Jetter, R.; Kunst, L.; Samuelsa, L. Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 2009, 21, 1230–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edstam, M.M.; Laurila, M.; Hoglund, A.; Raman, A.; Dahlstrom, K.M.; Salminen, T.A.; Edqvist, J.; Blomqvist, K. Characterization of the GPI-anchored lipid transfer proteins in the moss Physcomitrella patens. Plant Physiol. Biochem. 2014, 75, 55–69. [Google Scholar] [CrossRef]
- Kim, H.; Lee, S.B.; Kim, H.J.; Min, M.K.; Hwang, I.; Suh, M.C. Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 1391–1403. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zheng, X.; Song, Y.; Zhu, L.; Yu, Z.; Gan, L.; Zhou, S.; Liu, H.; Wen, F.; Zhu, C. NtLTP4, a lipid transfer protein that enhances salt and drought stresses tolerance in Nicotiana tabacum. Sci. Rep. 2018, 8, 8873. [Google Scholar] [CrossRef] [Green Version]
- Park, C.J.; Shin, R.; Park, J.M.; Lee, G.J.; Youl, J.S.; Paek, K.H. Induction of pepper cDNA encoding a lipid transfer protein during the resistance response to tobacco mosaic virus. Plant Mol. Biol. 2002, 48, 243–254. [Google Scholar] [CrossRef]
- Pan, Y.; Li, J.; Jiao, L.; Li, C.; Zhu, D.; Yu, J. A non-specific setaria italica lipid transfer protein gene plays a critical role under abiotic stress. Front. Plant Sci. 2016, 7, 1752. [Google Scholar] [CrossRef] [Green Version]
- Jang, C.S.; Yim, W.C.; Moon, J.-C.; Jung, J.H.; Lee, T.G.; Lim, S.D.; Cho, S.H.; Lee, K.K.; Kim, W.; Seo, Y.W.; et al. Evolution of non-specific lipid transfer protein (nsLTP) genes in the Poaceae family: Their duplication and diversity. Mol. Genet. Genom. 2008, 279, 481–497. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Zhong, X. Non-specific lipid transfer proteins in maize. BMC Plant Biol. 2014, 14, 281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kouidri, A.; Whitford, R.; Suchecki, R.; Kalashyan, E.; Baumann, U. Genome-wide identification and analysis of non-specific lipid transfer proteins in hexaploid wheat. Sci. Rep. 2018, 8, 17087. [Google Scholar] [CrossRef]
- Zhang, M.; Kim, Y.; Zong, J.; Lin, H.; Dievart, A.; Li, H.; Zhang, D.; Liang, W. Genome-wide analysis of the barley non-specific lipid transfer protein gene family. Crop J. 2019, 7, 65–76. [Google Scholar] [CrossRef]
- Li, J.; Gao, G.; Xu, K.; Chen, B.; Yan, G.; Li, F.; Qiao, J.; Zhang, T.; Wu, X. Genome-wide survey and expression analysis of the putative non-specific lipid transfer proteins in Brassica rapa L. PLoS ONE 2014, 9, e84556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, J.; Lv, H.; Yang, L.; Fang, Z.; Zhuang, M.; Zhang, Y.; Liu, Y.; Li, Z. Genome-wide identification and characterization of non-specific lipid transfer proteins in cabbage. PeerJ 2018, 6, e5379. [Google Scholar] [CrossRef]
- Li, G.; Hou, M.; Liu, Y.; Pei, Y.; Ye, M.; Zhou, Y.; Huang, C.; Zhao, Y.; Ma, H. Genome-wide identification, characterization and expression analysis of the non-specific lipid transfer proteins in potato. BMC Genom. 2019, 20, 375. [Google Scholar] [CrossRef]
- D’Agostino, N.; Buonanno, M.; Ayoub, J.; Barone, A.; Monti, S.M.; Rigano, M.M. Identification of non-specific lipid transfer protein gene family members in Solanum lycopersicum and insights into the features of Sola l 3 protein. Sci. Rep. 2019, 9, 1607. [Google Scholar] [CrossRef]
- Li, F.; Fan, K.; Ma, F.; Yue, E.; Bibi, N.; Wang, M.; Shen, H.; Hasan, M.M.-U.; Wang, X. Genomic identification and comparative expansion analysis of the non-specific lipid transfer protein gene family in Gossypium. Sci. Rep. 2016, 6, 38948. [Google Scholar] [CrossRef]
- Song, S.; You, J.; Shi, L.; Sheng, C.; Zhou, W.; Dossou, S.S.K.; Dossa, K.; Wang, L.; Zhang, X. Genome-wide analysis of nsLTP gene family and identification of SiLTPs contributing to high oil accumulation in sesame (Sesamum indicum L.). Int. J. Mol. Sci. 2021, 22, 5291. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Li, E.; Song, H.; Du, G.; Li, S.; Zhu, H.; Chen, G.; Zhao, C.; Qiao, L.; Wang, J.; et al. Genome-wide identification and characterization of nonspecific lipid transfer protein (nsLTP) genes in Arachis duranensis. Genome 2020, 112, 4332–4341. [Google Scholar] [CrossRef] [PubMed]
- van Loon, L.C.; van Strien, E.A. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef]
- Zribi, I.; Ghorbel, M.; Brini, F. Pathogenesis related proteins (PRs): From cellular mechanisms to plant defense. Curr. Protein Pept. Sci. 2021, 22, 396–412. [Google Scholar] [CrossRef] [PubMed]
- Fahlberg, P.; Buhot, N.; Johansson, O.N.; Andersson, M.X. Involvement of lipid transfer proteins in resistance against a non-host powdery mildew in Arabidopsis thaliana. Mol. Plant Pathol. 2019, 20, 69–77. [Google Scholar] [CrossRef] [Green Version]
- Kreis, M.; Forde, B.G.; Rahman, S.; Miflin, B.J.; Shewry, P.R. Molecular evolution of the seed storage proteins of barley, rye and wheat. J. Mol. Biol. 1985, 183, 499–502. [Google Scholar] [CrossRef]
- Shewry, P.R.; Beaudoin, F.; Jenkins, J.; Griffiths-Jones, S.; Mills, E.N. Plant protein families and their relationships to food allergy. Biochem. Soc. Trans. 2002, 30, 906–910. [Google Scholar]
- Edqvist, J.; Blomqvist, K.; Nieuwland, J.; Salminen, T.A. Plant lipid transfer proteins: Are we finally closing in on the roles of these enigmatic proteins? J. Lipid Res. 2018, 59, 1374–1382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Li, P.; Liu, C.; Wang, P.; Cao, P.; Ye, X.; Li, Q. Systematic analysis of the non-specific lipid transfer protein gene family in Nicotiana tabacum reveal its potential roles in stress responses. Plant Physiol. Biochem. 2022, 172, 33–47. [Google Scholar] [CrossRef]
- Boutrot, F.; Chantret, N.; Gautier, M.-F. Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genom. 2008, 9, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odintsova, T.I.; Slezina, M.P.; Istomina, E.A.; Korostyleva, T.V.; Kovtun, A.S.; Kasianov, A.S.; Shcherbakova, L.A.; Kudryavtsev, A.M. Non-specific lipid transfer proteins in Triticum kiharae Dorof. et Migush.: Identification, characterization and expression profiling in response to pathogens and resistance inducers. Pathogens 2019, 8, 221. [Google Scholar] [CrossRef] [Green Version]
- Duo, J.; Xiong, H.; Wu, X.; Li, Y.; Si, J.; Zhang, C.; Duan, R. Genome-wide identification and expression profile under abiotic stress of the barley non-specific lipid transfer protein gene family and its qingke orthologues. BMC Genom. 2021, 22, 674. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Chen, B.; Wang, R.; Win, A.N.; Li, J.; Chai, Y. Genome-wide survey and characterization of fatty acid desaturase gene family in Brassica napus and its parental species. Appl. Biochem. Biotech. 2018, 184, 582–598. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Jiang, J.; Yang, X.; Jiang, H.; Du, Y.; Liu, X.; Xie, R.; Chai, Y. Genome-wide mining and comparative analysis of fatty acid elongase gene family in Brassica napus and its progenitors. Gene 2020, 747, 144674. [Google Scholar] [CrossRef]
- Blein, J.P.; Coutos-Thevenot, P.; Marion, D.; Ponchet, M. From elicitins to lipid-transfer proteins: A new insight in cell signalling involved in plant defence mechanisms. Trends Plant Sci. 2002, 7, 293–296. [Google Scholar] [CrossRef]
- Jia, Z.; Gou, J.; Sun, Y.; Yuan, L.; Tang, Q.; Yang, X.; Pei, Y.; Luo, K. Enhanced resistance to fungal pathogens in transgenic Populus tomentosa Carr. by overexpression of an nsLTP-like antimicrobial protein gene from motherwort (Leonurus japonicus). Tree Physiol. 2010, 30, 1599–1605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Gao, H.; Chu, Z.; Ji, C.; Xu, Y.; Cao, W.; Zhou, S.; Song, Y.; Liu, H.; Zhu, C. A nonspecific lipid transfer protein, StLTP10, mediates resistance to Phytophthora infestans in potato. Mol. Plant Pathol. 2021, 22, 48–63. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, J.E.; Darwish, N.I.; Garcia-Sanchez, J.; Tyagi, N.; Trick, H.N.; McCormick, S.; Dill-Macky, R.; Tumer, N.E. A lipid transfer protein has antifungal and antioxidant activity and suppresses Fusarium head blight disease and DON accumulation in transgenic wheat. Phytopathology 2021, 111, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Safi, H.; Saibi, W.; Alaoui, M.M.; Hmyene, A.; Masmoudi, K.; Hanin, M.; Brini, F. A wheat lipid transfer protein (TdLTP4) promotes tolerance to abiotic and biotic stress in Arabidopsis thaliana. Plant Physiol. Biochem. 2015, 89, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, Z.; Xu, H.; Zhou, M.; Du, L.; Zhang, Z. Overexpression of wheat lipid transfer protein gene TaLTP5 increases resistances to Cochliobolus sativus and Fusarium graminearum in transgenic wheat. Funct. Integr. Genom. 2012, 12, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.W.; Kim, K.D.; Hwang, B.K. Identification of pathogen-responsive regions in the promoter of a pepper lipid transfer protein gene (CALTPI) and the enhanced resistance of the CALTPI transgenic Arabidopsis against pathogen and environmental stresses. Planta 2005, 221, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Du, K.; Gao, Y.; Kong, Y.; Chu, C.; Sokolov, V.; Wang, Y. Transformation of LTP gene into Brassica napus to enhance its resistance to Sclerotinia sclerotiorum. Genetika 2013, 49, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Fu, X.; Wen, M.; Wang, F.; Tang, Q.; Tian, Q.; Luo, K. Overexpression of an nsLTPs-like antimicrobial protein gene (LJAMP2) from motherwort (Leonurus japonicus) enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Physiol. Mol. Plant Pathol. 2013, 82, 81–87. [Google Scholar] [CrossRef]
- Guo, C.; Ge, X.; Ma, H. The rice OsDIL gene plays a role in drought tolerance at vegetative and reproductive stages. Plant Mol. Biol. 2013, 82, 239–253. [Google Scholar] [CrossRef]
- Gangadhar, B.H.; Sajeesh, K.; Venkatesh, J.; Baskar, V.; Abhinandan, K.; Yu, J.W.; Prasad, R.; Mishra, R.K. Enhanced tolerance of transgenic potato plants over-expressing non-specific lipid transfer protein-1 (StnsLTP1) against multiple abiotic stresses. Front. Plant Sci. 2016, 7, 1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.; Yang, H.; Zhang, X.; Yang, S. Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J. Exp. Bot. 2013, 64, 1755–1767. [Google Scholar] [CrossRef] [PubMed]
- Edstam, M.M.; Edqvist, J. Involvement of GPI-anchored lipid transfer proteins in the development of seed coats and pollen in Arabidopsis thaliana. Physiol. Plant. 2014, 152, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Muench, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Truksa, M.; Shah, S.; Weselake, R.J. A survey of quantitative real-time polymerase chain reaction internal reference genes for expression studies in Brassica napus. Anal. Biochem. 2010, 405, 138–140. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jiang, C.; Ren, J.; Dong, J.; Shi, X.; Zhao, X.; Wang, X.; Wang, J.; Zhong, C.; Zhao, S.; et al. An advanced lipid metabolism system revealed by transcriptomic and lipidomic analyses plays a central role in peanut cold tolerance. Front. Plant. Sci. 2020, 11, 1110. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).