Alteration of the Nucleotide Excision Repair (NER) Pathway in Soft Tissue Sarcoma
Abstract
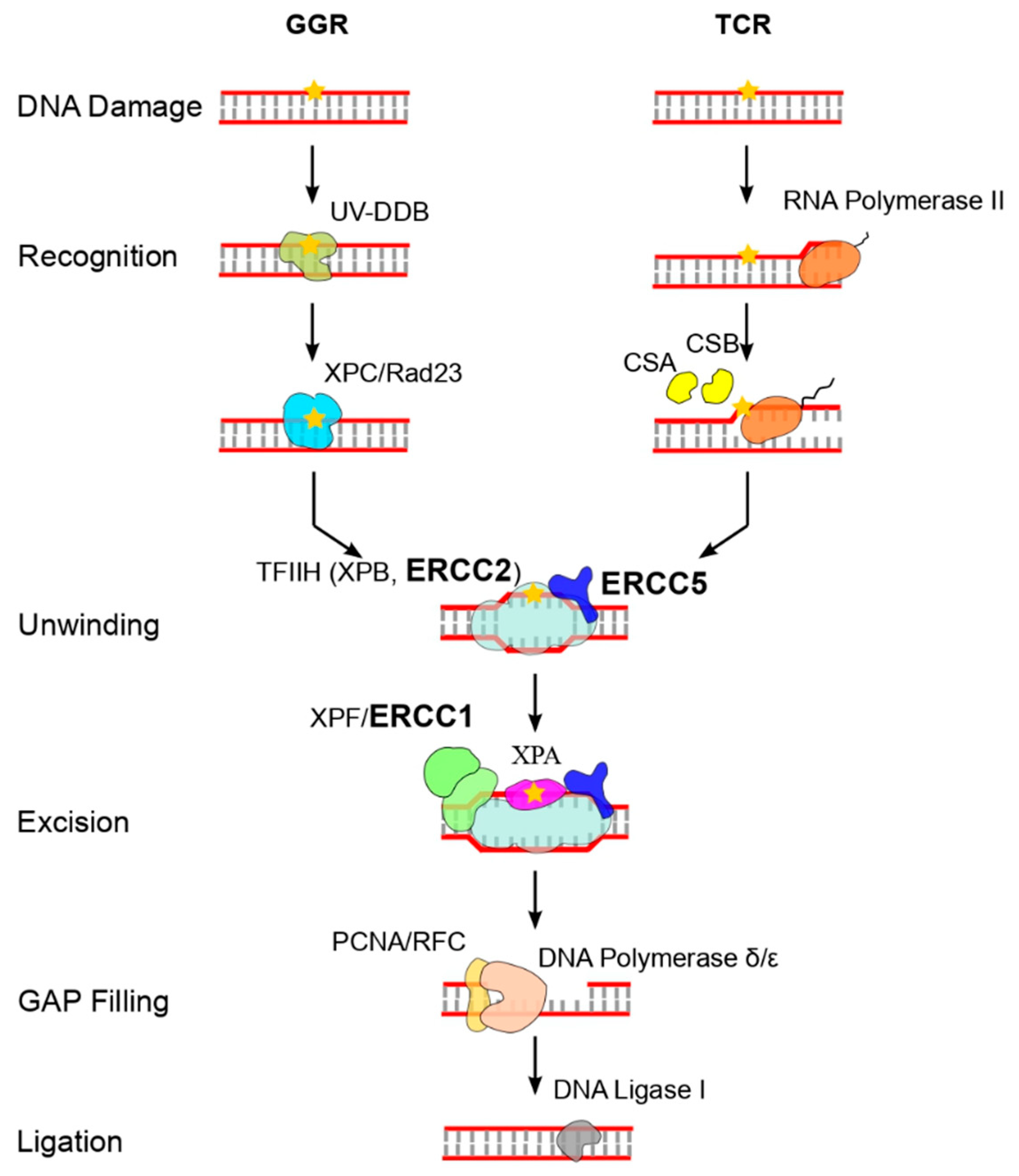
1. Introduction
2. Materials and Methods
2.1. STS Patients and Samples
2.2. Genomic and Somatic DNA Extraction and Genotyping by SNP Assay
2.3. RNA Extraction and Quantitative Real-Time Reverse Transcription PCR (qPCR)
2.4. Molecular Dynamics Simulations
2.5. Statistical Analysis
3. Results
3.1. Clinicopathological Features of OS Patients
3.2. Evaluation of Germinal SNP Panel in NER Genes
3.3. Linkage Disequilibrium and Haplotypes
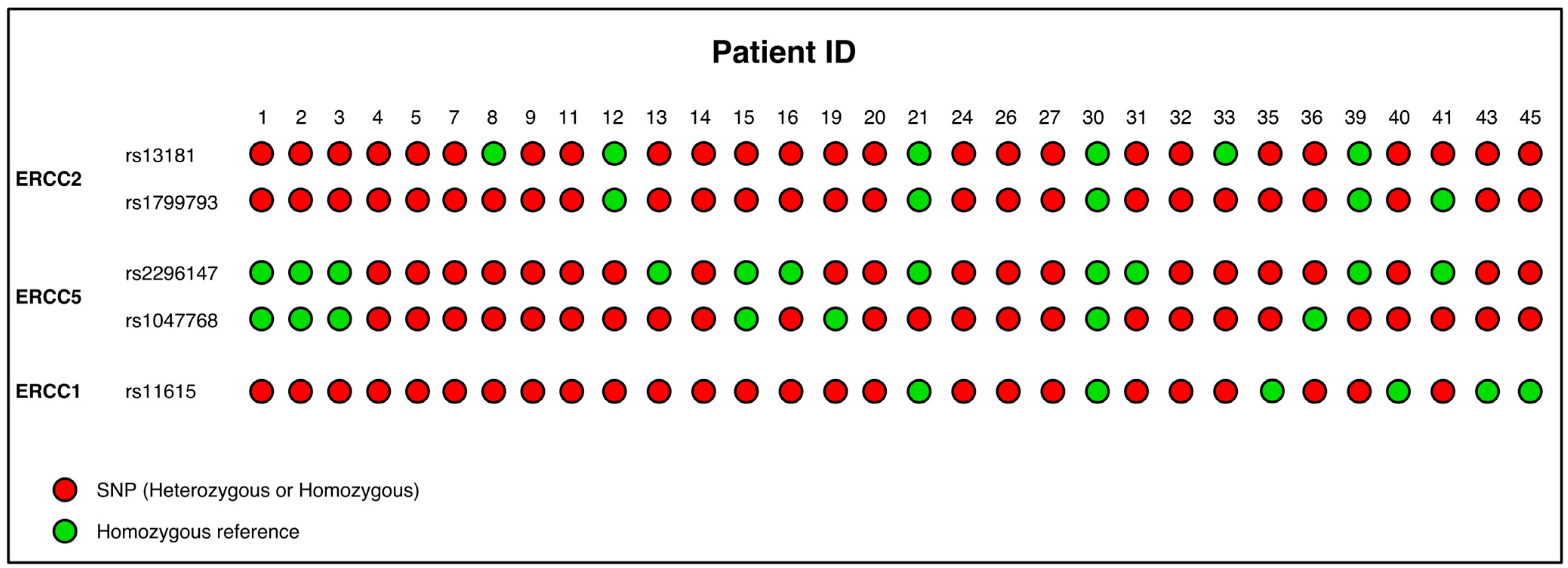
3.4. Evaluation of Somatic SNP Panel in NER Genes in STS Patients
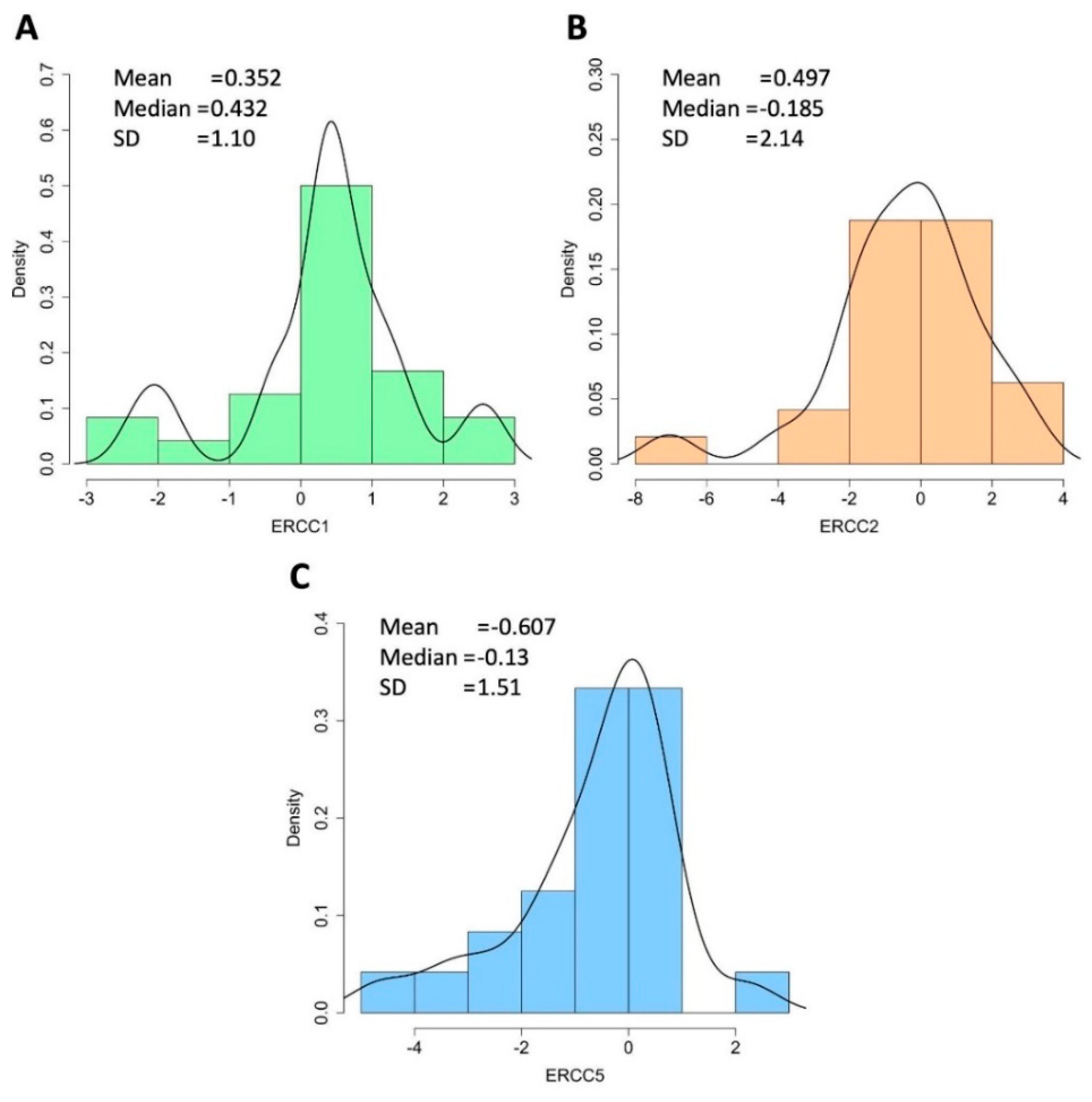
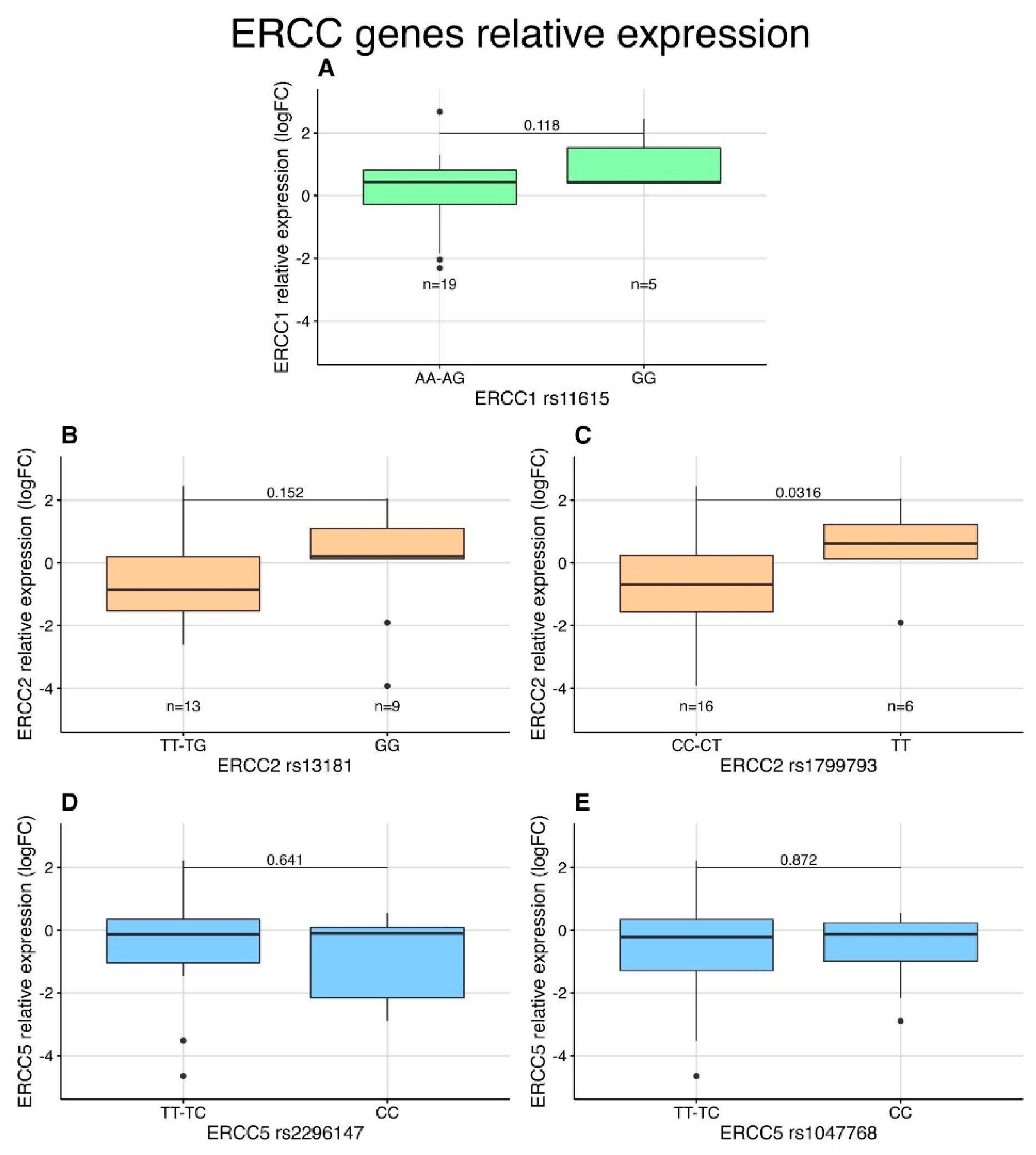
3.5. Expression Analysis of NER Genes in STS Patients by qPCR
3.6. Correlation between TP53 and NER Genes
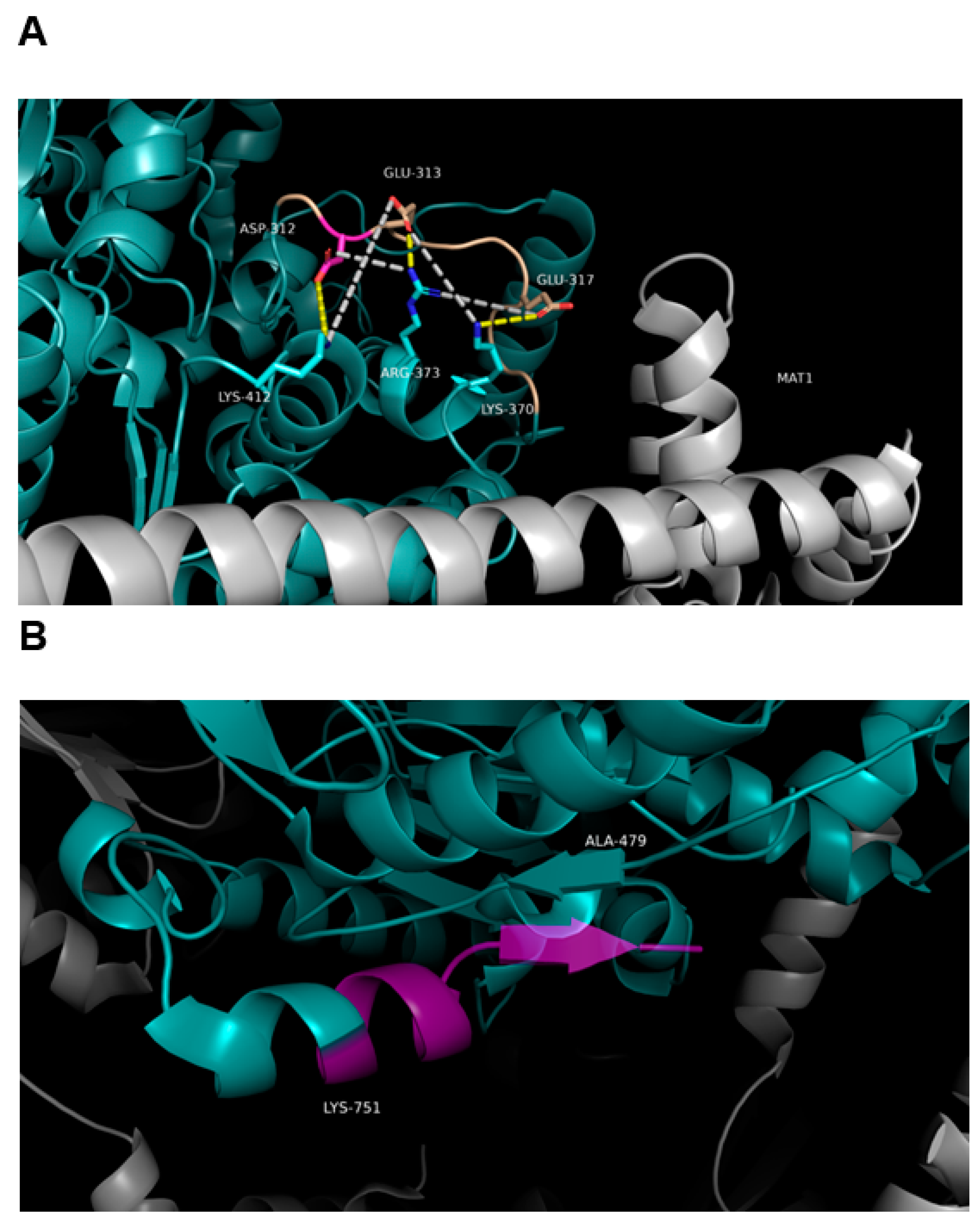
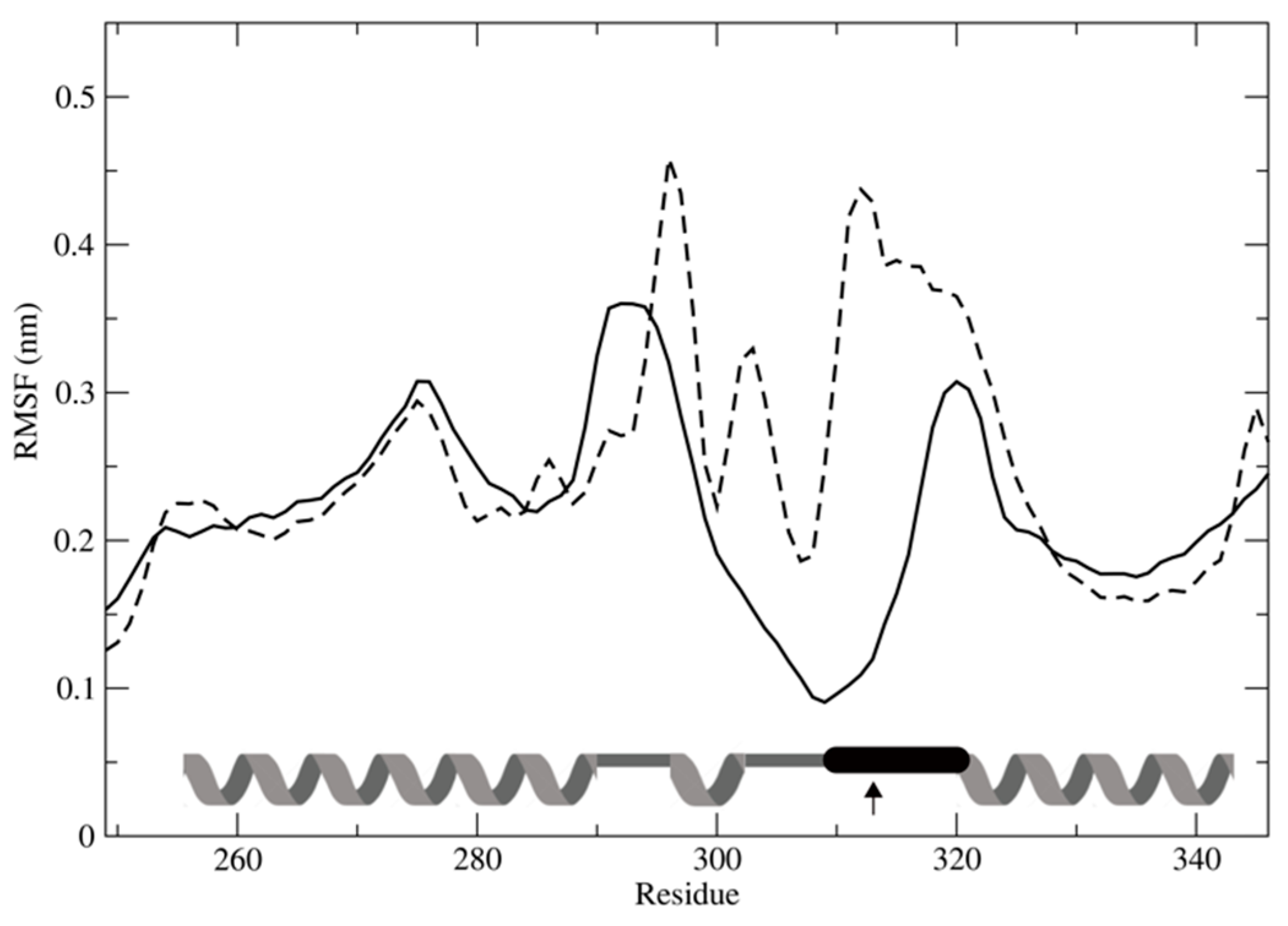
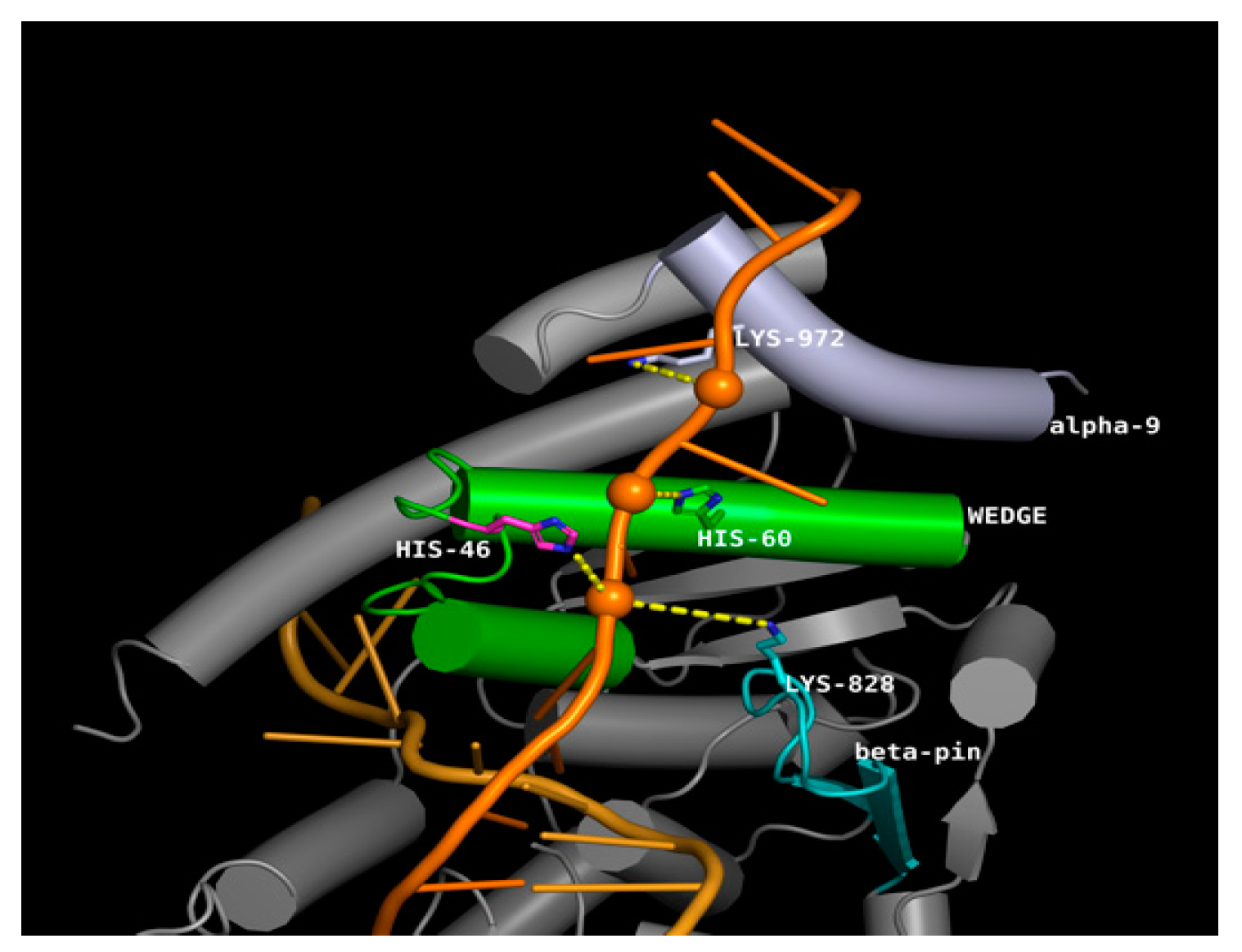
3.7. Effect of SNPs on the Tertiary Structure of NER Proteins
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stiller, C.A.; Trama, A.; Serraino, D.; Rossi, S.; Navarro, C.; Chirlaque, M.D.; Casali, P.G.; RARECARE Working Group. Descriptive epidemiology of sarcomas in Europe: Report from the RARECARE project. Eur. J. Cancer 2013, 49, 684–695. [Google Scholar] [CrossRef]
- Sbaraglia, M.; Bellan, E.; Tos, A.P.D. The 2020 WHO Classification of Soft Tissue Tumours: News and perspectives. Pathologica 2020, 113, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Casali, P.G.; Abecassis, N.; Bauer, S.; Biagini, R.; Bielack, S.; Bonvalot, S.; Boukovinas, I.; Bovee, J.V.M.G.; Brodowicz, T.; Broto, J.; et al. ESMO Guidelines Committee and EURACAN. Soft tissue and visceral sarcomas: ESMO-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29, iv51–iv67. [Google Scholar] [CrossRef] [PubMed]
- Antman, K.; Crowley, J.; Balcerzak, S.P.; E Rivkin, S.; Weiss, G.R.; Elias, A.; Natale, R.B.; Cooper, R.M.; Barlogie, B.; Trump, D.L. An intergroup phase III randomized study of doxorubicin and dacarbazine with or without ifosfamide and mesna in advanced soft tissue and bone sarcomas. J. Clin. Oncol. 1993, 11, 1276–1285. [Google Scholar] [CrossRef] [PubMed]
- Judson, I.; Verweij, J.; Gelderblom, H.; Hartmann, J.T.; Schöffski, P.; Blay, J.-Y.; Kerst, J.M.; Sufliarsky, J.; Whelan, J.; Hohenberger, P.; et al. Doxorubicin alone versus intensified doxorubicin plus ifosfamide for first-line treatment of advanced or metastatic soft tissue sarcoma: A randomised controlled phase 3 trial. Lancet Oncol. 2014, 15, 415–423. [Google Scholar] [CrossRef]
- D’Ambrosio, L.; Touati, N.; Blay, J.; Grignani, G.; Flippot, R.; Czarnecka, A.M.; Piperno-Neumann, S.; Martin-Broto, J.; Sanfilippo, R.; Katz, D.; et al. European Organization for Research and Treatment of Cancer Soft Tissue and Bone Sarcoma Group. Doxorubicin plus dacarbazine, doxorubicin plus ifosfamide, or doxorubicin alone as a first-line treatment for advanced leiomyosarcoma: A propensity score matching analysis from the European Organization for Research and Treatment of Cancer Soft Tissue and Bone Sarcoma Group. Cancer 2020, 126, 2637–2647. [Google Scholar] [CrossRef] [PubMed]
- Stacchiotti, S.; Palassini, E.; Sanfilippo, R.; Vincenzi, B.; Arena, M.; Bochicchio, A.M.; De Rosa, P.; Nuzzo, A.; Turano, S.; Morosi, C.; et al. Gemcitabine in advanced angiosarcoma: A retrospective case series analysis from the Italian Rare Cancer Network. Ann. Oncol. 2012, 23, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Seddon, B.; Strauss, S.J.; Whelan, J.; Leahy, M.; Woll, P.J.; Cowie, F.; Rothermundt, C.; Wood, Z.; Benson, C.; Ali, N.; et al. Gemcitabine and docetaxel versus doxorubicin as first-line treatment in previously untreated advanced unresectable or metastatic soft-tissue sarcomas (GeDDiS): A randomised controlled phase 3 trial. Lancet Oncol. 2017, 18, 1397–1410. [Google Scholar] [CrossRef]
- Penel, N.; Bui, B.N.; Bay, J.-O.; Cupissol, D.; Ray-Coquard, I.; Piperno-Neumann, S.; Kerbrat, P.; Fournier, C.; Taieb, S.; Jimenez, M.; et al. Phase II trial of weekly paclitaxel for unresectable angiosarcoma: The ANGIOTAX Study. J. Clin. Oncol. 2008, 26, 5269–5274. [Google Scholar] [CrossRef] [PubMed]
- Gambale, E.; Boddi, A.; Pasqui, A.; Campanacci, D.A.; Scoccianti, G.; Palchetti, I.; Bernini, A.; Antonuzzo, L.; Pillozzi, S. Pharmacogenomics of soft tissue sarcomas: New horizons to understand efficacy and toxicity. Cancer Treat. Res. Commun. 2022, 31, 100528. [Google Scholar] [CrossRef] [PubMed]
- Nagel, Z.D.; Kitange, G.J.; Gupta, S.K.; Joughin, B.A.; Chaim, I.A.; Mazzucato, P.; Lauffenburger, D.A.; Sarkaria, J.N.; Samson, L.D. DNA Repair Capacity in Multiple Pathways Predicts Chemoresistance in Glioblastoma Multiforme. Cancer Res. 2017, 77, 198–206. [Google Scholar] [CrossRef]
- Lindahl, T.; Wood, R.D. Quality control by DNA repair. Science 1999, 286, 1897–1905. [Google Scholar] [CrossRef] [PubMed]
- Hoeijmakers, J.H. Genome maintenance mechanisms for preventing cancer. Nature 2001, 411, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Moura, D.S.; Peña-Chilet, M.; Varela, J.A.C.; Alvarez-Alegret, R.; Agra-Pujol, C.; Izquierdo, F.; Ramos, R.; Ortega-Medina, L.; Martin-Davila, F.; Castilla-Ramirez, C.; et al. A DNA damage repair gene-associated signature predicts responses of patients with advanced soft-tissue sarcoma to treatment with trabectedin. Mol. Oncol. 2021, 15, 3691–3705. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.; Mallick, A.B.; Augustine, T.; Daroqui, C.; Jiffry, J.; Merla, A.; Chaudhary, I.; Seetharam, R.; Sood, A.; Gajavelli, S.; et al. Excision repair cross-complementing group-1 (ERCC1) induction kinetics and polymorphism are markers of inferior outcome in patients with colorectal cancer treated with oxaliplatin. Oncotarget 2019, 10, 5510–5522. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Sirimangkalakitti, N.; Baba, A.; Toyoshima-Nagasaki, R.; Enomoto, Y.; Saito, N.; Ogasawara, Y. Characterization of the nucleotide excision repair pathway and evaluation of compounds for overcoming the cisplatin resistance of non-small cell lung cancer cell lines. Oncol. Rep. 2022, 47, 70. [Google Scholar] [CrossRef]
- Biason, P.; Hattinger, C.; Innocenti, F.; Talamini, R.; Alberghini, M.; Scotlandi, K.; Zanusso, C.; Serra, M.; Toffoli, G. Nucleotide excision repair gene variants and association with survival in osteosarcoma patients treated with neoadjuvant chemotherapy. Pharmacogenomics J. 2011, 12, 476–483. [Google Scholar] [CrossRef]
- Han, P.; Gao, F.; Liu, H.; Liu, Z.; Shi, Q.; Troy, J.D.; Owzar, K.; Lee, W.; Zevallos, J.P.; Sturgis, E.M.; et al. Reduced mRNA expression of nucleotide excision repair genes in lymphocytes and risk of squamous cell carcinoma of the head and neck. Carcinogenesis 2017, 38, 504–510. [Google Scholar] [CrossRef]
- Ren, P.; Niu, X.; Liu, C.; Liu, J.; Li, H.; Zhao, Q.; Xing, J.; Bai, Y.; Liang, Y.; Han, P. Associations between expression levels of nine core nucleotide excision repair genes in lymphocytes and risk of head and neck squamous cell carcinomas in a Chinese population. Int. J. Clin. Oncol. 2019, 25, 660–669. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, S.; Zhou, X.; Huang, F.; Chen, R.; Wang, Y.; Wu, J. Association between nucleotide excision repair gene polymorphism and colorectal cancer risk. J. Clin. Lab. Anal. 2019, 33, e22956. [Google Scholar] [CrossRef]
- Ibrahim, O.M.; Sobeai, H.M.A.; Grant, S.; Latimer, J.J. Nucleotide excision repair is a predictor of early relapse in pediatric acute lymphoblastic leukemia. BMC Med Genom. 2018, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Aiello, M.M.; Solinas, C.; Santoni, M.; Battelli, N.; Restuccia, N.; Latteri, F.; Paratore, S.; Verderame, F.; Albanese, G.V.; Bruzzi, P.; et al. Excision Repair Cross Complementation Group 1 Single Nucleotide Polymorphisms and Nivolumab in Advanced Non-Small Cell Lung Cancer. Front. Oncol. 2020, 10, 1167. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Mouw, K.W.; Kim, P.; Iyer, G.; Wagle, N.; Al-Ahmadie, H.; Zhu, C.; Ostrovnaya, I.; Kryukov, G.V.; O’Connor, K.W.; et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 2014, 4, 1140–1153. [Google Scholar] [CrossRef]
- Zhang, X.; Crawford, E.L.; Blomquist, T.M.; Khuder, S.A.; Yeo, J.; Levin, A.M.; Willey, J.C. Haplotype and diplotype analyses of variation in ERCC5 transcription cis-regulation in normal bronchial epithelial cells. Physiol. Genom. 2016, 48, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Marinescu, V.D.; Kohane, I.S.; Riva, A. The MAPPER database: A multi-genome catalog of putative transcription factor binding sites. Nucleic Acids Res. 2004, 33, D91–D97. [Google Scholar] [CrossRef]
- Orans, J.; McSweeney, E.A.; Iyer, R.R.; Hast, M.A.; Hellinga, H.W.; Modrich, P.; Beese, L.S. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell 2011, 145, 212–223. [Google Scholar] [CrossRef] [PubMed]
- González-Corrochano, R.; Ruiz, F.M.; Taylor, N.M.I.; Huecas, S.; Drakulic, S.; Spínola-Amilibia, M.; Fernández-Tornero, C. The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair. Nucleic Acids Res. 2020, 48, 9943–9958. [Google Scholar] [CrossRef]
- Devos, J.M.; Tomanicek, S.J.; Jones, C.E.; Nossal, N.G.; Mueser, T.C. Crystal structure of bacteriophage T4 5′ nuclease in complex with a branched DNA reveals how flap endonuclease-1 family nucleases bind their substrates. J. Biol. Chem. 2007, 282, 31713–31724. [Google Scholar] [CrossRef] [PubMed]
- Maki, R.G.; Wathen, J.K.; Patel, S.R.; Priebat, D.A.; Okuno, S.H.; Samuels, B.; Fanucchi, M.; Harmon, D.C.; Schuetze, S.M.; Reinke, D.; et al. Randomized phase II study of gemcitabine and docetaxel compared with gemcitabine alone in patients with metastatic soft tissue sarcomas: Results of sarcoma alliance for research through collaboration study 002. J. Clin. Oncol. 2007, 25, 2755–2763. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Laetsch, T.W.; Kummar, S.; Dubois, S.G.; Lassen, U.N.; Demetri, G.D.; Nathenson, M.; Doebele, R.C.; Farago, A.F.; Pappo, A.S.; et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N. Engl. J. Med. 2018, 378, 731–739. [Google Scholar] [CrossRef]
- van der Graaf, W.T.; Blay, J.-Y.; Chawla, S.P.; Kim, D.-W.; Bui-Nguyen, B.; Casali, P.G.; Schöffski, P.; Aglietta, M.; Staddon, A.P.; Beppu, Y.; et al. Pazopanib for metastatic soft-tissue sarcoma (PALETTE): A randomised, double-blind, placebo-controlled phase 3 trial. Lancet 2012, 379, 1879–1886. [Google Scholar] [CrossRef]
- Schöffski, P.; Chawla, S.; Maki, R.G.; Italiano, A.; Gelderblom, H.; Choy, E.; Grignani, G.; Camargo, V.; Bauer, S.; Rha, S.Y.; et al. Eribulin versus dacarbazine in previously treated patients with advanced liposarcoma or leiomyosarcoma: A randomised, open-label, multicentre, phase 3 trial. Lancet 2016, 387, 1629–1637. [Google Scholar] [CrossRef]
- Frezza, A.M.; Jones, R.L.; Vullo, S.L.; Asano, N.; Lucibello, F.; Ben-Ami, E.; Ratan, R.; Teterycz, P.; Boye, K.; Brahmi, M.; et al. Anthracycline, Gemcitabine, and Pazopanib in Epithelioid Sarcoma: A Multi-institutional Case Series. JAMA Oncol. 2018, 4, e180219. [Google Scholar] [CrossRef] [PubMed]
- Gentile, F.; Tuszynski, J.A.; Barakat, K.H. New design of nucleotide excision repair (NER) inhibitors for combination cancer therapy. J. Mol. Graph. Model. 2016, 65, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Bowden, N.A. Nucleotide excision repair: Why is it not used to predict response to platinum-based chemotherapy? Cancer Lett. 2014, 346, 163–171. [Google Scholar] [CrossRef]
- Duan, M.; Ulibarri, J.; Liu, K.J.; Mao, P. Role of Nucleotide Excision Repair in Cisplatin Resistance. Int. J. Mol. Sci. 2020, 21, 9248. [Google Scholar] [CrossRef]
- Tse, D.; Zhai, R.; Zhou, W.; Heist, R.S.; Asomaning, K.; Su, L.; Lynch, T.J.; Wain, J.C.; Christiani, D.C.; Liu, G. Polymorphisms of the NER pathway genes, ERCC1 and XPD are associated with esophageal adenocarcinoma risk. Cancer Causes Control 2008, 19, 1077–1083. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, J.; Deng, N.; Xu, Q.; Sun, L.; Tu, H.; Wang, Z.; Xing, C.; Yuan, Y. Polymorphisms of multiple genes involved in NER pathway predict prognosis of gastric cancer. Oncotarget 2016, 7, 48130–48142. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Fu, W.; Jia, W.; Xia, H.M.; Liu, G.-C.; He, J. Association between NER Pathway Gene Polymorphisms and Wilms Tumor Risk. Mol. Ther. Nucleic Acids 2018, 12, 854–860. [Google Scholar] [CrossRef] [PubMed]
- Italiano, A.; Laurand, A.; Laroche, A.; Casali, P.G.; Sanfilippo, R.; Le Cesne, A.; Judson, I.; Blay, J.-Y.; Ray-Coquard, I.; Bui, B.; et al. ERCC5/XPG, ERCC1, and BRCA1 gene status and clinical benefit of trabectedin in patients with soft tissue sarcoma. Cancer 2011, 117, 3445–3456. [Google Scholar] [CrossRef] [PubMed]
- Börcsök, J.; Sztupinszki, Z.; Bekele, R.; Gao, S.P.; Diossy, M.; Samant, A.S.; Dillon, K.M.; Tisza, V.; Spisák, S.; Rusz, O.; et al. Identification of a Synthetic Lethal Relationship between Nucleotide Excision Repair Deficiency and Irofulven Sensitivity in Urothelial Cancer. Clin. Cancer Res. 2020, 27, 2011–2022. [Google Scholar] [CrossRef] [PubMed]
- Le Morvan, V.; Longy, M.; Bonaïti-Pellié, C.; Bui, B.; Houédé, N.; Coindre, J.M.; Robert, J.; Pourquier, P. Genetic polymorphisms of the XPG and XPD nucleotide excision repair genes in sarcoma patients. Int. J. Cancer 2006, 119, 1732–1735. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Liu, Y.; Yu, T.; Xiao, S.; Bao, X.; Pan, L.; Zhu, G.; Cai, Y.; Liu, Q.; Jin, C.; et al. ERCC1 and ERCC2 haplotype modulates induced BPDE-DNA adducts in primary cultured lymphocytes. PLoS ONE 2013, 8, e60006. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Khrunin, A.V.; Firsov, S.Y.; Limborska, S.A. Polymorphisms of DNA repair genes ERCC2 and XRCC1 in populations of Russia. Russ. J. Genet. 2011, 47, 1391–1393. [Google Scholar] [CrossRef]
- Liu, J.; Li, H.; Sun, L.; Feng, X.; Wang, Z.; Yuan, Y.; Xing, C. The Differential Expression of Core Genes in Nucleotide Excision Repair Pathway Indicates Colorectal Carcinogenesis and Prognosis. BioMed Res. Int. 2018, 2018, 9651320. [Google Scholar] [CrossRef]
- Bajpai, D.; Banerjee, A.; Pathak, S.; Jain, S.K.; Singh, N. Decreased expression of DNA repair genes (XRCC1, ERCC1, ERCC2, and ERCC4) in squamous intraepithelial lesion and invasive squamous cell carcinoma of the cervix. Mol. Cell. Biochem. 2013, 377, 45–53. [Google Scholar] [CrossRef]
- Huang, M.-Y.; Lee, H.-H.; Huang, C.-W.; Huang, C.-M.; Ma, C.-J.; Yin, T.-C.; Tsai, H.-L.; Chai, C.-Y.; Chen, Y.-T.; Wang, J.-Y. ERCC overexpression associated with a poor response of cT4b colorectal cancer with FOLFOX-based neoadjuvant concurrent chemoradiation. Oncol. Lett. 2020, 20, 212. [Google Scholar] [CrossRef] [PubMed]
- Kirkilevsky, S.I.; Krakhmalev, P.S.; Malyshok, N.V.; Zadvornyi, T.V.; Borikun, T.V.; Yalovenko, T.M. Prognostic significance of microRNA-200b and ERCC1 expression in tumor cells of patients with esophageal cancer. Exp. Oncol. 2020, 42, 167–171. [Google Scholar] [CrossRef]
- Moura, D.S.; Sanchez-Bustos, P.; Fernandez-Serra, A.; Lopez-Alvarez, M.; Mondaza-Hernandez, J.L.; Blanco-Alcaina, E.; Gavilan-Naranjo, A.; Martinez-Delgado, P.; Lacerenza, S.; Santos-Fernandez, P.; et al. CUL4A, ERCC5, and ERCC1 as Predictive Factors for Trabectedin Efficacy in Advanced Soft Tissue Sarcomas (STS): A Spanish Group for Sarcoma Research (GEIS) Study. Cancers 2020, 12, 1128. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell 2017, 171, 950–965e28. [Google Scholar] [CrossRef] [PubMed]
- Peissert, S.; Sauer, F.; Grabarczyk, D.B.; Braun, C.; Sander, G.; Poterszman, A.; Egly, J.-M.; Kuper, J.; Kisker, C. In TFIIH the Arch domain of XPD is mechanistically essential for transcription and DNA repair. Nat. Commun. 2020, 11, 1667. [Google Scholar] [CrossRef] [PubMed]
- Rump, A.; Benet-Pages, A.; Schubert, S.; Kuhlmann, J.D.; Janavičius, R.; Machackova, E.; Foretova, L.; Kleibl, Z.; Lhota, F.; Zemánková, P.; et al. Identification and Functional Testing of ERCC2 Mutations in a Multi-national Cohort of Patients with Familial Breast- and Ovarian Cancer. PLoS Genet. 2016, 12, e1006248. [Google Scholar] [CrossRef] [PubMed]


| Clinicopathological Features | Cohort (N = 32) |
|---|---|
| Gender | Male = 62.5% |
| Female = 37.5% | |
| Age at surgery (years) | Mean = 68.4 |
| Max = 93 | |
| Min = 39 | |
| BMI (kg/m2) | Mean = 25.5 |
| Max = 44.3 | |
| Min = 19.2 | |
| Surgical margins | Large = 90.6% |
| Marginal = 9.4% | |
| Primary/Relapse | Primary = 81% |
| Relapse = 19% | |
| Grade | High = 78.0% |
| Low = 22.0% | |
| Hystotype | Liposarcoma = 28.1% |
| Pleomorphic sarcoma = 37.5% | |
| Leiomyosarcoma = 9.4% | |
| Myxofibrosarcoma = 12.5% | |
| Fibrosarcoma = 3.1% | |
| Fibromyxoid sarcoma = 3.1% | |
| Dermatofibrosarcoma protuberans = 3.1% | |
| Undifferentiated pleomorphic cell sarcoma = 3.1% | |
| Anatomical site | Thigh = 62.5% |
| Foot = 3.1% | |
| Gluteal-sacral region = 6.25% | |
| Scapular region = 3.1% | |
| Deltoid region = 3.1% | |
| Abdominal region = 3.1% | |
| Pectoral region = 6.25% | |
| Upper limb = 12.5% |
| Gene Name | Primer Forward | Primer Reverse | Lenght (bp) |
|---|---|---|---|
| ERCC1 | 5′ CTCAAGGAGCTGGCTAAGATGT 3′ | 5′ CATAGGCCTTGTAGGTCTCCAG 3′ | 90 |
| ERCC2 | 5′ CTGGAGGTGACCAAACTCATCTA 3′ | 5′ CCTGCTTCTCATAGAAGTTGAGC 3′ | 100 |
| ERCC5 | 5′ CAGACACAGCTCCGAATTGA 3′ | 5′ TTCTGGGTTTTTCGTTTTGC 3′ | 209 |
| 18S | 5′ AACCCGTTGAACCCCAT 3′ | 5′ CCATCCAATCGGTAGTAGCG 3′ | 151 |
| Gene | ercc1 | ercc2 | ercc5 | |||
|---|---|---|---|---|---|---|
| SNP ID | rs11615 | rs13181 | rs1799793 | rs1047768 | rs2296147 | |
| Assay ID | C_2532959_20 | C_3145033_10 | C_3145050_10 | C_1891769_20 | C_1891782_20 | |
| Chromosome Location | chr19:45420395 | chr19:45351661 | chr19:45364001 | chr13:102852167 | chr13:102846025 | |
| Variant | A>G | T>G | C>T | T>C | T>C | |
| SNP Type | Synonymous variant | Stop gained variant | Missense variant | Missense variant | Upstream variant | |
| Genotype | Homozygous Reference | AA = 6 (19%) | TT = 6 (29%) | CC = 6 (19%) | TT = 6 (19%) | TT = 10 (32%) |
| Heterozygotes | AG = 18 (58%) | TG = 14 (45%) | CT = 17 (55%) | TC = 14 (45%) | TC = 15 (49%) | |
| Alternative Homozygotes | GG = 7 (23%) | GG = 10 (36%) | TT = 7 (26%) | CC = 11 (36%) | CC = 6 (19%) | |
| STS cohort alleles frequencies | Reference | 48% | 42% | 47% | 42% | 56% |
| Alternative | 52% | 58% | 53% | 58% | 44% | |
| General population alleles frequencies * | Reference | 59% | 64% | 68% | 42% | 54% |
| Alternative | 41% | 36% | 32% | 58% | 46% | |
| χ² Test | χ² | 2.43 | 9.72 | 9.02 | 0.00 | 0.08 |
| p | 0.1190 | 0.0018 | 0.0027 | 1.0000 | 0.7762 | |
| Gene | SNP ID | Homozygotes Reference (%) | Heterozygotes (%) | Homozygotes Alternative (%) |
|---|---|---|---|---|
| ERCC1 | rs11615 | 100 | 0 | 0 |
| ERCC5 | rs1047768 | 80 | 20 | 0 |
| ERCC5 | rs2296147 | 87.5 | 12.5 | 0 |
| ERCC2 | rs13181 | 100 | 0 | 0 |
| ERCC2 | rs1799793 | 100 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pasqui, A.; Boddi, A.; Campanacci, D.A.; Scoccianti, G.; Bernini, A.; Grasso, D.; Gambale, E.; Scolari, F.; Palchetti, I.; Palomba, A.; et al. Alteration of the Nucleotide Excision Repair (NER) Pathway in Soft Tissue Sarcoma. Int. J. Mol. Sci. 2022, 23, 8360. https://doi.org/10.3390/ijms23158360
Pasqui A, Boddi A, Campanacci DA, Scoccianti G, Bernini A, Grasso D, Gambale E, Scolari F, Palchetti I, Palomba A, et al. Alteration of the Nucleotide Excision Repair (NER) Pathway in Soft Tissue Sarcoma. International Journal of Molecular Sciences. 2022; 23(15):8360. https://doi.org/10.3390/ijms23158360
Chicago/Turabian StylePasqui, Adriano, Anna Boddi, Domenico Andrea Campanacci, Guido Scoccianti, Andrea Bernini, Daniela Grasso, Elisabetta Gambale, Federico Scolari, Ilaria Palchetti, Annarita Palomba, and et al. 2022. "Alteration of the Nucleotide Excision Repair (NER) Pathway in Soft Tissue Sarcoma" International Journal of Molecular Sciences 23, no. 15: 8360. https://doi.org/10.3390/ijms23158360
APA StylePasqui, A., Boddi, A., Campanacci, D. A., Scoccianti, G., Bernini, A., Grasso, D., Gambale, E., Scolari, F., Palchetti, I., Palomba, A., Fancelli, S., Caliman, E., Antonuzzo, L., & Pillozzi, S. (2022). Alteration of the Nucleotide Excision Repair (NER) Pathway in Soft Tissue Sarcoma. International Journal of Molecular Sciences, 23(15), 8360. https://doi.org/10.3390/ijms23158360








