Abstract
The nonsense-mediated mRNA decay (NMD) pathway rapidly detects and degrades mRNA containing premature termination codons (PTCs). UP-frameshift 1 (UPF1), the master regulator of the NMD process, has two alternatively-spliced isoforms; one carries 353-GNEDLVIIWLR-363 insertion in the ‘regulatory loop (involved in mRNA binding)’. Such insertion can induce catalytic and/or ATPase activity, as determined experimentally; however, the kinetics and molecular level information are not fully understood. Herein, applying all-atom molecular dynamics, we probe the binding specificity of UPF1 with different GC- and AU-rich mRNA motifs and the influence of insertion to the viable control over UPF1 catalytic activity. Our results indicate two distinct conformations between 1B and RecA2 domains of UPF1: ‘open (isoform_2; without insertion)’ and ‘closed (isoform_1; with insertion)’. These structural movements correspond to an important stacking pattern in mRNA motifs, i.e., absence of stack formation in mRNA, with UPF1 isoform_2 results in the ‘open conformation’. Particularly, for UPF1 isoform_1, the increased distance between 1B and RecA2 domains has resulted in reducing the mRNA–UPF1 interactions. Lower fluctuating GC-rich mRNA motifs have better binding with UPF1, compared with AU-rich sequences. Except CCUGGGG, all other GC-rich motifs formed a 4-stack pattern with UPF1. High occupancy R363, D364, T627, and G862 residues were common binding GC-rich motifs, as were R363, N535, and T627 for the AU-rich motifs. The GC-rich motifs behave distinctly when bound to either of the isoforms; lower stability was observed with UPF1 isoform_2. The cancer-associated UPF1 variants (P533L/T and A839T) resulted in decreased protein–mRNA binding efficiency. Lack of mRNA stacking poses in the UPF1P533T system significantly decreased UPF1-mRNA binding efficiency and increased distance between 1B-RecA2. These novel findings can serve to further inform NMD-associated mechanistic and kinetic studies.
Keywords:
UPF1; GC-rich; AU-rich; isoform; regulatory loop; mRNA; PTC; molecular dynamics; degradation; stability; NMD; alternatively spliced; motifs 1. Introduction
Aberrant, misfolded, and mislocalized proteins are dangerous to cell viability due to their toxicity, which can be the cause of multiple human diseases such as Parkinson’s and Alzheimer’s diseases, frontotemporal dementia, cystic fibrosis, and several others [1,2,3,4,5,6,7]. Natural selection pressures have led to a cellular quality control pathway that prevents aberrant proteins at the ribosome or that senses the aberrations in the mRNA templates, resulting in their degradation. Many quality control or protective mechanisms are engaged cotranslationally, while proteins are being synthesized from mRNA [1,7,8]. Protein synthesis, along with the degradation of defective proteins, is an energetically demanding processes, and ribosome-associated quality control can hinder futile aberrant protein synthesis. The mRNA surveillance quality control pathways such as the nonsense-mediated mRNA decay (NMD) can detect as well as eliminate the defective mRNAs with premature termination codons (PTC), the no-go decay (NGD) pathway can trace truncated and stalled in translation mRNAs, and the non-stop decay (NSD) pathway can detect mRNAs without natural stop codons [7,9,10,11,12].
Particularly at the mRNA level, two essential features are observed: (i) either it contains the correct set of proteins bound to a particular mRNA, or (ii) the coding potential of the mRNA is intact. Here, the NMD pathway copes with the latter one; by degrading the PTC-containing mRNAs, it consequently reduces the accumulation of potentially toxic truncated proteins [10,13,14]. The NMD process is one of the best studied translation-dependent mRNA surveillance mechanisms, which typifies all eukaryotes examined to date and helps to regulate the quality of gene expression. Due to its capacity of detecting a PTC for accelerating the degradation of the aberrant mRNA, NMD is an important modulator of genetic disease phenotypes in humans [15,16]. In addition, one-third of inherited genetic diseases are caused by mRNAs harboring PTC as a result of nonsense mutations [15,16]. By degrading transcriptome, the NMD process can harbor distinct contexts at translation termination (e.g., uORF (upstream open reading frames)-containing mRNAs, long 3′UTRs), suggesting that it is a critical regulatory pathway [17,18,19,20,21,22]. In relation to this, the NMD pathway can exert a beneficial or a detrimental effect: the former if it prevents the synthesis of toxic truncated proteins and the latter if it prevents the production of proteins having some residual or partial functions [14,15,23,24,25].
UP-frameshift 1 (UPF1; Figure 1a,b), UPF2, and UPF3(a/b) proteins are the core components crucial for the NMD process in all organisms. Among these, UPF1 is primarily a cytoplasmic RNA-binding protein exhibiting RNA-dependent ATPase and RNA helicase activities for the NMD [26,27,28,29,30,31,32,33,34]. These three UPF proteins together form a ‘surveillance complex’ that triggers RNA-dependent ATPase activity of UPF1 [31,34,35,36,37,38,39,40]. Various models explaining how these NMD components recognize a PTC and recruit RNA degradation proteins have been proposed (a highly accepted model is shown in Figure 1c) [31,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56]. A PTC is defined relative to the position of a downstream cis-acting signal, and it varies across the species [45,57,58]. The majority of models require that RNA decay be triggered once a stop codon is followed by a second signal, which specifies the stop codon as premature [31,59,60]. In mammals, the cis-acting signal is an exon–exon junction (EJC). For organisms or mRNAs in which EJCs do not contribute to substrate recognition in NMD, the PTC has been postulated to be distinct due to an absence of termination promoting signals from the faux 3′ untranslated region (UTR), particularly from poly(A)-binding protein 1 (PAB1 or PABPC1; poly(A)-binding protein cytoplasmic 1) [45,57,58,61,62,63,64]. This scheme is referred to as the ‘faux 3′-UTR’ model, according to which the translation termination at a normal stop codon is considered to be essentially different from translation termination at a PTC; the aberrant nature of premature termination activates the mRNA decay [31,44]. Moreover, an interaction between a terminating ribosome and a specific messenger ribonucleoprotein (mRNP) structure localized 3′ to the stop codon is required for proper termination [44,65,66,67]. The ‘faux 3′-UTR’ model also suggests that the proximity of the PAB1 to the PTC is essential for NMD activation [31,44,68,69].
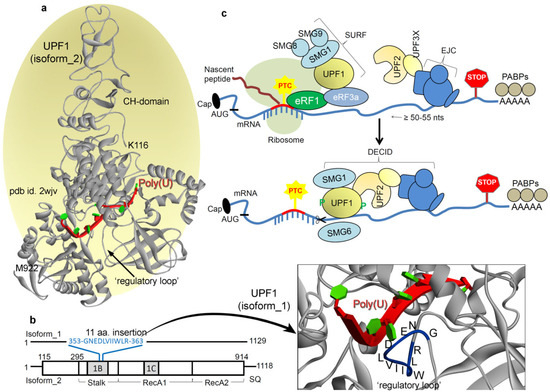
Figure 1.
The master regulator of the NMD (nonsense-mediated mRNA decay) pathway: UPF1 (UP-frameshift 1). (a) Optimized UPF1 isoform_2 model structure [40] with the poly(U) mRNA motif, constructed based on the pdb (protein data bank) id: 2wjv [67] and 2xzo [37]. The crystal structure of UPF1 (pdb id: 2wjv [67]) is with the UPF2 protein, but since UPF1 adopts a different conformation when bound with UPF2, we removed it from the presentation. In addition, the poly(U) mRNA motif has been inserted into the structural representation (from pdb id: 2xzo [37]) as a reference since it is not present in the original crystal structure (pdb id: 2wjv [67]). (b) Isoform_1 structure of the UPF1 (build considering the pdb id: 2wjv [67]) protein, with the 11 aa (amino acids) insertion (353-GNEDLVIIWLR-363) in the ‘regulatory loop’. These 353-GNEDLVIIWLR-363 residues are highlighted over the protein structure in blue color. This UPF1 isoform_1 structure with inserting 11 aa was modeled considering the isoform_2 crystal structure as the template (pdb id: 2wjv) [67]. (c) In the NMD pathway, recognition of PTC-containing mRNA transcript requires UPF1, UPF2, and UPF3 proteins as the core machinery, along with various suppressors with morphogenetic effect on genitalia (SMG) proteins [7,61]. There is a general agreement that NMD substrate recognition relies upon differences in mRNA ribonucleoprotein (mRNP) composition between normal mRNAs and PTC-containing mRNA transcripts. In mammalian cells, an exon–exon junction complex (EJC) of proteins deposited at exon–exon boundaries during pre-mRNA splicing is considered to be a primary determinant of NMD [62]. Translation termination at a nonsense codon located ~50–55 nucleotides upstream of an exon–exon junction usually triggers NMD. It is suggested that translation termination events leading to NMD entail the SURF complex, consisting of SMG1, UPF1, eRF1 (eukaryotic translation termination factor 1), and eRF3 [41]. Once translation terminates, UPF1 then interacts with UPF2 that is bound to the EJC together with additional UPF3 proteins. These interactions result in the assembly of the surveillance complex, UPF1 phosphorylation by SMG1, and eventually mRNA degradation [63]. In principle, if an EJC-related exon–exon junction resides ≥50–55 nucleotides downstream of the termination event, then NMD initiates. Concerning the dynamics of NMD components, the binding of UPF2 with UPF1 causes a large conformational change in the UPF1 CH-domain (cysteine-histidine-rich domain), which triggers the UPF1 helicase activity. Furthermore, within the resulting decay-inducing complex (DECID), the SMG1 protein phosphorylates UPF1 that inhibits further translation termination initiation at the AUG codon [56].
The UPF1 protein is a key component in the NMD pathway, also termed as the master regulator. Deletion of UPF1 in yeast not only stabilizes the nonsense-containing mRNAs but also enhances nonsense codon read-through as well and inhibits degradation of prematurely terminated polypeptides [36,37,39,46]. Moreover, the work by Chan et al. [70] has shown that deletion of UPF1 induces the production of (approximately6- to 8-fold increase) novel peptide read-through. Gowravaram et al. [71] examined the UPF1 core features and identified a structural element that adopts various conformations in the nucleotide as well as RNA-bound states of UPF1. Through biochemical and single molecule assays, they have shown that a structural element modulates UPF1 catalytic activity, referred to as the ‘regulatory loop’. Interestingly, two alternatively spliced isoforms of UPF1 that differ only in length of the ‘regulatory loop’ exist in mammals [71]. UPF1 isoform_1 contains 11 aa (amino acids) insertion (353-GNEDLVIIWLR-363) in the domain 1B, which extends the ‘regulatory loop’ to 22 aa. Whereas, the UPF1 isoform_2 (the more abundant UPF1 short loop isoform) is composed of only an 11 aa ‘regulatory loop’ which weakens the UPF1 affinity for mRNA. In addition, this insertion of 11 amino acids to the ‘regulatory loop’ can considerably increase the catalytic and/or ATPase activity and result in a higher affinity for mRNA [71].
Considering such influence of the ‘regulatory loop’ in the structural dynamics of UPF1 and its control over the catalytic and/or ATPase activity, we investigated both isoforms of UPF1 in the presence or absence of the poly(U) mRNA by applying the molecular dynamics simulation (MDS) technique. Additionally, it is known that UPF1 can bind with different mRNAs efficiently [41,43,56,61,63,72,73,74], but the molecular details are still not clear. Hence, in this study, we also studied the UPF1 isoform_1 (having a longer ‘regulatory loop’) and isoform_2 binding with different mRNA motifs. The significant structural changes upon inserting the most frequently occurring cancer mutations in the UPF1 isoform_1 bound with mRNA motifs were investigated. Furthermore, preferential binding affinity of UPF1 with mRNAs in a 3’UTR length-dependent manner has been reported [17,75]. David et al. identifiedUPF1-mRNA interactions in vivo [76] and proposed that the UPF1 protein promiscuously interacts with mRNA before translation. In addition, the traced preferential binding with 3′UTRs for UPF1 at steady state originate from the selective displacement of UPF1 from coding regions by translating ribosomes [76,77]. The phosphorylated UPF1 mRNA footprint data and CLIP (cross-linking immunoprecipitation)-Seq of UPF1 in vivo identified UPF1 binding sites across transcriptome, describing UPF1 targets 3′ UTR GC-rich motifs [17,18,19,20]. Particularly, Naoto et al. [19] suggested that in 3′ UTR the GC-rich motifs (5′-CCUGGGG-3′, 5′-CCUGGGA-3′, 5′-CCUGGAA-3′, and 5′-CCUGAGA-3′) can be the target motifs in UPF1-dependent mRNA decay. Due to such specificity of UPF1 towards a particular class of mRNA motifs, and to understand the UPF1-mRNA dynamics, we performed MD simulation of UPF1 protein with differentGC-rich RNA motifs, as well as performing a comparative analysis with the AU-rich motifs (5′-UUUUUUU-3′, 5′-UUAAUUU-3′, 5′-UUAGUUU-3′, and 5′-UUGAUUU-3′) [67]. Such novel perspectives from mRNA–protein-binding pairs identified in this work may contribute to understanding the selectivity of respective partners, along with advancing NMD-associated structural dynamics and kinetics.
2. Results and Discussion
2.1. Different UPF1 Isoforms and Their Selectivity towards mRNA Motifs
In mammals, two alternatively spliced isoforms for theUPF1 protein were identified that differed in length of the ‘regulatory loop’ from 1B domain, positioned at the mRNA binding interface. Such insertion of 11 aa (353-GNEDLVIIWLR-363) in the ‘regulatory loop’ made the UPF1 isoform_1 capable of a 2-fold increase in UPF1 translocation and ATPase activities [71] compared withUPF1 isoform_2. Applying the MDS approach, we identified the selectivity of amino acids from two different UPF1 isoforms with poly(U) mRNA motifs (Figure 2a,b). Binding interfaces and the number of protein–mRNA interactions (hydrogen bond; H-bond) theorized that UPF1 isoform_1 would havea higher number of interactions with poly(U) mRNA compared with that of the UPF1 isoform_2 (Figure 2b). We further traced the residues resulting instable hydrogen bonding with an individual mRNA (occupancy ≥ 10 %/ns; Figure 2a). The UPF1 isoform_1 was found forming interactions with each nucleotide of poly(U) mRNA, whereas the isoform_2 lacked such a binding pattern (Figure 2a). Apart from several isoform-specific UPF1-mRNA interactions (Figure 2a), the following amino acids (isoform_2/isoform_1) were common making interactions with mRNAs from both UPF1 isoforms: G851/G862, E645/E656, D622/D633, R422/R433, N524/N535, and T616/T627 (Figure 2a). Moreover, the presence of 11 aa insertion in the ‘regulatory loop’ for the UPF1 isoform_1 formed an interface for several residues to form H-bond interactions with the mRNA motifs—particularly the E355, N354, and R363 residues were involved in such binding (Figure 2a).
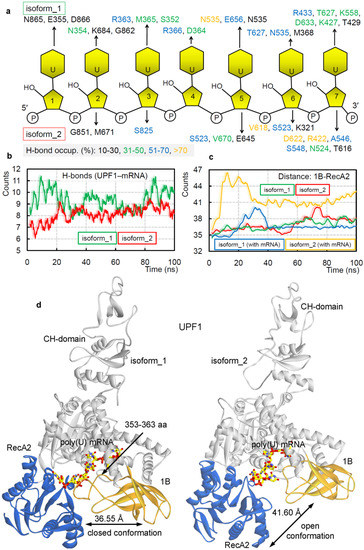
Figure 2.
Diversity in the binding of UPF1 isoforms with poly(U) mRNA, reflected in their structural dynamics. (a) Individual UPF1 amino acids making long-lasting interactions with the nucleotides having occupancy ≥10%/ns. (b) Intermolecular hydrogen bond interactions between UPF1 isoform_1 or isoform_2 with the poly(U) mRNA motifs. (c) The distance center of mass between 1B and RecA2 domains from both isoforms in the presence/absence of the poly(U) mRNA. (d) The structural dynamics of the ‘regulatory loop’ in presence (isoform_1) and absence of (isoform_2) of the 11 aa insertion (353-GNEDLVIIWLR-363). The averaged UPF1-mRNA coordinates extracted from 100 ns (nanosecond) MD (molecular dynamics) simulations, with labeled average distance marked based on data from panel (c).
The dynamics of different UPF1 isoforms over MD simulation (100 ns) time, demonstrates that the 11 aa insertion in the ‘regulatory loop’ induces movements between 1B and RecA2 domains. Considering such conformational switch, we computed the distance center of mass between 1B and RecA2 domains (Figure 2c). The findings highlight that in the absence of the insertion, UPF1 isoform_2 (with mRNA) has the highest distance, compared with that in theUPF1 isoform_1 (Figure 2c). On the contrary, the apo-form (UPF1 isoform_1 or isoform_2) systems lacked any such distinctive behavior (Figure 2c). These data suggest that intact binding between 1B and RecA2 domains in the presence of 11 aa insertion (isoform_1) within the ‘regulatory loop’ may be responsible for the induced catalytic and/or ATPase activity, as observed by Gowravaram et al. [71]. The averaged UPF1–mRNA coordinates extracted from 100 ns (nanosecond) MD simulations (Figure 2d) defines a ‘closed conformation (for isoform_1)’ and ‘open conformation (for isoform_2)’ between the 1B and RecA2 domains. In addition, similar conformational dynamics have been previously reported for one of the ATPases: DEAH-box adenosine triphosphatases by Florian et al. [73], their crystal structures [73] revealed that the RecA2 domain in presence of RNA formed an ‘open conformation’ and that the apo-form resembled a ‘closed conformation’. However, such two distinct conformations for the UPF1 protein were observed in our analysis having poly(U) mRNA in the system and in the presence or absence of the insertion in the ‘regulatory loop’ (Videos S1–S2).
2.2. GC- and AU-Rich mRNA Motifs Binding Pattern with UPF1 Isoforms
PTC-containing mRNAs are rapidly degraded via the NMD pathway (Figure 1), and it has been proposed that UPF1 (the master regulator) is often associated with these mRNAs [74,78]. In addition, the CLIP-Seq studies suggest that UPF1 binds mRNA rather non-specifically, but tends to accumulate in GC-rich motifs of 3′ UTRs [17,18,19]. Therefore, to investigate the binding pattern and selectivity of UPF1 with different mRNA motifs, we performed MD simulations on different GC-rich mRNA motifs with the UPF1protein, as well as made a comparative analysis with AU-rich sequences. For constructing different GC- and AU-rich mRNA motifs, model structures for the MD simulations with UPF1, the poly(U) mRNA (5′-UUUUUUU-3′) from the X-ray structure with UPF1 (pdb id: 2xzo [37]), was considered as the template. These eight investigated mRNA motifs (GC- (5′-CCUGGGG-3′, 5′-CCUGGGA-3′, 5′-CCUGGAA-3′, 5′-CCUGAGA-3′) and AU-rich motifs (5′-UUUUUUU-3′, 5′-UUAAUUU-3′, 5′-UUAGUUU-3′, 5′-UUGAUUU-3′), by our MD simulation approach, can be some of the many mRNAs with whichUPF1 can interact.
The intermolecular interactions of different protein–mRNA systems revealed that different mRNA motifs vary in binding affinity with UPF1 (isoform_1; Figure 3a). Explicitly, the 5′-CCUGGGG-3′, 5′-CCUGGGA-3′, 5′-CCUGGAA-3′, and 5′-CCUGAGA-3′mRNA motifs that represent 246 UPF1 targets [19] individually have slightly different affinity with the UPF1 isoform_1 (Figure 3a; left panel). Particularly, 5′-CCUGAGA-3′ and 5′-CCUGGAA-3′ have higher interactions with the UPF1 protein compared with other GC-rich motifs, whereas 5′-CCUGGGG-3′ has the lowest protein–mRNA interactions (Figure 3a; left panel). Among the AU-rich RNA motifs, the 5′-UUAAUUU-3′, 5′-UUAGUUU-3′, and 5′-UUGAUUU-3′ sequences represented a reduced binding with the UPF1 compared with that of the poly(U) mRNA (Figure 3a; right panel). The 5′-UUAGUUU-3′ motif has the least binding with UPF1 among AU-rich mRNA motifs (Figure 3a; right panel). The binding affinity of GC- and AU-rich mRNA motifs with UPF1 suggest that comparatively the GC-rich mRNA motifs have better binding in the majority of cases (Figure 3a). A corresponding trend of difference in their RMSDs (root-mean-square deviation of atomic positions) was traced, i.e., the 5′-CCUGGAA-3′ and poly(U) mRNA motifs were found to be highly stable throughout the MD simulations (Figure 3b). In addition, the 5′-CCUGGGG-3′ and 5′-UUGAUUU-3′ motifs with the least binding with UPF1 isoform_1 have higher flexibility (Figure 3b). Overall, the GC-rich motifs in the presence of UPF1 were found to show less fluctuation (more stable) in structure compared with that of the AU-rich motifs (isoform_1; Figure 3b).
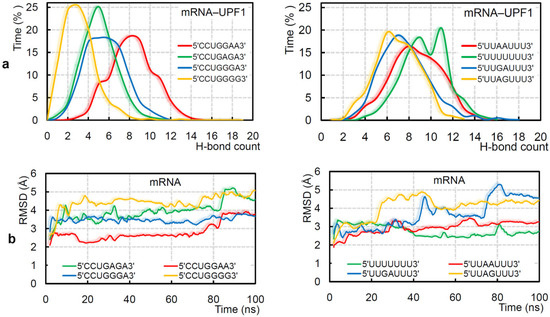
Figure 3.
Adopted binding patterns by UPF1 (isoform_1) with different mRNA motifs. (a) Frequency of the number of hydrogen bond interactions formed between UPF1 (isoform_1) and different GC-rich (5′-CCUGGGG-3′, 5′-CCUGGGA-3′, 5′-CCUGGAA-3′, and 5′-CCUGAGA-3′) or AU-rich (5′-UUUUUUU-3′, 5′-UUAAUUU-3′, 5′-UUAGUUU-3′, and 5′-UUGAUUU-3′) motifs. (b) Root-mean-square deviation (RMSD) of atomic positions (excluding hydrogen atoms) for the GC- and AU-rich mRNA motifs computed in the presence of the UPF1 protein.
Over the MD simulation time, it was observed that the poly(U) mRNA motif forms a 4-stack pattern when making interactions with UPF1 isoform_1 (Figure 4a,b), whereas the mRNA motif lacks such conformation in the presence of UPF1 isoform_2 (Figure 4b, bottom panel). These differences in the binding pattern of mRNA with two isoforms of UPF1 may explain the cause of induced catalytic activity for the UPF1 isoform_1 [71]. Particularly, among the GC-rich motifs, except 5′-CCUGGGG-3′, all other studied mRNA motifs formed a 4-stack pattern of binding with UPF1 (Figure 4a). Among the AU-rich motifs, the 5′-UUAAUUU-3′ and 5′-UUAGUUU-3′ formed a 3-stack and 4-stack pattern, respectively, whereas 5′-UUGAUUU-3′ lacked such formation (Figure 4b). Correlating the distance center of mass between 1B and RecA2 domains (Figure 4c; left panel) with the mRNA–UPF1 interactions (Figure 4c; right panel) suggests that as the distance between both domains increases (open conformation), the hydrogen bonds between mRNA–UPF1 (isoform_1) decreases (Figure 4c). For example, the system with 5′-CCUGGAA-3′ motif has less distance between 1B and RecA2 domains (closed conformation) and higher mRNA–UPF1 interactions, whereas a contrary behavior is observed for the 5′-CCUGGGG-3′ motif (isoform_1; open conformation; Figure 4c). Furthermore, we studied GC-rich mRNA motifs with UPF1 isoform_2, which is found to be abundant in cells, and compared findings with the UPF1 isoform_1 (Figures S2 and S3). The RMSD of individual GC-rich mRNA motifs with the both UPF1 isoforms, suggests that the same mRNA motifs behave differently when bound with isoform_1 or isoform_2 (Figure S2)—i.e., mRNA motifs with UPF1 isoform_2 were found to be highly flexible when compared with that of the UPF1 isoform_1. The 5′-CCUGGGG-3′ motif showed the least binding with UPF1 isoform_2, similar as with the UPF1 isoform_1 (Figure S2). These weak-binding mRNA motifs with the UPF1 isoform_2 resulted in an increased distance between 1B and RecA2 domains (open conformation; Figure S2).
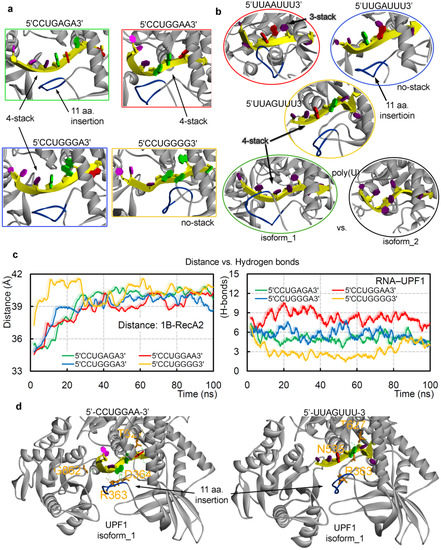
Figure 4.
The dynamics of different UPF1-mRNA simulated systems. The (a,b) stacking pattern interactions formed by different GC- and AU-rich mRNA motifs with UPF1 isoforms and averaged structures retrieved from the 100 ns MD simulations are presented. For UPF1 isoform_1, the insertion 353-GNEDLVIIWLR-363 is marked in blue color. (c) The distance center of mass computed between 1B and RecA2 domains of UPF1 isoform_1 with GC-rich mRNA motifs (left panel), as well as the UPF1-mRNA intermolecular hydrogen bond interactions (right panel). (d) UPF1 isoform_1 residues that were found common, as well as those making high occupancy (≥50 ns) interactions with GC-rich motifs (R363, D364, T627, and G862) or AU-rich motifs (R363, N535, and T627). As an example, the 5′-UUAGUUU-3′ and 5′-CCUGGAA-3′ motifs are represented with the UPF1 protein. Nucleotide color scheme; adenine (A) in red color, uracil (U) in dark purple, cytosine (C) in light purple, and guanine (G) in green.
Additionally, UPF1 isoform_1 with the 5′-UUGAUUU-3′ motif formed an ‘open conformation’ between 1B-RecA2 domains, whereas the other two AU-rich systems (5′-UUAAUUU-3′ and 5′-UUAGUUU-3′) showed a ‘closed conformation’ (similar to when complexed with the poly(U) mRNA motif). These ‘open conformations’ of the UPF1 (isoform_1) protein in the presence of the 5′-CCUGGGG-3′ and 5′-UUGAUUU-3′ mRNA motifs correlated with the conformation dynamics of UPF1 isoform_2 which is suggested to have comparatively less catalytic activity [71]. Our findings propose that to induce the catalytic activity of the UPF1 helicase, a ‘closed conformation’ between 1B and RecA2 domains, as well as a specific stacking pattern in the mRNA transcript may be needed (Figure 4). Furthermore, tracing the interacting residues of the UPF1 protein with different mRNA motifs (Tables S1 and S2) demonstrated that a few amino acids were found to be common, resulting in a high occupancy (≥50 ns; Figure 4d) binding with GC-rich motifs (R363, D364, T627, and G862) or AU-rich motifs (R363, N535, and T627).
2.3. UPF1A839T and UPF1P533L/T Cancer Mutants Influencing the UPF1-mRNA Binding
Point mutation often alters the normal functioning of proteins. Therefore, to investigate such an effect on the UPF1 (isoform_1) helicase activity, we performed MD simulations upon inserting the commonly occurring UPF1 cancer variants in the presence of poly(U) mRNA—A839T and P533L/T (cBioPortal [79])—positioned at the mRNA-binding interface. Upon inserting mutations in the UPF1 protein, a reduction in the protein–mRNA bindings was observed, and such significant changes were traced for the UPF1P533T system. The change in binding motifs of UPF1 upon mutating with poly(U) mRNA was identified, and amino acids from the insertion (353-GNEDLVIIWLR-363; isoform_1) in the ‘regulatory loop’ were found making stable interactions with the mRNA motifs (Figure 5a,b). Interestingly, the mutated UPF1P533L/T residue itself was found interacting with mRNA in the mutated as well as the wild-type systems, whereas the protein with mutated residue A839T lacked such behavior with the mRNA motif. Particularly, the UPF1A839T system had a distinctive interaction with mRNA compared with other simulated systems (Figure 5b). For example, the residue E614 formed stable (highest occupancy) interactions with mRNA in UPF1A839T, whereas other all simulated systems lacked such binding (Figure 5b). Moreover, residues Y316, Q369, and T429 were binding with mRNA only in the wild-type systems, and similarly, residues K321, G353, G630, G681, and Q841 formed interactions only in the mutated systems (Figure 5b).
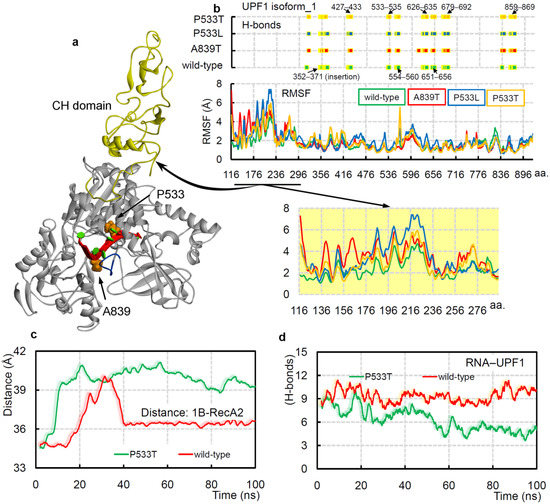
Figure 5.
The cancer variants UPF1A839T and UPF1P533L/T can influence the protein–mRNA binding dynamics. (a) UPF1 isoform_1 structure representing mutated residues in its RNA-binding pocket, and the CH domain is colored in yellow. The 11 aa insertion (353-GNEDLVIIWLR-363) for UPF1 isoform_1 is also highlighted in blue. (b) The protein–poly(U) mRNA intermolecular hydrogen bond interactions in wild-type and different mutated systems. Amino acids from the following range of the UPF1 isoform_1 protein were involved in poly(U) mRNA binding: S352–D371, K427–R433, P533–N535, R554–R560, C626–R635, Q651–E656, P679–S692, S836–Q841, and Q859–R869. The bottom panel represents RMSFs (root-mean-square fluctuations) of individual amino acids of UPF1 (isoform_1) under different conditions. The regions formed by the CH-domain showing high RMSF fluctuations are highlighted. (c) The distance center of mass between 1B and RecA2 domains of UPF1 isoform_1 with poly(U) mRNA in the UPF1P533Tmutated and wild-type systems. (d) The UPF1–poly(U) mRNA intermolecular interactions from the UPF1P533T and wild-type systems.
Overall, the RMSF (root-mean-square fluctuations) findings suggest that the UPF1 (isoform_1) amino acids have higher flexibility in the mutated systems (Figure 5b, bottom panel) compared with that of the wild-type form. In addition, significant RMSF fluctuation differences were observed in the CH domain (cysteine-histidine-rich domain), which could be an allosteric effect upon inserting mutations in the RNA-binding sites (Figure 5b). The conformational dynamics of protein upon inserting cancer variants in the presence or absence of mRNA suggest that the UPF1P533T system in both conditions constitute an ‘open conformation’ for the 1B and RecA2 domains, whereas the UPF1P533L and UPF1A839T lack such domain displacements. Moreover, in the UPF1P533T system, the mRNA does not form a stable stacking pattern, as is observed in the wild-type system (isoform_1). Comparing the distance center of mass between the 1B and RecA2 domains with the intermolecular UPF1-mRNA interactions revealed that as the distance between domains gradually increases, the protein–mRNA interactions decreased (Figure 5c,d, and Video S3). These data could support the hypothesis that the ‘open conformation’ between 1B and RecA2 domains may reduce the catalytic activity, whereas the ‘closed conformation’ can induce such activity.
3. Materials and Methods
Systems Build-Up for Different mRNA Motifs with UPF1
Different structures of UPF1 with ATP or mRNA are available in the Protein Data Bank database (pdb; http://www.rcsb.org/pdb; accessed on March 2021); however, the majority of them lack the CH domain, which is crucial for the proper functioning of the UPF1 protein. Therefore, in this study, we used the complete optimized UPF1structure (pdb id: 2wjv [67]) retrieved from our previous study [40]. The UPF1 (pdb id: 2wjv [67]) is an isoform_2 crystal structure having 11 aa less (353-GNEDLVIIWLR-363) in the ‘regulatory loop’ compared with the isoform_1 (http://www.rcsb.org/pdb; accessed on March 2021). The UPF1 isoform_1 structure was modeled by considering the isoform_2 crystal structure as the template (pdb id: 2wjv [67]; Figure 1a,b). The missing amino acids in the tertiary structures of UPF1 in both isoforms were built using the Swiss model, as well as using the ‘prepare protein’ protocol implemented in the macromolecules module of the BIOVIA Discovery Studio Client v18.1 (Dassault Systèmes, BIOVIA Corp., San Diego, CA, USA) program [41,80]. Superimposing both UPF1 isoform modeled structures (build based on the pdb id: 2wjv [67]) with different structures available (UPF1 isoform_2 (pdb id: 2xzp [37]) and UPF1_isoform_1 (pdb id: 6ej5 [71])) suggests a strong correlation between conformations (Figure S1). The UPF1 crystal structure containing the CH domain (pdb id: 2wjv [67]) lacks the mRNA, and therefore, the position of the poly(U) mRNA motif was used from the pdb id: 2xzo X-ray structure (UPF1 without the CH domain) [37]. Using the ‘superimpose module’ in BIOVIA Discovery Studio v18.1 package, the position of poly(U) mRNA with both UPF1 isoforms was defined. The minimization of the modeled UPF1–poly(U) mRNA systems were performed using the ‘smart minimizer’ algorithm and applying the CHARMm (Chemistry at Harvard Macromolecular Mechanics) forcefield [81], with other parameters set as default in the BIOVIA Discovery Studio v18.1 package.
Systems for the MD simulations of UPF1 with GC-rich (5′-CCUGGGG-3′, 5′-CCUGGGA-3′, 5′-CCUGGAA-3′, and 5′-CCUGAGA-3′) and AU-rich (5′-UUUUUUU-3′, 5′-UUAAUUU-3′, 5′-UUAGUUU-3′, and 5′-UUGAUUU-3′) mRNA motifs were generated using the Molecular Operating Environment (MOE; Chemical Computing Group Inc., Montreal, QC, Canada) package. A similar position as that of the poly(U) mRNA motif with UPF1 from the X-ray structure (pdb id: 2xzo) [37] was used for these GC/AU-rich mRNA–UPF1 systems. Subsequently, the UPF1 protein with different mRNA motifs were optimized in the MOE package applying the CHARMM27 forcefield [82]. The cancer mutations retrieved from the cancer genomics database cBioPortal [79] were inserted in the UPF1 structure using the MOE modules. Tertiary coordinates of mRNA as input parameters for the GROMACS 4.6.5 package [83] were retrieved using the CHARMM-GUI server [84]. Applying CHARMM27 forcefield, the following 21 different protein–RNA systems were simulated using GROMACS 4.6.5 [85]: UPF1 isoform_1 apo-form and with poly(U) mRNA; UPF1 isoform_2 apo-form and with poly(U) mRNA; UPF1 isoform_1 and isoform_2 with GC-rich mRNA motifs (5′-CCUGGGG-3′, 5′-CCUGGGA-3′, 5′-CCUGGAA-3′, and 5′-CCUGAGA-3′); UPF1 isoform_1 with AU-rich mRNA motifs (5′-UUAAUUU-3′, 5′-UUAGUUU-3′, and 5′-UUGAUUU-3′) and the mutated UPF1 isoform_1 (A839T, P533L, or P533T) apo-form and with poly(U) mRNA. These protein–mRNA systems were simulated in a 10 Å thick dodecahedron box and solvated using simple point charge (SPC) water molecules [86]. Net charge of the individual system was neutralized by adding the appropriate number of Na+ and Cl− counter ions in the MD simulation box. Periodic boundary conditions were employed in all three dimensions, and to relax possible steric crashes, the energy of each system was minimized for 50,000 steps of the steepest descent algorithm. Different electrostatic interactions were treated using the particle mesh Ewald (PME) method [87], and the LINCS (linear constraint solver) algorithm [88] was used to constrain the bond lengths for each simulated system. In addition, the cutoff distance for van der Waals and Coulomb interactions were set to 10 Å. Subsequently, all modeled systems were equilibrated in an NpT-ensemble (isobaric–isothermal) simulation for 1000 ps. The temperature and pressure were maintained at 300 K and 1 bar using V-rescale thermostat [88] and Parrinello–Rahman barostat [89], respectively. A leapfrog integrator [90] was used to propagate the dynamics of each system. The MDS production run was carried out for 100 ns, and the trajectories/coordinates were saved every 10 ps (picosecond), which were analyzed using GROMACS tools. Hydrogen bonds (H-bonds) were calculated using a donor-acceptor atom cutoff distance of 3.5 Å and intermolecular donor-H-acceptor angle cutoff ≥160°–180°. The VMD (Visual Molecular Dynamics) tool [91] was used to visualize the MD simulated trajectories, and an MOE/BIOVIA Discovery Studio visualizer was used for the protein–mRNA structure representation.
4. Conclusions
In this work, we investigated two alternatively-spliced UPF1 isoforms with different mRNA motifs, and we traced significant structural dynamics by inserting cancer-derived mutations in the UPF1 mRNA binding pocket. To this end, we suggest that 11 aa insertion (353-GNEDLVIIWLR-363) in the ‘regulatory loop’ could be responsible for the ‘open (isoform_2)’ and ‘closed (isoform_1)’ conformations between 1B and RecA2 domains, which may impact the catalytic activity of the UPF1 helicase. These two distinct conformations of UPF1 correspond to the important stacking pattern observed in the mRNA motifs, i.e., the absence of a stacking formation in isoform_2 results in an ‘open conformation’. The amino acids E355, N354, and R363 from the insertion region for UPF1 isoform_1 induced interactions with the mRNA motif.
Binding affinities of GC- and AU-rich mRNA motifs with UPF1 highlighted that GC-rich mRNAs have better binding with the protein in the majority of studied cases. In addition, when complexed with the UPF1 protein, the GC-rich motifs have lower fluctuations in their structure compared with that of the AU-rich motifs. Except for 5′-CCUGGGG-3′, all other simulated GC-rich mRNA motifs formed a 4-stack binding pattern with the UPF1 protein. Additionally, it was observed that as the distance between 1B-RecA2 domain increases forming an ‘open conformation’, it has a decline in the UPF1-mRNA intermolecular interactions. For example, the system with 5′-CCUGGAA-3′ motif had less distance between 1B-RecA2 domains (closed conformation) and higher mRNA–UPF1 interactions, whereas contrary behavior was observed for the 5′-CCUGGGG-3′ (open conformation) sequence. Among the AU-rich motifs, the poly(U) mRNA had the highest binding affinity with the UPF1 and that with least binding affinity was the 5′-UUAGUUU-3′ motif. Particularly, 5′-UUAAUUU-3′ and 5′-UUAGUUU-3′ formed a 3-stack and 4-stack pattern, respectively, whereas 5′-UUGAUUU-3′ lacked such formation, resulting in an ‘open conformation’ between 1B-RecA2 domains.
A few high occupancy R363, D364, T627, and G862 residues from UPF1 isoform_1 were common binders among the GC-rich motifs, as were residues R363, N535, and T627 among the AU-rich motifs. Comparing different GC-rich mRNA motifs binding with both UPF1 isoforms highlighted that mRNA motifs behave slightly differently when bound with either of the isoforms, and particularly, mRNA motifs with UPF1 isoform_2 were more flexible. The amino acid positioned at 533 in the UPF1 isoform_1 protein was found interacting with mRNA in the mutated (UPF1P533L/T) as well as in the wild-type systems, whereas the UPF1A839T systems have distinctive interactions with the mRNA motif. Particularly, in the cancer-associated mutated systems, the mRNA motif with UPF1P533T lacks the stacking interactions, due to which the distance between 1B-RecA2 domains increased and the intermolecular protein–mRNA interactions showed a declining trend. From these data it could be proposed that the increased affinity betweenUPF1-mRNA components, should contribute to the enhanced RNA-dependent ATPase/helicase activity of the UPF1 protein that is necessary for the NMD pathway. These novel perspectives from identified mRNA–UPF1 binding pairs can contribute to understanding the selectivity of respective partners, as well as advancing NMD-associated dynamics and kinetics.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms222312744/s1.
Author Contributions
Conceptualization, M.P., T.H. and U.K.; data curation, M.P. and U.K.; formal analysis, M.P. and U.K.; funding acquisition, U.K.; investigation, U.K.; methodology, M.P. and U.K.; resources, U.K.; supervision, U.K.; visualization, M.P. and U.K.; writing—original draft, M.P. and U.K.; writing—review and editing, M.P., R.F., T.H. and U.K. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was funded by grant 2020/36/C/NZ2/00108 (The National Science Centre, Poland). U.K. is supported by grant 2020/36/C/NZ2/00108 from The National Science Centre, Poland (Narodowe Centrum Nauki, Krakow, Poland).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or supplementary material.
Acknowledgments
The International Centre for Cancer Vaccine Science project was carried out within the International Research Agendas programme of the Foundation for Polish Science, co-financed by the European Union under the European Regional Development Fund. Authors would also like to thank the PL-Grid Infrastructure, Poland, for providing their hardware and software resources.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| aa | amino acids |
| CLIP | cross-linking immunoprecipitation |
| CH domain | cysteine-histidine-rich domain |
| CHARMm | Chemistry at Harvard Macromolecular Mechanics |
| DECID | decay-inducing complex |
| EJC | exon–exon junction complex |
| eRF1 | eukaryotic translation termination factor 1 |
| H-bond | hydrogen bond |
| LINCS | linear constraint solver |
| mRNP | messenger ribonucleoprotein |
| MDS | molecular dynamics simulation |
| MOE | Molecular Operating Environment |
| NGD | no-go decay |
| NMD | nonsense-mediated mRNA decay |
| NSD | non-stop decay |
| ns | nanosecond |
| PAB1 | poly(A)-binding protein cytoplasmic 1 |
| pdb | protein data bank |
| PME | particle mesh Ewald |
| PTC | premature termination codon |
| ps | picosecond |
| RMSD | root-mean-square deviation |
| RMSF | root-mean-square fluctuations |
| SMG | suppressors with morphogenetic effect on genitalia |
| SPC | simple point charge |
| uORF | upstream open reading frames |
| UPF1 | UP-frameshift 1 |
| UTR | untranslated region |
| VMD | Visual Molecular Dynamics |
References
- Stefani, M.; Dobson, C.M. Protein aggregation and aggregate toxicity: New insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. 2003, 81, 678–699. [Google Scholar] [CrossRef]
- Gregersen, N.; Bross, P.; Vang, S.; Christensen, J.H. Protein Misfolding and Human Disease. Annu. Rev. Genom. Hum. Genet. 2006, 7, 103–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, R.; Müller, L.; Wullich, B. Protein transport into the endoplasmic reticulum: Mechanisms and pathologies. Trends Mol. Med. 2006, 12, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Hebert, D.N.; Molinari, M. In and Out of the ER: Protein Folding, Quality Control, Degradation, and Related Human Diseases. Physiol. Rev. 2007, 87, 1377–1408. [Google Scholar] [CrossRef]
- Jarjanazi, H.; Savas, S.; Pabalan, N.; Dennis, J.W.; Ozcelik, H. Biological implications of SNPs in signal peptide domains of human proteins. Proteins Struct. Funct. Bioinform. 2007, 70, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Hipp, M.S.; Park, S.-H.; Hartl, F.U. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014, 24, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Karamyshev, A.L.; Karamysheva, Z.N. Lost in Translation: Ribosome-Associated mRNA and Protein Quality Controls. Front. Genet. 2018, 9, 431. [Google Scholar] [CrossRef] [Green Version]
- Karamyshev, A.L.; Tikhonova, E.B.; Karamysheva, Z.N. Translational Control of Secretory Proteins in Health and Disease. Int. J. Mol. Sci. 2020, 21, 2538. [Google Scholar] [CrossRef] [Green Version]
- Welch, E.M.; Jacobson, A. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 1999, 18, 6134–6145. [Google Scholar] [CrossRef] [Green Version]
- Doma, M.K.; Parker, R. RNA Quality Control in Eukaryotes. Cell 2007, 131, 660–668. [Google Scholar] [CrossRef] [Green Version]
- Shoemaker, C.J.; Green, R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012, 19, 594–601. [Google Scholar] [CrossRef]
- Popp, M.W.-L.; Maquat, L.E. Organizing Principles of Mammalian Nonsense-Mediated mRNA Decay. Annu. Rev. Genet. 2013, 47, 139–165. [Google Scholar] [CrossRef] [Green Version]
- Lewis, B.P.; Green, R.; Brenner, S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. USA 2002, 100, 189–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, P.; Mühlemann, O. Cutting the nonsense: The degradation of PTC-containing mRNAs. Biochem. Soc. Trans. 2010, 38, 1615–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frischmeyer, P.A.; Dietz, H.C. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999, 8, 1893–1900. [Google Scholar] [CrossRef] [Green Version]
- Mort, M.; Ivanov, D.; Cooper, D.N.; Chuzhanova, N.A. A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 2008, 29, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Hurt, J.A.; Robertson, A.D.; Burge, C.B. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013, 23, 1636–1650. [Google Scholar] [CrossRef] [Green Version]
- Colombo, M.; Karousis, E.; Bourquin, J.; Bruggmann, R.; Mühlemann, O. Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. RNA 2016, 23, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Imamachi, N.; Salam, K.A.; Suzuki, Y.; Akimitsu, N. A GC-rich sequence feature in the 3′ UTR directs UPF1-dependent mRNA decay in mammalian cells. Genome Res. 2016, 27, 407–418. [Google Scholar] [CrossRef] [Green Version]
- Karousis, E.D.; Gurzeler, L.A.; Annibaldis, G.; Dreos, R.; Mühlemann, O. Human NMD ensues independently of stable ribosome stalling. Nat. Commun. 2020, 11, 4134. [Google Scholar] [CrossRef]
- Yepiskoposyan, H.; Aeschimann, F.; Nilsson, D.; Okoniewski, M.; Mühlemann, O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 2011, 17, 2108–2118. [Google Scholar] [CrossRef] [Green Version]
- Thoren, L.A.; Nørgaard, G.A.; Weischenfeldt, J.; Waage, J.; Jakobsen, J.S.; Damgaard, I.; Bergström, F.C.; Blom, A.M.; Borup, R.; Bisgaard, H.C.; et al. UPF2 is a critical regulator of liver development, function and regeneration. PLoS ONE 2010, 5, e11650. [Google Scholar] [CrossRef] [PubMed]
- Kuzmiak, H.A.; Maquat, L.E. Applying nonsense-mediated mRNA decay research to the clinic: Progress and challenges. Trends Mol. Med. 2006, 12, 306–316. [Google Scholar] [CrossRef]
- Khajavi, M.; Inoue, K.; Lupski, J.R. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur. J. Hum. Genet. 2006, 14, 1074–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- A Holbrook, J.; Neu-Yilik, G.; Hentze, M.; E Kulozik, A. Nonsense-mediated decay approaches the clinic. Nat. Genet. 2004, 36, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Czaplinski, K.; Weng, Y.; Hagan, K.W.; Peltz, S.W. Purification and characterization of the Upf1 protein: A factor involved in translation and mRNA degradation. RNA 1995, 1, 610–623. [Google Scholar] [PubMed]
- Atkin, A.L.; Altamura, N.; Leeds, P.; Culbertson, M.R. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol. Biol. Cell 1995, 6, 611–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, Y.; Czaplinski, K.; Peltz, S.W. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol. Cell. Biol. 1996, 16, 5491–5506. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, A.; Czaplinski, K.; Trifillis, P.; He, F.; Jacobson, A.; Peltz, S.W. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 2000, 6, 1226–1235. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Perlick, H.A.; Dietz, H.C.; Maquat, L.E. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 1998, 95. [Google Scholar] [CrossRef] [Green Version]
- Lasalde, C.; Rivera, A.V.; León, A.J.; González-Feliciano, J.A.; Estrella, L.A.; Rodríguez-Cruz, E.N.; Correa, M.E.; Cajigas, I.J.; Bracho, D.P.; Vega, I.E.; et al. Identification and functional analysis of novel phosphorylation sites in the RNA surveillance protein Upf1. Nucleic Acids Res. 2013, 42, 1916–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, Y.; Czaplinski, K.; Peltz, S.W. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol. Cell. Biol. 1996, 16, 5477–5490. [Google Scholar] [CrossRef] [Green Version]
- Atkin, A.L.; Schenkman, L.R.; Eastham, M.; Dahlseid, J.N.; Lelivelt, M.J.; Culbertson, M.R. Relationship between Yeast Polyribosomes and Upf Proteins Required for Nonsense mRNA Decay. J. Biol. Chem. 1997, 272, 22163–22172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, Y.; Czaplinski, K.; Peltz, S.W. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA 1998, 4, 205–214. [Google Scholar]
- Kadlec, J.; Guilligay, D.; Ravelli, R.B.; Cusack, S. Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA 2006, 12, 1817–1824. [Google Scholar] [CrossRef] [Green Version]
- He, F.; Brown, A.H.; Jacobson, A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 1997, 17, 1580–1594. [Google Scholar] [CrossRef] [Green Version]
- Chakrabarti, S.; Jayachandran, U.; Bonneau, F.; Fiorini, F.; Basquin, C.; Domcke, S.; Le Hir, H.; Conti, E. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell 2011, 41, 693–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serin, G.; Gersappe, A.; Black, J.D.; Aronoff, R.; Maquat, L.E. Identification and Characterization of Human Orthologues to Saccharomyces cerevisiae Upf2 Protein and Upf3 Protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol. 2001, 21, 209–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamieh, H.; Ballut, L.; Bonneau, F.; Le Hir, H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 2007, 15, 85–93. [Google Scholar] [CrossRef]
- Kalathiya, U.; Padariya, M.; Pawlicka, K.; Verma, C.S.; Houston, D.; Hupp, T.R.; Alfaro, J.A. Hupp Insights into the Effects of Cancer Associated Mutations at the UPF2 and ATP-Binding Sites of NMD Master Regulator: UPF1. Int. J. Mol. Sci. 2019, 20, 5644. [Google Scholar] [CrossRef] [Green Version]
- Kashima, I.; Yamashita, A.; Izumi, N.; Kataoka, N.; Morishita, R.; Hoshino, S.; Ohno, M.; Dreyfuss, G.; Ohno, S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006, 20, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Echevarría, M.J.; González, C.I.; Peltz, S.W. Identifying the right stop: Determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998, 17, 575–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, J.; Sato, H.; Tang, Y.; Matsuda, D.; Maquat, L.E. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol. Cell 2010, 39, 396–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amrani, N.; Ganesan, R.; Kervestin, S.; Mangus, D.A.; Ghosh, S.; Jacobson, A. A faux 3′-UTR promotes aberrant termination and triggers nonsense- mediated mRNA decay. Nature 2004, 432, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Lejeune, F.; E Maquat, L. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 2005, 17, 309–315. [Google Scholar] [CrossRef]
- Kuroha, K.; Tatematsu, T.; Inada, T. Upf1 stimulates degradation of the product derived from aberrant messenger RNA containing a specific nonsense mutation by the proteasome. EMBO Rep. 2009, 10, 1265–1271. [Google Scholar] [CrossRef] [Green Version]
- Carter, M.S.; Li, S.; Wilkinson, M.F. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996, 15, 5965–5975. [Google Scholar] [CrossRef]
- Kervestin, S.; Li, C.; Buckingham, R.; Jacobson, A. Testing the faux-UTR model for NMD: Analysis of Upf1p and Pab1p competition for binding to eRF3/Sup35p. Biochimie 2012, 94, 1560–1571. [Google Scholar] [CrossRef] [Green Version]
- Ohnishi, T.; Yamashita, A.; Kashima, I.; Schell, T.; Anders, K.; Grimson, A.; Hachiya, T.; Hentze, M.; Anderson, P.; Ohno, S. Phosphorylation of hUPF1 Induces Formation of mRNA Surveillance Complexes Containing hSMG-5 and hSMG-7. Mol. Cell 2003, 12, 1187–1200. [Google Scholar] [CrossRef]
- Hosoda, N.; Kim, Y.K.; Lejeune, F.; E Maquat, L. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2005, 12, 893–901. [Google Scholar] [CrossRef] [PubMed]
- González, C.I.; Ruiz-Echevarría, M.J.; Vasudevan, S.; Henry, M.F.; Peltz, S.W. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell 2000, 5, 489–499. [Google Scholar] [CrossRef]
- Lykke-Andersen, J.; Shu, M.-D.; Steitz, J.A. Human Upf Proteins Target an mRNA for Nonsense-Mediated Decay When Bound Downstream of a Termination Codon. Cell 2000, 103, 1121–1131. [Google Scholar] [CrossRef] [Green Version]
- Shibuya, T.; Tange, T.; Stroupe, M.E.; Moore, M.J. Mutational analysis of human eIF4AIII identifies regions necessary for exon junction complex formation and nonsense-mediated mRNA decay. RNA 2006, 12, 360–374. [Google Scholar] [CrossRef] [Green Version]
- Gehring, N.H.; Kunz, J.B.; Neu-Yilik, G.; Breit, S.; Viegas, M.H.; Hentze, M.W.; Kulozik, A.E. Exon-Junction Complex Components Specify Distinct Routes of Nonsense-Mediated mRNA Decay with Differential Cofactor Requirements. Mol. Cell 2005, 20, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Ohnishi, T.; Kashima, I.; Taya, Y.; Ohno, S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001, 15, 2215–2228. [Google Scholar] [CrossRef] [Green Version]
- Popp, M.W.; Maquat, L.E. Leveraging Rules of Nonsense-Mediated mRNA Decay for Genome Engineering and Personalized Medicine. Cell 2016, 165, 1319–1322. [Google Scholar] [CrossRef] [Green Version]
- Conti, E.; Izaurralde, E. Nonsense-mediated mRNA decay: Molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 2005, 17, 316–325. [Google Scholar] [CrossRef]
- Amrani, N.; Sachs, M.; Jacobson, A.J. Early nonsense: mRNA decay solves a translational problem. Nat. Rev. Mol. Cell Biol. 2006, 7, 415–425. [Google Scholar] [CrossRef]
- Amrani, N.; Dong, S.; He, F.; Ganesan, R.; Ghosh, S.; Kervestin, S.; Li, C.; Mangus, D.A.; Spatrick, P.; Jacobson, A. Aberrant termination triggers nonsense-mediated mRNA decay. Biochem. Soc. Trans. 2006, 34, 39–42. [Google Scholar] [CrossRef]
- Rebbapragada, I.; Lykke-Andersen, J. Execution of nonsense-mediated mRNA decay: What defines a substrate? Curr. Opin. Cell Biol. 2019, 21, 394–402. [Google Scholar] [CrossRef]
- Behm-Ansmant, I.; Kashima, I.; Rehwinkel, J.; Saulière, J.; Wittkopp, N.; Izaurralde, E. mRNA quality control: An ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007, 581, 2845–2853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Hir, H.; Gatfield, D.; Izaurralde, E.; Moore, M.J. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001, 20, 4987–4997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isken, O.; Kim, Y.K.; Hosoda, N.; Mayeur, G.L.; Hershey, J.W.; Maquat, L.E. Upf1 Phosphorylation Triggers Translational Repression during Nonsense-Mediated mRNA Decay. Cell 2008, 133, 314–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huntzinger, E.; Braun, J.E.; Heimstädt, S.; Zekri, L.; Izaurralde, E. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J. 2010, 29, 4146–4160. [Google Scholar] [CrossRef] [Green Version]
- Kebaara, B.W.; Atkin, A.L. Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res. 2009, 37, 2771–2778. [Google Scholar] [CrossRef] [Green Version]
- Muhlrad, D.; Parker, R. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 1999, 5, 1299–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clerici, M.; Mourão, A.; Gutsche, I.; Gehring, N.H.; Hentze, M.; Kulozik, A.; Kadlec, J.; Sattler, M.; Cusack, S. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 2009, 28, 2293–2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safaee, N.; Kozlov, G.; Noronha, A.M.; Xie, J.; Wilds, C.; Gehring, K. Interdomain Allostery Promotes Assembly of the Poly(A) mRNA Complex with PABP and eIF4G. Mol. Cell 2012, 48, 375–386. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Sternberg, M.J.E. Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Roy, B.; He, F.; Jacobson, A. Nonsense suppression position effect implicates poly(A)-binding protein in the regulation of translation termination. SSRN Electron. J. 2019. [Google Scholar] [CrossRef]
- Gowravaram, M.; Bonneau, F.; Kanaan, J.; Maciej, V.D.; Fiorini, F.; Raj, S.; Croquette, V.; Le Hir, H.; Chakrabarti, S. A conserved structural element in the RNA helicase UPF1 regulates its catalytic activity in an isoform-specific manner. Nucleic Acids Res. 2018, 46, 2648–2659. [Google Scholar] [CrossRef] [Green Version]
- Kurosaki, T.; Maquat, L.E. Rules that govern UPF1 binding to mRNA 3′ UTRs. Proc. Natl. Acad. Sci. USA 2013, 110, 3357–3362. [Google Scholar] [CrossRef] [Green Version]
- Hamann, F.; Enders, M.; Ficner, R. Structural basis for RNA translocation by DEAH-box ATPases. Nucleic Acids Res. 2019, 47, 4349–4362. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Zhang, H.; Wang, L.; Zhu, C.; Sheng, K.; Du, Y.; Wang, K.; Dias, A.; Chen, S.; Whitman, M.; et al. Premature termination codons are recognized in the nucleus in a reading-frame-dependent manner. Cell Discov. 2015, 1, 15001. [Google Scholar] [CrossRef] [Green Version]
- Hogg, J.R.; Goff, S.P. Upf1 Senses 3′UTR Length to Potentiate mRNA Decay. Cell 2010, 143, 379–389. [Google Scholar] [CrossRef] [Green Version]
- Zünd, D.; Gruber, A.R.; Zavolan, M.; Mühlemann, O. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat. Struct. Mol. Biol. 2013, 20, 936–943. [Google Scholar] [CrossRef]
- Lee, S.R.; Pratt, G.A.; Martinez, F.J.; Yeo, G.W.; Lykke-Andersen, J. Target Discrimination in Nonsense-Mediated mRNA Decay Requires Upf1 ATPase Activity. Mol. Cell 2015, 59, 413–425. [Google Scholar] [CrossRef] [Green Version]
- Hoernes, T.P.; Clementi, N.; Juen, M.A.; Shi, X.; Faserl, K.; Willi, J.; Gasser, C.; Kreutz, C.; Joseph, S.; Lindner, H.; et al. Atomic mutagenesis of stop codon nucleotides reveals the chemical prerequisites for release factor-mediated peptide release. Proc. Natl. Acad. Sci. USA 2018, 115, E382–E389. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Brooks, B.R.; Brooks, C.L., III; MacKerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef]
- Bjelkmar, P.; Larsson, P.; Cuendet, M.A.; Hess, B.; Lindahl, E. Implementation of the CHARMM Force Field in GROMACS: Analysis of Protein Stability Effects from Correction Maps, Virtual Interaction Sites, and Water Models. J. Chem. Theory Comput. 2010, 6, 459–466. [Google Scholar] [CrossRef]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [Green Version]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction Models for Water in Relation to Protein Hydration. In Intermolecular Forces. The Jerusalem Symposia on Quantum Chemistry and Biochemistry; Pullman, B., Ed.; Springer: Dordrecht, The Netherlands, 1981; Volume 14. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 19, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Van Gunsteren, W.F.; Berendsen, H.J.C. A Leap-frog Algorithm for Stochastic Dynamics. Mol. Simul. 1988, 1, 173–185. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).