Long Tandem Arrays of Cassandra Retroelements and Their Role in Genome Dynamics in Plants
Abstract
1. Introduction
2. Results
2.1. In Silico Identification of the Cassandra Retrotransposon and Long Tandem Arrays in Plant Genomes
2.2. Long Distance PCR for Complete Tandem Cassandra Cluster Isolation
2.3. Cassandra Tandem Repeats in mRNA and Genomic DNA
2.4. Cassandra Secondary Structure
2.5. Cassandra Copy Number Variation
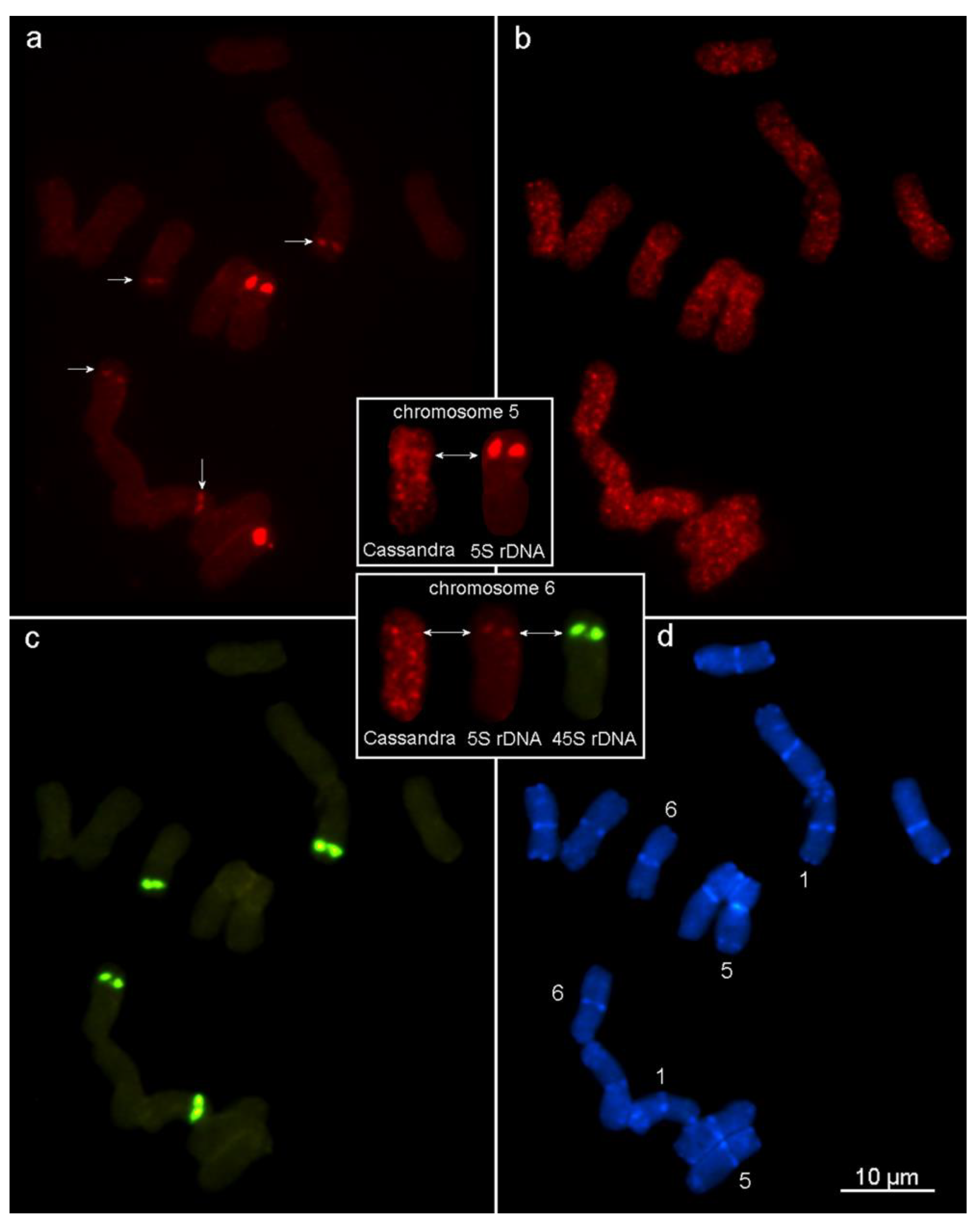
2.6. Chromosomal Distribution of Cassandra
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Computation Analysis and Alignment
4.3. Phylogenetic Analyses and Tree Building
4.4. In Silico Query for Cassandra in Plant Genomes
4.5. Primer Design
4.6. DNA Extraction
4.7. Long-Distance Inverted PCR to Isolate Cassandra Tandems
4.8. Gel Electrophoresis
4.9. Cloning PCR Fragments
4.10. Quantitative Real-Time PCR and Relative Quantification
4.11. Cassandra TRIM Element Copy Number Estimation by Dot Blot
4.12. Probe Labeling, In Situ Hybridization, and Differential Staining
Data Deposition
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| EC | Evolution Canyon |
| LARDs | LArge Retrotransposon Derivatives |
| LTR | long terminal repeat |
| MITEs | Miniature Inverted-repeat Transposable Elements |
| NFS | North-Facing Slope |
| PBS | Primer Binding Site |
| PPT | PolyPurine Tract |
| RLC | LTR retrotransposon of superfamily Copia |
| RLX | LTR retrotransposon |
| TE | transposable element |
| TRIMs | Terminal Repeats in Miniature |
References
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Schulman, A.H. Retrotransposon replication in plants. Curr. Opin. Virol. 2013, 3, 604–614. [Google Scholar] [CrossRef] [PubMed]
- Arkhipova, I.R.; Yushenova, I.A. Giant transposons in eukaryotes: Is bigger better? Genome Biol. Evol. 2019, 11, 906–918. [Google Scholar] [CrossRef]
- Sultana, T.; Zamborlini, A.; Cristofari, G.; Lesage, P. Integration site selection by retroviruses and transposable elements in eukaryotes. Nat. Rev. Genet. 2017, 18, 292–308. [Google Scholar] [CrossRef] [PubMed]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Schulman, A.H.; Tanskanen, J.; Spannagl, M.; Twardziok, S.; Mascher, M.; Springer, N.M.; Li, Q.; Waugh, R.; Li, C.; et al. The repetitive landscape of the 5100 mbp barley genome. Mobile DNA 2017, 8, 22. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. Helitrons on a roll: Eukaryotic rolling-circle transposons. Trends Genet. 2007, 23, 521–529. [Google Scholar] [CrossRef]
- Shams, I.; Raskina, O. Intraspecific and intraorganismal copy number dynamics of retrotransposons and tandem repeat in Aegilops speltoides tausch (poaceae, triticeae). Protoplasma 2018, 255, 1023–1038. [Google Scholar] [CrossRef]
- Presting, G.G. Centromeric retrotransposons and centromere function. Curr. Opin. Genet. Dev. 2018, 49, 79–84. [Google Scholar] [CrossRef]
- Pollak, Y.; Zelinger, E.; Raskina, O. Repetitive DNA in the architecture, repatterning, and diversification of the genome of aegilops speltoides tausch (poaceae, triticeae). Front. Plant Sci. 2018, 9, 1779. [Google Scholar] [CrossRef]
- Bilinski, P.; Han, Y.; Hufford, M.B.; Lorant, A.; Zhang, P.; Estep, M.C.; Jiang, J.; Ross-Ibarra, J. Genomic abundance is not predictive of tandem repeat localization in grass genomes. PLoS ONE 2017, 12, e0177896. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Wolfgruber, T.K.; Presting, G.G. Tandem repeats derived from centromeric retrotransposons. BMC Genom. 2013, 14, 142. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Liang, P. Transposable elements are a significant contributor to tandem repeats in the human genome. Comp. Funct. Genom. 2012, 2012, 947089. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, D.H.; Gaubert, H.; Drost, H.G.; Zabet, N.R.; Paszkowski, J. High-frequency recombination between members of an ltr retrotransposon family during transposition bursts. Nat. Commun. 2017, 8, 1283. [Google Scholar] [CrossRef]
- Belyayev, A.; Kalendar, R.; Brodsky, L.; Nevo, E.; Schulman, A.H.; Raskina, O. Transposable elements in a marginal plant population: Temporal fluctuations provide new insights into genome evolution of wild diploid wheat. Mobile DNA 2010, 1. [Google Scholar] [CrossRef]
- Hastings, P.J.; Lupski, J.R.; Rosenberg, S.M.; Ira, G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009, 10, 551–564. [Google Scholar] [CrossRef]
- Eickbush, T.H.; Eickbush, D.G. Finely orchestrated movements: Evolution of the ribosomal rna genes. Genetics 2007, 175, 477–485. [Google Scholar] [CrossRef]
- Michel, A.H.; Kornmann, B.; Dubrana, K.; Shore, D. Spontaneous rdna copy number variation modulates sir2 levels and epigenetic gene silencing. Genes Dev. 2005, 19, 1199–1210. [Google Scholar] [CrossRef]
- Lyckegaard, E.M.; Clark, A.G. Ribosomal DNA and stellate gene copy number variation on the y chromosome of drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1989, 86, 1944–1948. [Google Scholar] [CrossRef]
- Stults, D.M.; Killen, M.W.; Pierce, H.H.; Pierce, A.J. Genomic architecture and inheritance of human ribosomal rna gene clusters. Genome Res. 2008, 18, 13–18. [Google Scholar] [CrossRef]
- Paco, A.; Freitas, R.; Vieira-da-Silva, A. Conversion of DNA sequences: From a transposable element to a tandem repeat or to a gene. Genes 2019, 10, 1014. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Gundlach, H.; Spannagl, M.; Uauy, C.; Borrill, P.; Ramirez-Gonzalez, R.H.; De Oliveira, R.; International Wheat Genome Sequencing, C.; Mayer, K.F.X.; Paux, E.; et al. Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biol. 2018, 19, 103. [Google Scholar] [CrossRef] [PubMed]
- Zervudacki, J.; Yu, A.; Amesefe, D.; Wang, J.; Drouaud, J.; Navarro, L.; Deleris, A. Transcriptional control and exploitation of an immune-responsive family of plant retrotransposons. FEBS J. 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.K. Structural and sequence diversity of eukaryotic transposable elements. Genes Genet. Syst. 2018, 94, 233–252. [Google Scholar] [CrossRef]
- Kalendar, R.; Vicient, C.M.; Peleg, O.; Anamthawat-Jonsson, K.; Bolshoy, A.; Schulman, A.H. Large retrotransposon derivatives: Abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics 2004, 166, 1437–1450. [Google Scholar] [CrossRef]
- Witte, C.P.; Le, Q.H.; Bureau, T.; Kumar, A. Terminal-repeat retrotransposons in miniature (trim) are involved in restructuring plant genomes. Proc. Natl. Acad. Sci. USA 2001, 98, 13778–13783. [Google Scholar] [CrossRef]
- Satovic, E.; Luchetti, A.; Pasantes, J.J.; Garcia-Souto, D.; Cedilak, A.; Mantovani, B.; Plohl, M. Terminal-repeat retrotransposons in miniature (trims) in bivalves. Sci. Rep. 2019, 9, 19962. [Google Scholar] [CrossRef]
- Xia, C.; Zhang, L.; Zou, C.; Gu, Y.; Duan, J.; Zhao, G.; Wu, J.; Liu, Y.; Fang, X.; Gao, L.; et al. A trim insertion in the promoter of ms2 causes male sterility in wheat. Nat. Commun. 2017, 8, 15407. [Google Scholar] [CrossRef]
- Schorn, A.J.; Gutbrod, M.J.; LeBlanc, C.; Martienssen, R. Ltr-retrotransposon control by trna-derived small rnas. Cell 2017, 170, 61–71 e11. [Google Scholar] [CrossRef]
- Kalendar, R.; Tanskanen, J.; Chang, W.; Antonius, K.; Sela, H.; Peleg, O.; Schulman, A.H. Cassandra retrotransposons carry independently transcribed 5s rna. Proc. Natl. Acad. Sci. USA 2008, 105, 5833–5838. [Google Scholar] [CrossRef]
- Antonius-Klemola, K.; Kalendar, R.; Schulman, A.H. Trim retrotransposons occur in apple and are polymorphic between varieties but not sports. Theor. Appl. Genet. 2006, 112, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Vondrak, T.; Avila Robledillo, L.; Novak, P.; Koblizkova, A.; Neumann, P.; Macas, J. Characterization of repeat arrays in ultra-long nanopore reads reveals frequent origin of satellite DNA from retrotransposon-derived tandem repeats. Plant J. 2020, 101, 484–500. [Google Scholar] [CrossRef] [PubMed]
- Mitsuhashi, S.; Frith, M.C.; Mizuguchi, T.; Miyatake, S.; Toyota, T.; Adachi, H.; Oma, Y.; Kino, Y.; Mitsuhashi, H.; Matsumoto, N. Tandem-genotypes: Robust detection of tandem repeat expansions from long DNA reads. Genome Biol. 2019, 20, 58. [Google Scholar] [CrossRef] [PubMed]
- Nevo, E. “Evolution canyon,” a potential microscale monitor of global warming across life. Proc. Natl. Acad. Sci. USA 2012, 109, 2960–2965. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Tanskanen, J.; Immonen, S.; Nevo, E.; Schulman, A.H. Genome evolution of wild barley (hordeum spontaneum) by bare-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc. Natl. Acad. Sci. USA 2000, 97, 6603–6607. [Google Scholar] [CrossRef]
- Kimber, G.; Feldman, M. Wild Wheat, An Introduction; College of Agriculture University of Missouri: Columbia, MO, USA, 1987; Volume 353, p. 142. [Google Scholar]
- Raskina, O.; Belyayev, A.; Nevo, E. Quantum speciation in aegilops: Molecular cytogenetic evidence from rdna cluster variability in natural populations. Proc. Natl. Acad. Sci. USA 2004, 101, 14818–14823. [Google Scholar] [CrossRef]
- Raskina, O. Transposable elements in the organization and diversification of the genome of Aegilops speltoides tausch (poaceae, triticeae). Int. J. Genom. 2018, 2018, 4373089. [Google Scholar] [CrossRef]
- Belyayev, A. Bursts of transposable elements as an evolutionary driving force. J. Evol. Biol. 2014, 27, 2573–2584. [Google Scholar] [CrossRef]
- Hosid, E.; Brodsky, L.; Kalendar, R.; Raskina, O.; Belyayev, A. Diversity of long terminal repeat retrotransposon genome distribution in natural populations of the wild diploid wheat aegilops speltoides. Genetics 2012, 190, 263–412. [Google Scholar] [CrossRef]
- Vicient, C.M.; Jaaskelainen, M.J.; Kalendar, R.; Schulman, A.H. Active retrotransposons are a common feature of grass genomes. Plant Physiol. 2001, 125, 1283–1292. [Google Scholar] [CrossRef]
- Vicient, C.M.; Kalendar, R.; Schulman, A.H. Envelope-class retrovirus-like elements are widespread, transcribed and spliced, and insertionally polymorphic in plants. Genome Res. 2001, 11, 2041–2049. [Google Scholar] [CrossRef] [PubMed]
- Smykal, P.; Kalendar, R.; Ford, R.; Macas, J.; Griga, M. Evolutionary conserved lineage of angela-family retrotransposons as a genome-wide microsatellite repeat dispersal agent. Heredity 2009, 103, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Moisy, C.; Schulman, A.H.; Kalendar, R.; Buchmann, J.P.; Pelsy, F. The tvv1 retrotransposon family is conserved between plant genomes separated by over 100 million years. Theor. Appl. Genet. 2014, 127, 1223–1235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kalendar, R.; Amenov, A.; Daniyarov, A. Use of retrotransposon-derived genetic markers to analyse genomic variability in plants. Functional Plant Biology 2019, 46, 15–29. [Google Scholar] [CrossRef]
- Panaud, O. Horizontal transfers of transposable elements in eukaryotes: The flying genes. Comptes Rendus Biol. 2016, 339, 296–299. [Google Scholar] [CrossRef]
- Yoshida, S.; Kim, S.; Wafula, E.K.; Tanskanen, J.; Kim, Y.M.; Honaas, L.; Yang, Z.; Spallek, T.; Conn, C.E.; Ichihashi, Y.; et al. Genome sequence of striga asiatica provides insight into the evolution of plant parasitism. Curr. Biol. 2019, 29, 3041–3052 e3044. [Google Scholar] [CrossRef]
- Bernard, G.; Chan, C.X.; Ragan, M.A. Alignment-free microbial phylogenomics under scenarios of sequence divergence, genome rearrangement and lateral genetic transfer. Sci. Rep. 2016, 6, 28970. [Google Scholar] [CrossRef]
- Yin, H.; Du, J.; Li, L.; Jin, C.; Fan, L.; Li, M.; Wu, J.; Zhang, S. Comparative genomic analysis reveals multiple long terminal repeats, lineage-specific amplification, and frequent interelement recombination for cassandra retrotransposon in pear (pyrus bretschneideri rehd.). Genome Biol. Evol. 2014, 6, 1423–1436. [Google Scholar] [CrossRef]
- Gao, D.; Li, Y.; Kim, K.D.; Abernathy, B.; Jackson, S.A. Landscape and evolutionary dynamics of terminal repeat retrotransposons in miniature in plant genomes. Genome Biol. 2016, 17, 7. [Google Scholar] [CrossRef]
- Vicient, C.; Kalendar, R.; Schulman, A. Variability, recombination, and mosaic evolution of the barley bare-1 retrotransposon. J. Mol. Evol. 2005, 61, 275–291. [Google Scholar] [CrossRef][Green Version]
- Shang, Y.; Yang, F.; Schulman, A.H.; Zhu, J.; Jia, Y.; Wang, J.; Zhang, X.Q.; Jia, Q.; Hua, W.; Yang, J.; et al. Gene deletion in barley mediated by ltr-retrotransposon bare. Sci. Rep. 2017, 7, 43766. [Google Scholar] [CrossRef] [PubMed]
- Sabot, F.; Schulman, A.H. Template switching can create complex ltr retrotransposon insertions in triticeae genomes. BMC Genom. 2007, 8, 247. [Google Scholar] [CrossRef] [PubMed]
- Raskina, O.; Brodsky, L.; Belyayev, A. Tandem repeats on an eco-geographical scale: Outcomes from the genome of aegilops speltoides. Chromosome Res. 2011, 19, 607–623. [Google Scholar] [CrossRef] [PubMed]
- Raskina, O.; Belyayev, A.; Nevo, E. Activity of the en/spm-like transposons in meiosis as a base for chromosome repatterning in a small, isolated, peripheral population of aegilops speltoides tausch. Chromosome Res. 2004, 12, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Rose, R.; Golosova, O.; Sukhomlinov, D.; Tiunov, A.; Prosperi, M. Flexible design of multiple metagenomics classification pipelines with ugene. Bioinformatics 2019, 35, 1963–1965. [Google Scholar] [CrossRef]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. Ngphylogeny.Fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef]
- Kalendar, R.; Khassenov, B.; Ramanculov, E.; Samuilova, O.; Ivanov, K.I. Fastpcr: An in silico tool for fast primer and probe design and advanced sequence analysis. Genomics 2017, 109, 312–319. [Google Scholar] [CrossRef]
- Kalendar, R.; Tselykh, T.V.; Khassenov, B.; Ramanculov, E.M. Introduction on using the fastpcr software and the related java web tools for pcr and oligonucleotide assembly and analysis. Methods Mol. Biol. 2017, 1620, 33–64. [Google Scholar] [CrossRef]
- Kalendar, R.; Muterko, A.; Shamekova, M.; Zhambakin, K. In silico pcr tools for a fast primer, probe, and advanced searching. Methods Mol. Biol. 2017, 1620, 1–31. [Google Scholar] [CrossRef]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R. Universal DNA Isolation Protocol; Protocols.io: Berkeley, CA, USA, 2019. [Google Scholar] [CrossRef]
- Kalendar, R.; Shustov, A.V.; Seppänen, M.M.; Schulman, A.H.; Stoddard, F.L. Palindromic sequence-targeted (pst) pcr: A rapid and efficient method for high-throughput gene characterization and genome walking. Sci. Rep. 2019, 9, 17707. [Google Scholar] [CrossRef] [PubMed]
- Taketa, S.; Ando, H.; Takeda, K.; Harrison, G.E.; Heslop-Harrison, J.S. The distribution, organization and evolution of two abundant and widespread repetitive DNA sequences in the genus hordeum. Theor. Appl. Genet. 2000, 100, 169–176. [Google Scholar] [CrossRef]
- Baum, B.R.; Bailey, L.G. The 5s rrna gene sequence variation in wheats and some polyploid wheat progenitors (poaceae: Triticeae). Genet. Resour Crop Ev 2001, 48, 35–51. [Google Scholar] [CrossRef]
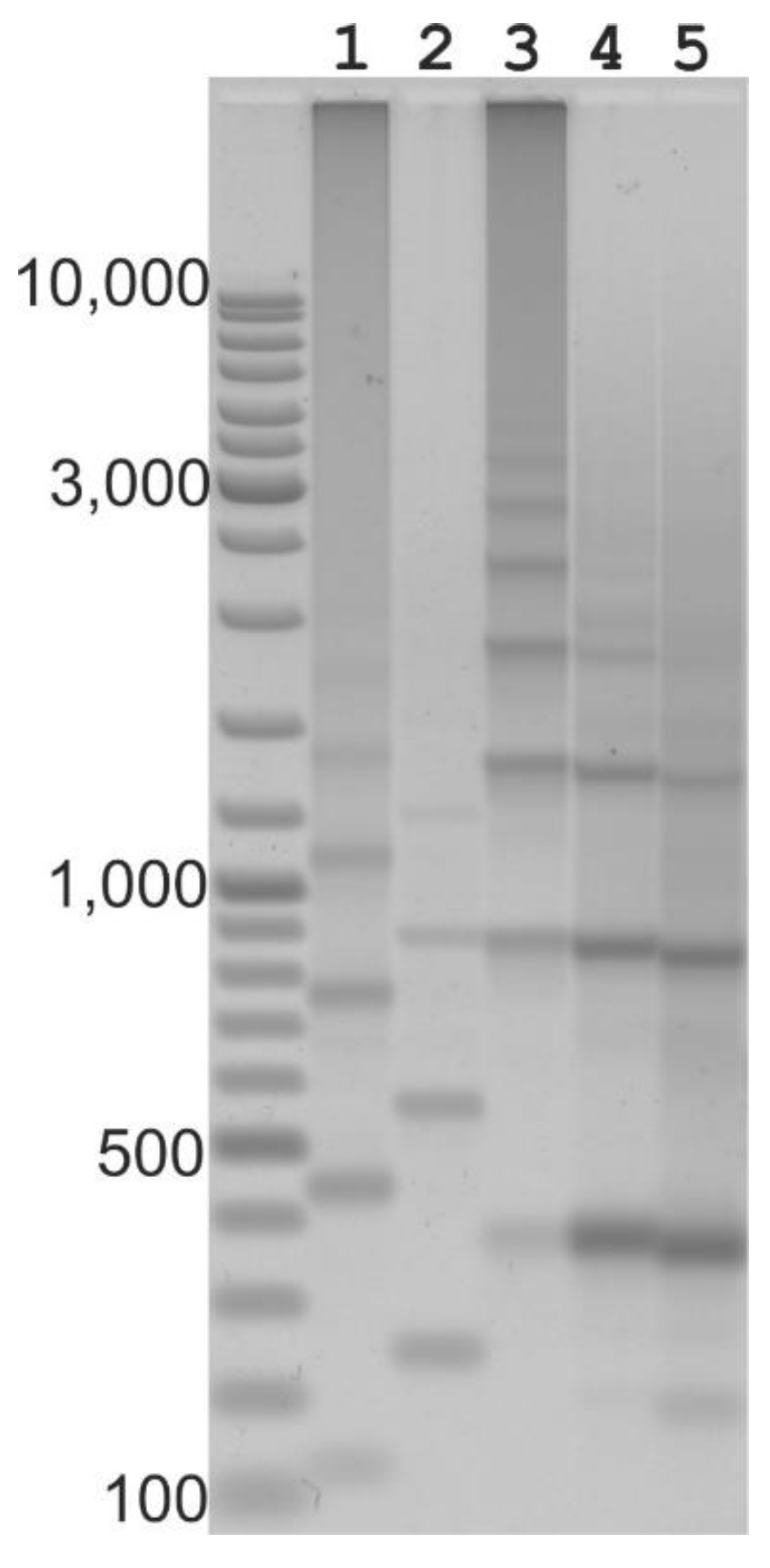
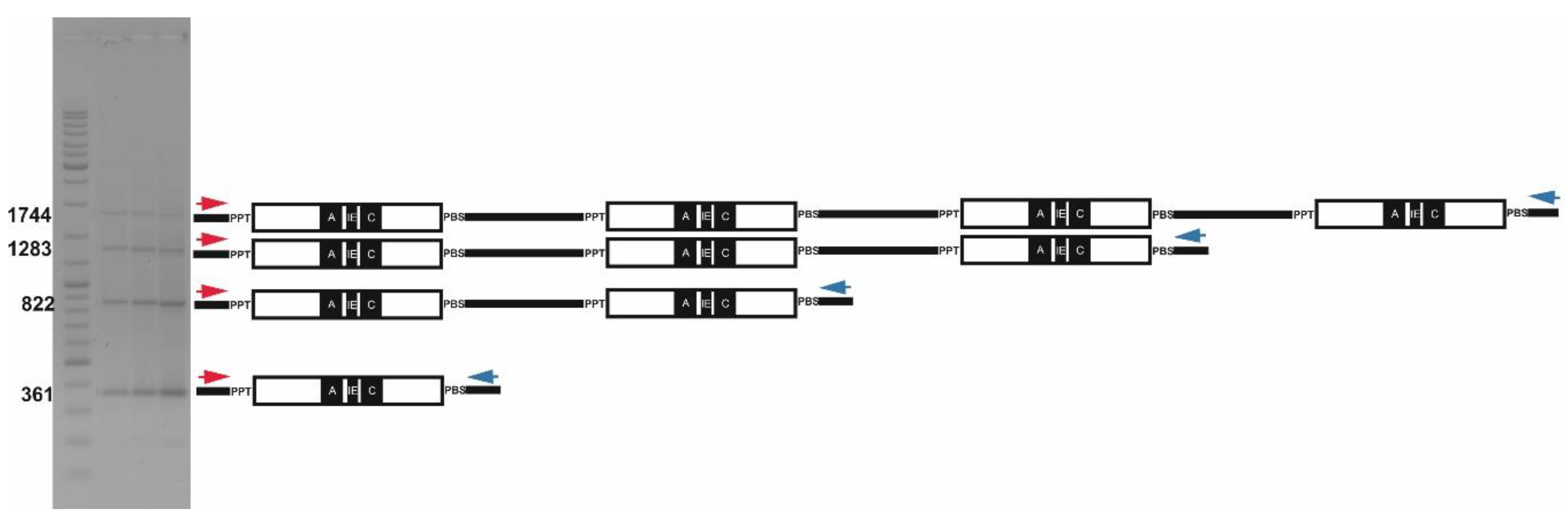
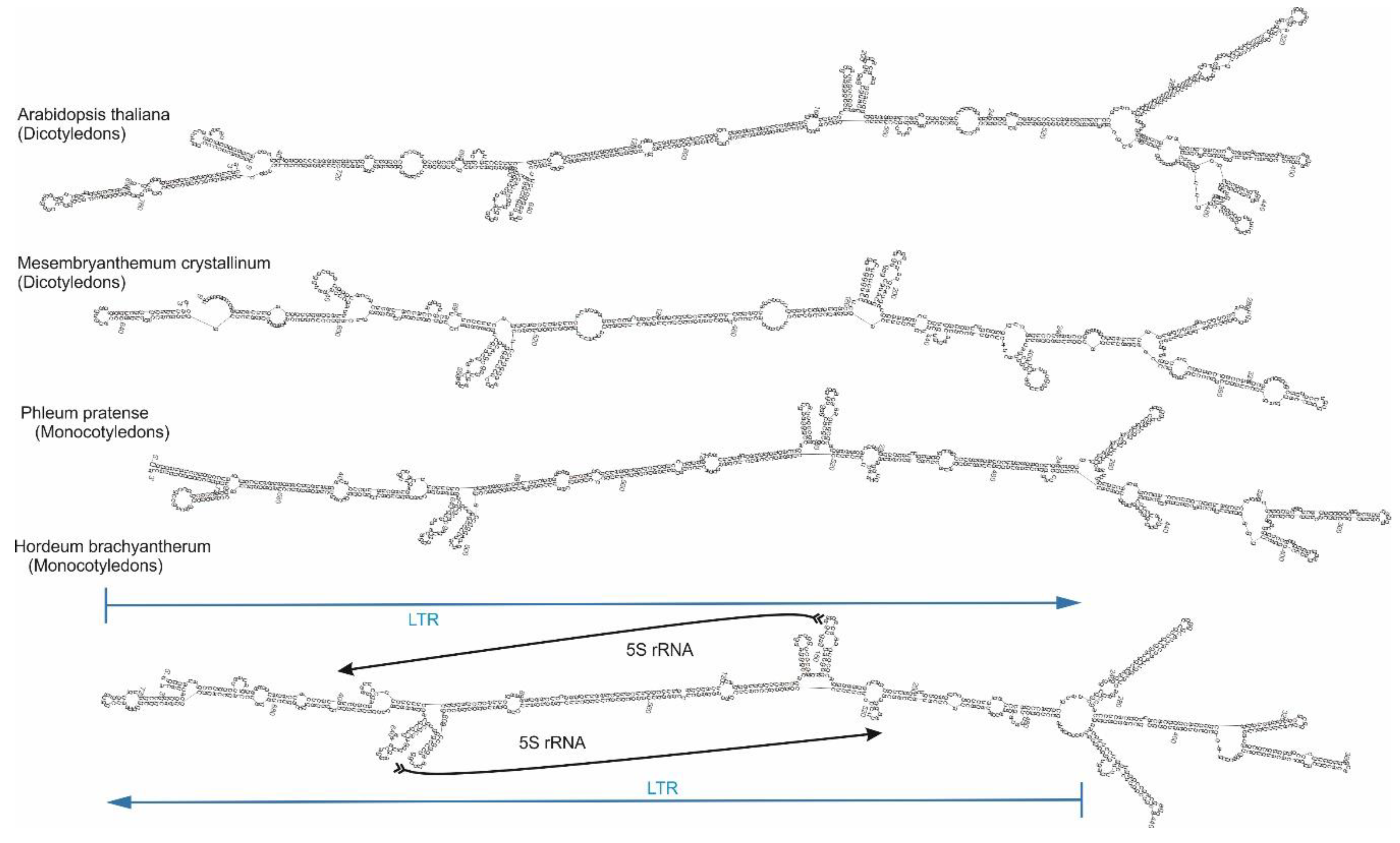
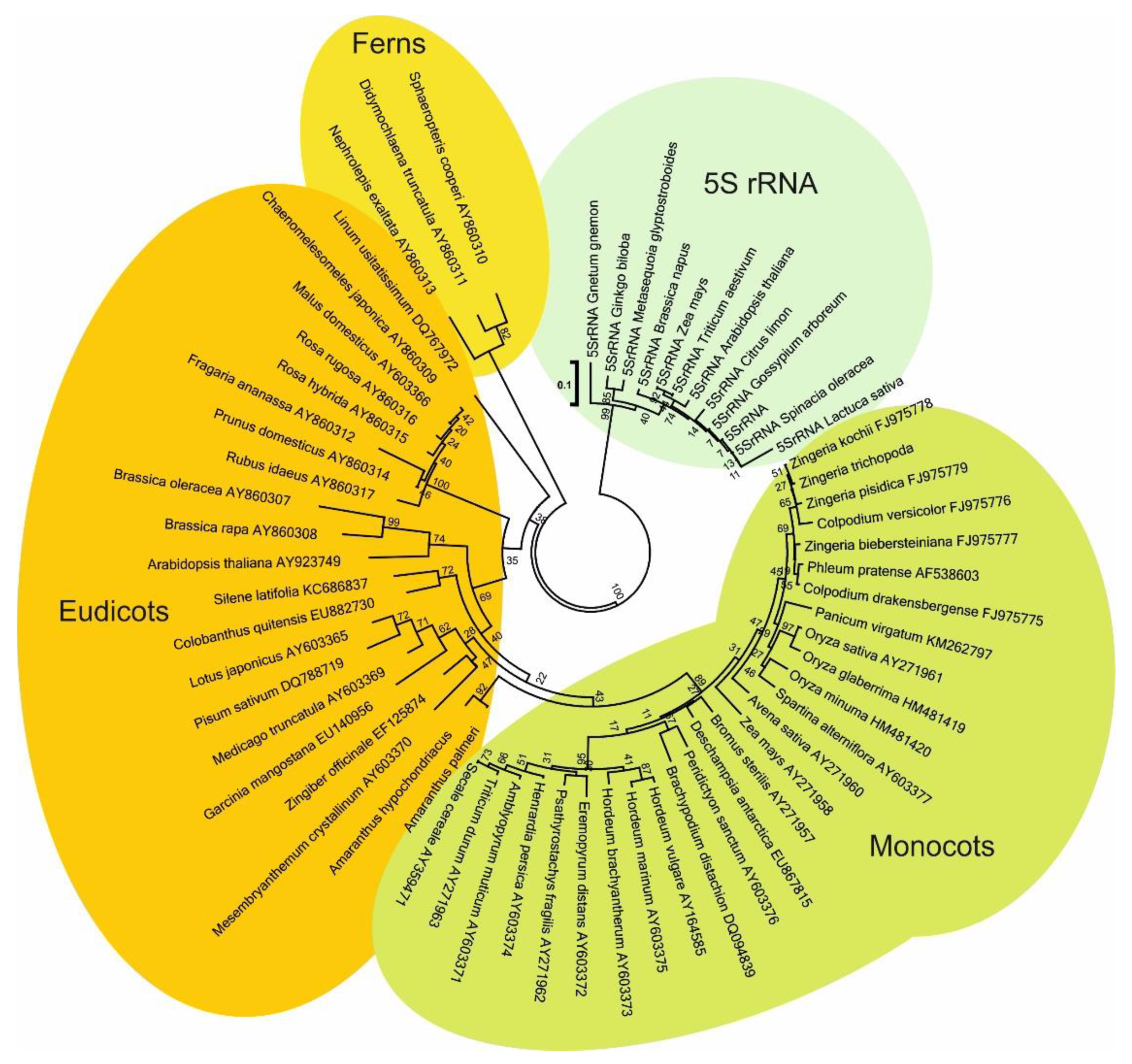
| Genome | Size in Mb | Predicted Copy Number |
|---|---|---|
| Arabidopsis thaliana | 126 | 4 |
| Brachypodium distachyon | 275 | 5 |
| Oryza sativa | 380 | 69 |
| Medicago truncatula | 391 | 32 |
| Vitis vinifera | 420 | 20 |
| Sorghum bicolor | 545 | 64 |
| Glycine max | 965 | 30 |
| Zea mays | 2100 | 701 |
| Homo sapiens | 3140 | 0 |
| Plant Species | Formula for Expected Ladder Lengths of PCR Fragments for Cassandra Tandems | Forward Primer ID | Reverse Primer ID |
|---|---|---|---|
| Hordeum vulgare L. | 361 + (461)n | 981 | 982 |
| Hordeum vulgare L. | 374 + (461)n | 2259 | 982 |
| Triticum durum Desf. | 326 + (460)n | 1032 | 982 |
| Triticum durum Desf. | 360 + (460)n | 981 | 982 |
| Triticum durum Desf. | 422 + (460)n | 1032 | 2258 |
| Secale cereal L. | 326 + (460)n | 1032 | 982 |
| Secale cereal L. | 348 + (460)n | 981 | 530 |
| Secale cereal L. | 360 + (460)n | 981 | 982 |
| Secale cereal L. | 422 + (460)n | 1032 | 2258 |
| Avena sativa L. | 357 + (481)n | 1032 | 3801 |
| Avena sativa L. | 361 + (481)n | 4170 | 4174 |
| Avena sativa L. | 368 + (481)n | 3802 | 3801 |
| Avena sativa L. | 494 + (481)n | 784 | 977 |
| Zea maize L. | 431 + (508)n | 1032 | 2263 |
| Zea maize L. | 501 + (508)n | 2262 | 2263 |
| Spartina alterniflora Loisel. | 358 + (537)n | 1032 | 3803 |
| Spartina alterniflora Loisel. | 408 + (537)n | 1032 | 3804 |
| Brachypodium distachyon (L.) P.Beauv. | 314 + (415)n | 1032 | 2261 |
| Brachypodium distachyon (L.) P.Beauv. | 414 + (415)n | 2260 | 2261 |
| Medicago truncatula Gaertn. | 468 + (459)n | 2070 | 2071 |
| Lotus corniculatus L. | 473 + (464)n | 2070 | 2071 |
| Malus domestica Borkh. | 364 + (377)n | 921 | 1611 |
| Vaccinium sp. | 414 + (418)n | 2016 | 622 |
| Garcinia mangostana L. | 525 + (509)n | 2495 | 2496 |
| Silene latifolia Poir. | 473 + (491)n | 623 | 629 |
| Nephrolepis exaltata (L.) Schott | 380 + (378)n | 1118 | 1120 |
| 5S rRNA Brassica rapa (LR031586) | 510..527 + (503..520)n | 2721 | 622 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalendar, R.; Raskina, O.; Belyayev, A.; Schulman, A.H. Long Tandem Arrays of Cassandra Retroelements and Their Role in Genome Dynamics in Plants. Int. J. Mol. Sci. 2020, 21, 2931. https://doi.org/10.3390/ijms21082931
Kalendar R, Raskina O, Belyayev A, Schulman AH. Long Tandem Arrays of Cassandra Retroelements and Their Role in Genome Dynamics in Plants. International Journal of Molecular Sciences. 2020; 21(8):2931. https://doi.org/10.3390/ijms21082931
Chicago/Turabian StyleKalendar, Ruslan, Olga Raskina, Alexander Belyayev, and Alan H. Schulman. 2020. "Long Tandem Arrays of Cassandra Retroelements and Their Role in Genome Dynamics in Plants" International Journal of Molecular Sciences 21, no. 8: 2931. https://doi.org/10.3390/ijms21082931
APA StyleKalendar, R., Raskina, O., Belyayev, A., & Schulman, A. H. (2020). Long Tandem Arrays of Cassandra Retroelements and Their Role in Genome Dynamics in Plants. International Journal of Molecular Sciences, 21(8), 2931. https://doi.org/10.3390/ijms21082931








