Abstract
Genetic analysis of “InochinoIchi,” an exceptionally large grain rice variety, was conducted through five continuous backcrosses with Koshihikari as a recurrent parent using the large grain F3 plant in Koshihikari × Inochinoichi as a nonrecurrent parent. Thorough the F2 and all BCnF2 generations, large, medium, and small grain segregated in a 1:2:1 ratio, indicating that the large grain is controlled by a single allele. Mapping by using simple sequence repeat (SSR) and single nucleotide polymorphism (SNP) markers with small grain homozygous segregants in the F2 of Nipponbare × Inochinoichi, revealed linkage with around 7.7 Mb markers from the distal end of the short arm of chromosome 2. Whole-genome sequencing on a large grain isogenic Koshihikari (BC4F2) using next-generation sequencing (NGS) identified a single nucleotide deletion in GW2 gene, which is located 8.1 Mb from the end of chromosome 2, encoding a RING protein with E3 ubiquitin ligase activity. The GW2-integrated isogenic Koshihikari showed a 34% increase in thousand kernel weight compared to Koshihikari, while retaining a taste score of 80. We further developed a large grain/semi-dwarf isogenic Koshihikari integrated with GW2 and the semidwarfing gene d60, which was found to be localized on chromosome 2. The combined genotype secured high yielding while providing robustness to withstand climate change, which can contribute to the New Green Revolution.
1. Introduction
There is a demand for a dramatic increase in the production of rice, as it is a staple food for over half of the world’s rapidly increasing population. In the period of the 20th century when most rice breeding took place, known as the “Green Revolution,” improving rice stems to be shorter decreased the likelihood of plant lodging, which made heavy manuring and dense planting possible, and radically increased yields. The “semidwarfness” led to a twofold increase in rice yields worldwide between the 1960s and the 1990s [1]. However, the trend of rice productively has now begun to plateau [2]. On top of that, the suppression of stem length is dependent on a single semidwarfing gene called sd1. In preparation for future increases in population and risk of crop damage due to climate change, there is a renewed demand for a “New Green Revolution” for genetic improvements to increase yields and make rice plants more robust.
Grain size is a major factor in affecting rice yield. Six quantitative trait loci (QTLs) genes related to rice grain size have been isolated [3,4,5,6,7,8]. For four of these genes, a loss of function causes the grains to be bigger, which shows that there is a mechanism that suppresses grain size. In general, Japonica rice has short and round grains, while Indica rice has long and thin grains. Whole-genome analysis of cultivated rice and wild rice shows that the domestication of rice started near the middle of the Pearl River in China, when Japonica was derived from a population of Oryza rufipogon [9]. Subsequently, Indica arose from hybrids between strains of wild rice in Southeast Asia and South Asia and Japonica. The qSW5 and GS3 genes confer grain size traits in Japonica and Indica, respectively [5,9].
The japonica rice Koshihikari is the leading variety in Japan, accounting for 36.1% of rice acreage in the country. The patent on the plant variety protection of Koshihikari, which was registered in 1956, has expired; therefore, global competition with Koshihikari produced in foreign countries is now of concern. The 2016 Trans-Pacific Partnership (TPP) eliminated tariffs on 82% (2135) of the 2594 agriculture, forestry, and fishery products that are imported by Japan [10]. The 341 JPY/kg (140%) tariffs on rice were maintained, but the simultaneous buy-and-sell (SBS) tender system with the US and Australia, which already produce Koshihikari, provides a special import framework for 78,400 t of rice. Foreign-produced Koshihikari is roughly 35% less expensive than Japanese Koshihikari, and it is genetically the same, with no difference in taste. Consequently, the influx of inexpensive Koshihikari produced in foreign countries is a concern to Japan. Furthermore, after the Trump administration began, the US pulled out of the TPP and has been pushing for a Trade Agreement on Goods (TAG) between the US and Japan to take its place. In the future, rice trade will inevitably be liberalized. Moreover, if Japan’s self-sufficiency collapses, the country will lose its paddy fields, which would not be ideal for maintaining national land conservation. In order to resolve this critical situation, there is a need to develop a low-cost and high-yield “super Koshihikari” variety that can compete in the international market.
Intensified climate change due to global warming is causing damage to crops on a global scale. Global contributions due to the New Green Revolution could uphold the innovation policy. In 2018, Japan was hit with the Western Japan heavy rain and floods [11], and seven large typhoons with wind speeds over 54 m/s, which were the worst in Japan’s history [12]. Typhoons Jebi and Trami were equivalent to the Isewan Typhoon. These extreme weather phenomena have caused marked damage to agriculture, forestry, and fisheries (totaling 436.5 billion yen) [13]. There is a need to genetically improve the sturdiness and robustness of rice plants to withstand the intensified climate change [14,15] and improve rice for the global market by lowering costs and increasing yield.
The grain weight of a large grain variety, “Inochinoichi,” is approximately 1.5 times that of Koshihikari. However, the genetic mode of the large grain is unclear, so it is not used for plant breeding at all. The production of “Inochinoichi” is also limited to the area around Gifu Prefecture. If the causative gene for the large grain of inochinoichi were to be identified, its possibility of application to improve varieties, including Koshihiakri, or to develop new varieties would be expanded. In this study, we identified from this unused and buried genetic resource Inochinoichi, the gene responsible for the large grain, and then by applying the gene to develop a large-grain Koshihikari. First, we conducted genetic analysis of the large grain through five continuous backcrosses with Koshihikari as a recurrent parent using the large grain segregant in the F2 generation of a Koshihikari × Inochinoichi as a nonrecurrent parent. Then, we conducted whole-genome analysis of the developed large grain isogenic Koshihikari and identified a gene responsible for the large grain. Furthermore, we developed a large grain/semi-dwarf isogenic line by integrating both the identified large grain gene and the semidwarfing gene d60 [16].
2. Results
2.1. Inheritance and Phenotypic Expression of Large Grain Gene in an Isogenic Background
As shown in Figure 1A, in the F2 generation of Koshihikari × Inochinoichi, grain diameters showed a bimodal distribution in 0.69–0.85 mm, which were comparable to both parenteral range. Namely large grain plants with grain diameters of 0.78–0.85 mm, the same as Inochinoichi, and small grain plants with grain diameters of 0.69–0.77 mm the same as Koshihikari were segregated in a ratio of 134 large grains:52 small grains, which fit to a 3:1 ratio, (χ2 = 0.87, df = 1, 0.35 < P < 0.40). Next, we conducted a progeny test using 50 F3 lines consisting of 50 randomly selected F2 plants of Koshihikari × Inochinoichi. As a results, the mean values of grain size in each F3 line were distributed as shown in Figure 2. Namely, the F3 progeny of large grain F2 plants (grain diameter: 0.78–0.85 mm) were classified into a line that had fixed in large grains with diameters of 0.8–0.83 mm and a line that segregated within the lines, whereas the F3 progeny of small grain F2 plants (grain diameter: 0.68–0.77 mm) fixed in small grain with diameters of 0.72–0.75 mm. In other words, F3 lines segregated in a ratio of 10 large grain homozygous lines:32 heterozygous lines:8 small grain homozygous lines, consistent with the theoretical single gene ratio (χ2 = 4.08, df = 1, 0.10 < P < 0.25). Using the fixed large grain homozygous plant in the F3 generation (grain diameter: 0.75 mm) as a nonrecurrent parent, five times of continuous backcrosses with Koshihikari as a recurrent parent were conducted. The BC1F2 plants segregated in a ratio of 10 large grain plants (grain area: 24.6–25.9 mm2):32 small and medium grain plants (grain area: 20.6–24.0 mm2) (Figure 1A). Furthermore, a large grain segregant in the BC1F2 generation (grain area: 25.9 mm2) was used in a second backcross with Koshihikari, which yielded a BC2F2 plants segregated in a ratio of 17 large grain plants (grain area: 23.6–25.1 mm2):39 medium plants (grain area: 20.6–23.5 mm2):14 small grain plants (grain area: 19.6–20.5 mm2); in both generations, segregation ratios fit to the theoretical single gene ratio (χ2 = 0.03, df = 1, 0.50 < P < 0.90; χ2 = 1.17, df = 2, 0.55 < P < 0.60) (Figure 1A). Subsequently, the BC3F2 plants segregated in a ratio of 15 large grain (grain area: 19.6–20.5 mm2):13 medium grain (grain area: 19.6–20.5 mm2):7 small grain (grain area: 19.6–20.5 mm2) (χ2 = 5.97, df = 2, 0.05 < P < 0.10). Next, a large grain BC3F2 segregant (grain area: 23.05 mm2) was used for the fourth backcross with Koshihikari, whose BC4F2 progenies segregated in a ratio of 8 large grain (grain area: 26.1–29.5 mm2):18 medium grain (grain area: 23.6–26.0 mm2):10 small grain (grain area: 21.1–23.5 mm2) plants, that fit well to a 1:2:1 ratio (χ2 = 0.20, df = 2, 0.85 < P < 0.90) (Figure 1A). As seen above, from genetic analyses of large grain through the four times of backcrosses with Koshihikari, each BC2F2 to BC4F2 progeny segregated in the theoretical ratio for single incomplete dominance gene, namely 1 large grain:2 medium grain:1 small grain (Figure 1A). This indicates that the large grain is definitely inherited as a single allele. Finally, the large grain isogenic Koshihikari (BC5F2), which produced by backcrossed with Koshihikari and a large grain BC4F2 segregant, showed a grain area 27.7% greater than that of Koshihikari (Koshihikari average grain area: 22.32 mm2, large grain phenotype average: 28.50 mm2), and the thousand kernel weight increased by 34%. Its taste score (80.0) was also equivalent to that of Niigata Koshihikari (81.0) (Table 1); thus, this isogenic Koshihikari holds promise as a Super Koshihikari, which is distinguishable from US-made Koshihikari.
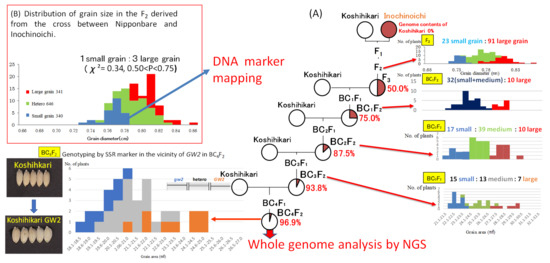
Figure 1.
Procedure to identify large grain gene in Inochinoichi. (A) Whole-genome analysis of isogeneic line developed by continuous back cross integrating the large grain gene. Genetic analyses for large grain through the four times of backcrosses with Koshihikari, each BC2F2 to BC4F2 progeny segregated in the theoretical ratio for single incomplete dominance gene, namely 1 large grain:2 medium grain:1 small grain. This indicates that the large grain is definitely inherited as a single allele. (B) Molecular linkage analysis by using small grain homozygous F2 derived from the cross of Nipponbare × Inochinoichi.
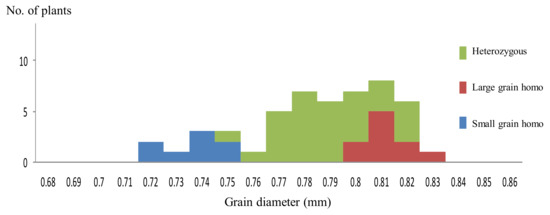
Figure 2.
Distribution of mean value of grain diameter in 50 F3 line derived from the cross, Koshihikari × Inochinoichi. F3 lines segregated in a ratio of 10 large grain homozygous lines:32 heterozygous lines:8 small grain homozygous lines, which fit to the theoretical single gene ratio.

Table 1.
Phenotypic expression of large-grain gene integrated isogenic Koshishikari.
2.2. Candidate Region of Large Grain Gene
Using small grain homozygous F2 segregants of a Nipponbare × Inochinoichi (Figure 1B), we genetically mapped the large grain gene by SSR and SNP markers across rice’s 12 chromosomes. Our results showed that the recombinant values between DNA markers and the large grain gene were detected on chromosome 2. Namely, from the distal end of the short arm of chromosome 2, 21.7 at J521 (7.6 Mb), 17.5 at RM3390 (7.7 Mb), 15.2 at J527 (8.2 Mb), 19.6 at J529 (8.6 Mb), 28.3 at J536(9.1 Mb), 30.0 at RM6375 (9.6 Mb), and 34.5 at RM1358 (10.2 Mb), respectively (Figure 3). The RM3390-homozygous plant by the diagnosis, which is linked with GW2, showed that the mean grain area with Inochinoichi alleles was 23.3 mm2, which is larger than 20.0 mm2 in Koshihikari (Figure 1A).
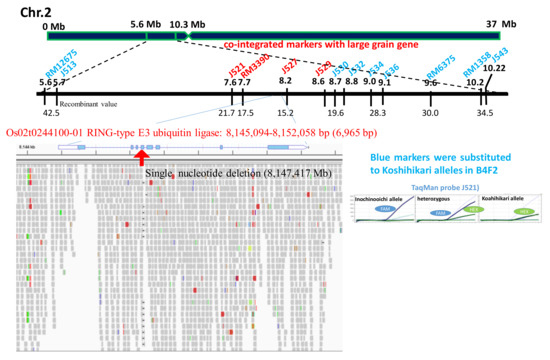
Figure 3.
Identification of single nucleotide deletion in GW2 responsible for large grain size of Inochinoichi. SNP allele-specific TaqMan probes were designed and labeled using the fluorescent dyes FAM or HEX. The real-time polymerase chain reaction (PCR) was used to amplify the allele-specific fluorescence. Blue DNA markers were substituted to Koshihikari alleles in B4F2. However, red DNA markers were inherited together with GW2.
2.3. Identification of DNA Variation Responsible for Large Grain Using Next-Generation Sequencing (NGS)
The read sequences of Koshihikari obtained by NGS were mapped using the Nipponbare genome as a reference sequence. The cover ratio was determined to be 99.05% and the mean depth was 32.43; Finally, a 372,912,445 bp long consensus sequence of the Koshihikari genome was constructed. Next, read sequences gained from the large grain isogenic Koshihikari (BC4F2) were mapped using the consensus sequence of Koshihikari as a reference sequence. In total, 187,159,213 reads were mapped, with a mapped read rate of 99.90%, a mean read length of 123.3 bp, and a 30.95× genome coverage.
Whole-genome sequencing detected a single nucleotide deletion (adenine) from the Koshihikari genome at the 8,147,417 bp position from the distal end of the short arm of chromosome 2 (Figure 3). This was the same as a single nucleotide deletion reported in the fourth exon of GW2 (Os02g024410), the QTL gene responsible for grain width in the large grain Chinese rice WY3 [4]. GW2 encodes a RING protein with E3 ubiquitin ligase activity, and a frame shift caused by a nucleotide deletion in this gene causes a loss-of-function [4]. GW2 derived from Inochinoichi is 6,965 bp with 100% identical to that of WY3, i.e., a single deletion in the fourth exon. There are two SNPs that flank the coding region of the hydroquinone glucosyltransferase gene (Os02g0242900). Our results show that the gene responsible for the large grain size of Inochinoichi, a promising gene source for increasing yield, is GW2. DNA markers around GW2, namely from the distal end of the short arm of chromosome 2, RM12675(5.6 Mb), J513(5.7 Mb), J530(8.7 Mb), J532(8.8 Mb), J534(9.0 Mb), J536(9.1 Mb), RM6375 (9.6 Mb), and RM1358 (10.2 Mb) were substituted to Koshihikari alleles in the B4F2 (Figure 3). On the contrary, Inochinoichi alleles of J521 (7.6 Mb), RM3390 (7.7 Mb), J527 (8.2 Mb), J529 (8.6 Mb) were tightly inherited together with GW2.
2.4. Linkage Relationship Between Semidwarfing Gene d60 and Large Grain Gene GW2
The first backcross with Koshihikari was conducted with a large grain semi-dwarf plant (stalk length: 76 cm, grain diameter: 0.8 mm) as the nonrecurrent parent segregated in the F2 between Koshihikari d60 (which was developed by integrating the Hokuriku 100-derived semidwarfing gene d60 into the Koshihikari genome through seven times of backcrosses) and Inochinoichi (Figure 4A,B). Here, regarding the genetics of d60, in the F1 hybrid (genotype D60d60Galgal) of Koshihikari (D60D60galgal) × Koshihikari d60(d60d60GalGal), male and female gametes having both gal and d60 become gamete lethal and the pollen and seed fertility decrease to 75%. As a results, the F2 progeny shows a unique mode of inheritance that is segregated into a ratio of 6 fertile long-culm (4D60D60:2D60d60GalGal: 2 partially sterile long-culm (D60d60Galgal = F1 type):1 dwarf(d60d60GalGal) [16] (Figure S1). In this study in the BC1F2 of Koshihikari/(Koshihikari d60 × Inochinoichi F2), the genotypic ratio for the D60/d60 allele was 11 d60 homozygous:26 partially sterile:75 long stem, which fit the theoretical ratio of 1:2:6 well (χ2 = 0.22, df = 2, 0.85 < P < 0.90) (Figure 4C). However, in the relationship between grain area and stem length, this contrasts with the Koshihikari*1/Koshihikari/Inochinoichi BC1F2, where there was an extremely small number of large grain segregants, a large number of small grain segregants, and no large grain long-stem segregants (Figure 4C). In other words, for BC1F2 as a whole, the ratio of (GW2 homozygous + heterozygous): gw2 homozygous was 73:39. This ratio should be close to 5:4, which arises when GW2 is completely linked with D60. Furthermore, while the segregation of the GW2 allele in d60 homozygous semi-dwarf plants was 10:1 for (large grain GW2 homozygous + hetero): small grain gw2 homozygous, in long-stem plants, the ratio of (GW2 large grain homozygous + heterozygous): small grain gw2 homozygous was 63:38. In other words, while large grain plants appeared at a higher rate in the semidwarf phenotype plants, they appeared at a lower rate in the long-stem phenotype plants (Figure 4C). Considerably deviated segregation in the GW2 locus occurred while opposing to each genotype of d60 allele. Furthermore, if GW2 and d60 are inherited independently, then the appearance rates of GW2 homozygous long stem plants and that of GW2 homozygous long stem partially sterile plants should be 6/36 (=(4D60D60 + 2D60d60)/9 × 1GW2GW2/4)) and 2/36(=2D60d60/9 × 1GW2GW2/4), respectively. However, actually there were no GW2 homozygotes among long stem or long stem partially sterile plants, respectively (Figure 4C). Thus, the fact that the segregation of the GW2 allele was considerably deviated in each genotype of d60 suggests linkage between GW2 and d60.
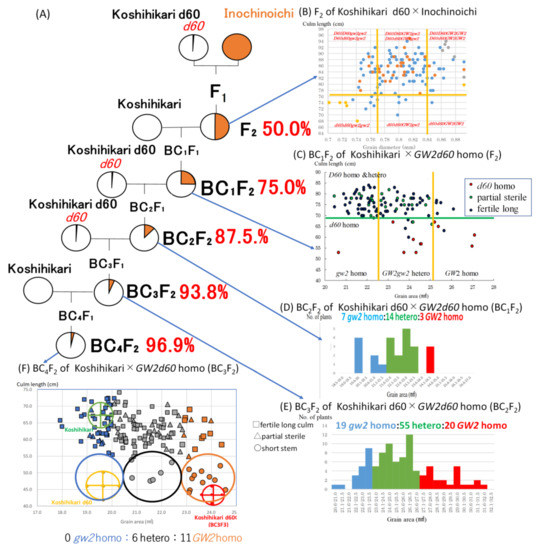
Figure 4.
Linkage relationship between GW2 and d60 identified by the deviated segregation ratio through the continuous backcross process by using Koshihikari or Koshihikari d60 as recurrent parents. (A) Backcross procedure combining d60 and GW2. (B) Distribution of grain size and culm length in the F2 of Koshihikari d60 × Inochinoichi. (C) Deviated segregation for grain size in each d60/D60 genotype in BC1F2. (D,E) Mendelian segregation for grain size in the d60 homo background in BC2F2 and BC3F2. (F) Recombination value (17.6) between GW2 and d60 was by segregation ratio in GW2 allele in d60 homozygous in BC4F2.
A large grain semi-dwarf segregant in BC1F2 was backcrossed with Koshihikari d60 as a nonrecurrent parent to make Koshihikari d60//Koshihikari/((Koshihikarid60 × Inochinoichi)F2) BC2F2, which segregated in a ratio of 3 large (grain area: 24.1–24.5 mm2):14 medium:7 small (grain area: 19.6–22.0 mm2) grain phenotypes = 1:2:1(χ2 = 2.00, df = 2, 0.30 < P < 0.50) (Figure 4D). Then, a GW2d60 homozygous large grain semi-dwarf segregant in the BC2F2 was thirdly backcrossed with a Koshihikari d60 to make Koshihikari d60*2//Koshihikari/((Koshihikarid60 × Inochinoichi)F2)BC3F2, which segregated in a ratio of 20 large (grain area: 27.1–32.5 mm2):55 medium:19 small (grain area: 20.6–23.5 mm2) grain phenotypes well fit to a 1:2:1 ratio (χ2 = 2.74, df = 2, 0.20 < P < 0.30) (Figure 4E). Above all, the affect of the linkage between GW2 and d60 disappeared in the d60 homozygous genetic background, so the large grain GW2 locus segregated according to a Mendelian ratio of 1 GW2 homozygous:2 heterozygous:1 gw2 homozygous.
As above, a large grain semi-dwarf plant (BC3F2), namely Koshihikari d60 integrated with GW2, was fourthly backcrossed with Koshihikari to make a Koshihikari///Koshihikari d60*2//Koshihikari/((Koshihikari d60 × Inochinoichi)F2 Gg genotype) BC4F2 generation (Figure 4A). The distribution of stem length in the BC4F2 was 17 (semi-dwarf 44.1–53.9 cm):36 (partially sterile):104 (long stem 53.9–74.1 cm) ≈ 1d60d60GalGal:2 D60d60Galgal:6(1D60D60GglGal + 2D60D60Galgal + 1D60D60galgal + 2D60d60GalGal)(χ2 = 0.01, df = 2, 0.9 < P < 0.95) (Figure 4F). Additionally, in each genotype for stem length, large (grain area: 22.9–24.6 mm2), medium (grain area: 20.1–22.8 mm2), and small grain (grain area: 17.9–20.0 mm2) were segregated in the ratio of 11 large grain:6 medium grain:0 small grain in the short stem phenotype, 4 large grain:23 medium grain:9 small grain in the partially sterile phenotype, and 10 large grain:60 medium grain:34 small grain for the long stem phenotype. In other words, as is the case in the F2 generation, while large grain genotypes segregated at a high rate in the d60 homozygous semi-dwarf plants, the small grain genotypes segregated at a high rate in the long stem plants; this indicates a linkage relationship between GW2 and d60. The recombinant value was calculated from the segregation ratio of 11 GW2 homozygous:6 GW2gw2:0 gw2 homozygous plants in the semi-dwarf phenotype, as follows: ((0 × 2 small grain + 6 heterozygous specimens)/17 × 2 total semi-dwarf specimens) × 100 = 17.6 (Figure 4F). The grain weight of the GW2d60 homozygous large-grain semidwarf isogenic line (BC4F2) increased by approximately 31% over that of Koshihikari, and the stem length decreased by 26% (Table 1, Figure 5). The robust and high yielding isogenic Koshihikari line developed in this study by integration with d60 and GW2 is a new rice variety could bring about the “New Green Revolution” and overcome difficulties in this age of climate change and globalization.
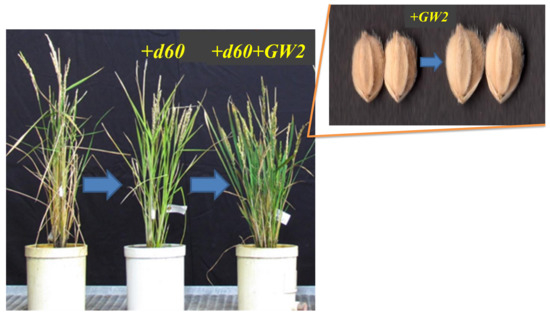
Figure 5.
Phenotypic alteration of isogenic Koshihikari by integration of d60 and GW2, derived from “Inochinoichi”.
3. Discussion
Crops in Japan and the world are being damaged by climate change caused by global warming. In 2017, the Japanese government put an innovation policy into place to contribute to the world through the development of high-yield crops for a “New Green Revolution.” The rice variety “Inochinoichi” is highly rated at both the consumer and producer levels, but the genes that control its large grain size have not been elucidated, so the buried genetic resources never have been used in breeding. In order to identify the genes that control large grain characteristics in Inochinoichi, we crossed a large grain rice variety with Koshihikari and, then, conducted a backcross with Koshihikari, which showed that the large grain size is controlled by a single gene. At the same time, we conducted a linkage analysis of a large grain gene using SSR markers and SNP markers across the 12 rice chromosomes that show polymorphisms between Nipponbare and Inochinoichi. As a result, we detected a linkage between the large grain gene and a DNA marker located 7.7 Mb from the distal end of the short arm of chromosome 2. Furthermore, we established a large grain isogenic line through five continuous back crosses with Koshihikari as the recurrent parent and then analyzed its whole genome using next-generation DNA sequencing. We successfully identified the target gene for large grains integrated in the genetic background of Koshihikari. There was no public information on the consensus sequence of Koshihikari, thus, we conducted a high-coverage whole-genome analysis, by first determining a consensus sequence for Koshihikari. We found that the responsible mutation for the large grain is a single nucleotide deletion located at 8.1 Mb from the distal end of the short arm of chromosome 2, in the GW2 gene. GW2 was identified as the causative gene at a QTL involved in grain width in large grain Japonica rice WY3; in Indica rice FAZ1, this allele (Os02g024410) encodes a new RING protein with E3 ubiquitin ligase activity [4]. E3 ubiquitin ligase is involved in the breakdown of proteins in the ubiquitin proteasome pathway [17]. RING-type E3 ubiquitin ligase can control seed development by catalyzing the ubiquitination of expansin-like 1 (EXPLA1), a cell wall-loosening protein that increases cell growth [18]. In contrast, the GW2 gene in WY3 lost function due to a frame shift caused by a single nucleotide deletion. It has long been known that the size of the awn covering the grain is one of the factors determining grain size [19]. In a FAZ1 near-isogenic line with GW2 from WY3 rice, the width of the awn was extended by 26.2% because of the increased cell number [4]. In this study, GW2, which was identified as a loss of function by the single nucleotide deletion, increased grain weight by 34% in the genetic background of Koshihikari.
In addition, it has been shown, by isolating genes affecting grain size, that cell number of awn is a factor that determines grain size and that grains get larger due to the loss-of-function of such genes [3,4,5,6,7,8]. GS3 (Os03g0407400), the first gene to be reported, encodes a transmembrane protein consisting of 232 amino acids [3]. It was shown through a functional complementation test that a loss-of-function by a mutation in the second exon caused an increase in grain size [20]. A gene coding for a new nuclear protein, qSW5 (Os05g0187500), makes grain width thin in Indica rice Kasalath [5]. The qSW5 allele in Kasalath reduces cell number in the width direction, which narrows the awn, and in turn suppresses the elongation of endosperm cells, resulting in narrowed grain width. On the contrary, the Nipponbare qSW5 allele has a 1212 bp deletion in its coding region, which results in a loss-of-function and allows for increase in grain width. GS5 (Os05g0158500) found in Indica rice Zhenshan97 is a regulatory factor, which encodes serine carboxypeptidase and controls positively for grain size [6]; the difference of GS5 expression affects grain size. GW8 derived from a high-yielding rice variety HJX74 is responsible for the QTL involved in grain width [7]; the HJX74 allele also increases grain width. GW8 is OsSPL16 (Os08g0531600), a gene that codes for a protein that positively controls cell proliferation. The GW8 allele in HJX74 also increases cell number, enlarging the awn and, consequently, causing increase in grain size. These past studies have shown that loss-of-function mutations such as GW2, GS3, GS5, qSW5, and GW8 cause an increase in cell number and subsequently, cause larger grain size.
TGW6 (Os06g0623700), a gene that increases the thousand kernel weight of Kasalath, has also been isolated [8]. TWG6 increases the cell number in the endosperm. The twg6 allele in Nipponbare codes for a protein that hydrolyzes indole-3-acetic acid (IAA)-glucose and synthesizes IAA, which promotes transition into the cell division stage. The Nipponbare tgw6 allele reduces the cell number in the endosperm as well as grain length. The TGW6 allele in Kasalath has a loss-of-function due to a single nucleotide deletion in its coding region, which means that grain length suppression via IAA does not occur, and grains elongate into a long phenotype characteristic of Indica rice. The distribution of TGW6 was investigated in the genetic stock and the Kasalath TGW6 allele was only found in one line of Oryza perennis and four local varieties in Indonesia [8]. This indicates that TGW6 was not a target of selection and thought to have been discarded during the domestication process. The Japanese large grain rice variety Oochikara is reported to have the same GW2 allele as those of WY3 and Inochinoichi. However, there is no historical relationship between Inochinoichi and Oochikara through their pedigree. Consequently, GW2 is thought to be an extremely rare allele.
As discussed earlier, large grain genes that confer Indica rice its characteristics have been identified in Indica or Chinese varieties. However, breeding to enlarge grain size has never been fully explored in the Japanese leading variety Koshihikari. We now have the opportunity to utilize a large grain gene that was ignored during the domestication process to develop a high-yield rice variety. In our study, we showed evidence that an isogenic large grain Koshihikari integrated with GW2 derived from Japanese rice via five times of backcrosses with Koshihikari has a 34% increased grain size compared to Koshihikari, and a taste score of 80.0, which is comparable to that of Niigata Koshihikari (81.0). Grain size-enlarged Koshihikari has the potential to become advantageously differentiated from US-made Koshihikari.
Rice yields around the world have doubled through the breeding of semi-dwarf varieties, which are representative of the Green Revolution in the mid-20th century, but the increase in yield is now leveling off. Additionally, there has been increased damage from lodging caused by severe weather events like the Western Japan floods and multiple typhoons under the recently intensified climate change; thus there is a need to develop rice plants that are sturdier and more robust. Also, with market liberalization through the Comprehensive and Progressive Agreement for Trans-Pacific Partnership (CPTPP) and negotiations for a Trade Agreement on Goods (TAG), there will soon be international competition in the rice market, so there is a need for low-cost and high-yielding rice. Thus, in order to make a breakthrough in high-yield breeding, which is dependent on a conventional semidwarfing gene sd1, we believe that to give rise to the New Green Revolution, semidwarfing should be used as a foundation with integrating/addition of genes related to high-yield including large grain and increased biomass. In this study, we combined the novel semidwarfing gene d60 and the large grain gene GW2 in the isogenic background of Koshihikari. In the BC4F2 by a cross Koshihikari × Koshihikari d60Gg (BC3F2), gametes with both d60 and the gametic lethal gene gal are not viable, so the segregation ratio was (1D60D60GglGal + 2D60D60Galgal + 1D60D60galgal + 2D60d60GalGal):2D60d60Galgal:1d60d60GalGal. Through this genetic process, a linkage between d60 and GW2 on chromosome 2 was discovered with a recombination value of 17.6, according to the deviated segregation ratio of the large grain allele, namely 11 GW2 homozygous:6 GW2gw2:0 gw2 homozygous in the semidwarf d60Gal homozygotes. The integration of GW2 and d60 resulted in a 20.0% increase in grain size and a 19.2 cm reduction in stem length compared to Koshihikari, which is effective to reduce the lodging risk that accompanies the increased panicle/grain weight. We obtained genetic achievement of the effective integration of genes for large grain and robustness. We made a breakthrough in breeding, which conventionally relied only on a single gene sd1, by combining the semidwarfing gene with a gene for a high yields-related factor. Such a combined genotype could secure high yields while providing robustness required to withstand climate change. In other words, this idea could contribute to the New Green Revolution. We have designated the large grain isogenic line with a 34% increased grain weight due to GW2, and the large grain semi-dwarf isogenic line due to GW2 + d60, which is capable of stable production with 31% increased grain weight/20 cm (26%) reduction in stem length, as “Koshihikari Suruga Gg” and “Koshihikari Suruga d60Gg”, respectively [21,22]. The two lines have been applied for plant variety registration. The taste and grain quality of these new varieties compared favorably with Niigata Koshihikari.
In China, grain size is being increased through the knockout of GW2 by genome editing [23]. On the contrary, in the countries under the ratification of the Cartagena Act, including European countries and Japan, there are barriers to the social implementation of genetically modified plants. In our study, we developed a large grain semi-dwarf isogenic variety for stable production that withstands climate change though smart breeding. This was done by identifying the gene responsible for large grain size by NGS and, then, integrating it into the reference Koshihikari genome by continuous backcrossing, to finally construct a targeted gene-integrated isogenic genotype. The variety “Koshihikari Suruga d60Gg” has an epoch-making phenotype as it integrates the large grain gene GW2, which increases grain weight by 34%, as well as the semidwarfing gene d60, which reduces lodging risk, into the Koshihikari genome. This new variety could potential be a Super Koshihikari that could replace the leading variety Koshihikari which currently has a 36% share but suffers from considerable damage by abnormal weather. Our breakthrough rice plant type that integrates both semidwarfing and large grain phenotype should be a key resource for the New Green Revolution.
4. Materials and Methods
4.1. Genetic Analysis
We focused on the large grain characteristics of Inochinoichi as a genetic resource for high yields. First, we analyzed the mode of inheritance of grain size in the F2 generation of Koshihikari × Inochinoichi. Furthermore, we conducted a progeny test using 50 F3 lines derived from 50 randomly selected F2 plants from Koshihikari × Inochinoichi. We then conducted five times of backcrosses with Koshihikari as a recurrent parent by using a large grain homozygous F3 plant (grain diameter: 0.75 mm) of Koshihikari × Inochinoichi as a nonrecurrent parent. We conducted genetic analysis of large grain in each BCnF2 generation through five times of backcrosses, and the large grain homozygous segregants in each BCnF2 were used as pollen parents for backcrosses with Koshihikari.
In order to develop an isogenic line that is both a semi-dwarf and large grains, the large grain semidwarf segregant in the F2 generation of Koshihikari d60 × Inochinoichi was used as a nonrecurrent parent to backcross with Koshihikari once, then Koshihikari d60 twice, then Koshihikari once again. Koshihikari d60 is an isogenic Koshihikari integrated with semidwarfing gene d60 derived from Hokuriku100 by seven times of continuous backcrosses, namely Koshihikari*7//(Koshihikari/Hokuriku 100 F2) [16]. Genetic analyses of the large grain gene and d60 were conducted in each backcross generation. The resulting genetically segregating populations were transplanted into Shizuoka University Ohya Field, and phenotypic traits (heading date, stem length, plant type, grain length, and grain width) of all plants were investigated. For grain characteristics, we evaluated the grain diameter at the early generation of F2, and grain area (grain length/2 × grain width/2 × π) in the near isogenic backgrounds through backcrossing.
4.2. Mapping of Large Grain Gene by DNA Markers
In order to map the large grain gene, 1328 F2 plants of Nipponbare × Inochinoichi were used. We sampled leaves from 371 small grain homozygous F2 plants. The leaves were powdered while being frozen by liquid nitrogen using a Precellys 24 high-throughput bead-mill homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France), and then genomic DNA was extracted using the cetyl trimethylammonium bromide (CTAB) method. A linkage analysis of large grain genes was conducted across the 12 rice chromosomes using SSR markers and SNP markers, which are polymorphic between Nipponbare and Inochinoichi. For the PCR reactions used to detect SSR markers, the mixtures were first heated to 95 °C for two minutes to denature the DNA, followed by 35 cycles of denaturing at 95 °C for 30 s, annealing at 50 °C or 55 °C for 30 s, and extension at 72 °C for 30 s. The SSR polymorphisms in the PCR products were analyzed by electrophoresis using a cartridge QIAxcel DNA Screening Kit (2400) in a QIAxcel electrophoresis apparatus (Qiagen, Hilden, Germany) at 5 kV for 10 min. SNP allele-specific TaqMan probes were designed and labeled using the fluorescent dyes FAM or HEX. The real time PCR reaction was used to amplify allele-specific fluorescence, by first heating the material to 95 °C for 30 s to denature the DNA, followed by 40 cycles of denaturing at 95 °C for 15 s, and annealing at 48 °C to 53.5 °C for 30 s.
4.3. Next-generation Sequencing (NGS) Analysis
Whole-genome sequencing of both Koshihikari and a large grain isogenic Koshihikari line (BC4F2), which was integrated with large grain gene derived from Inochinoichi by four times of back crosses into the genetic background of Koshihikari, were conducted. The leaves were powdered using a mortar and pestle while being frozen by liquid nitrogen. The DNA was then extracted using the CTAB method. Genomic DNA was fragmented and simultaneously tagged with the Nextera® transposome (Illumina, Rockville, MD, USA) such that the peak size of fragments was approximately 500 bp. Adapter sequences, including the sequencing primers, were synthesized in both ends via PCR. After the size selection of DNA fragments using magnetic beads, the DNA library was prepared following qualitative check by Bioanalyzer 2100 system (Agilent Technologies, Inc., Palo Alto, USA), and quantitative measurement by Qubit® Fluorometer (Life Technologies; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The sequencing data were gained with paired-end reads using a HiSeq next-gen sequencer. The read sequences obtained were mapped using Burrows-Wheeler Aligner (BWA) software to the Nipponbare genome as a reference.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/21/5442/s1. Figure S1. Complementary gamete lethal between semidwarfing gene d60 and gamete lethal gene gal.
Author Contributions
Conceptualization—M.T.; Methodology—M.T.; Investigation—M.T., S.Y., Y.U.; Resources—M.T.; Writing—Original draft preparation—M.T.; Writing—Review and editing—M.T.; Project administration, M.T.; Funding acquisition—M.T.
Funding
This work is founded by the Adaptable and Seamless Technology Transfer Program (A-STEP), Industry-Academia Joint Promotion Stage, High Risk Challenge Type grant by Japan Science and Technology Agency (JST) to Motonori Tomita, project ID14529973, entitled “Development of super high yield/large grain/early and late ripening rice suitable to the era of globalization and global warming by Next-Generation Sequencer/genome-wide analysis”, since 2014 to 2018.
Acknowledgments
The author thanks Rieko Kamioka; Tomoaki Tsuchiya; Hideumi Ebata for their assistance of phenotypic investigation.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Khush, G.S. Green revolution: Preparing for the 21st century. Genome 1999, 42, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, B.S.; Jabran, K.; Mahajan, G. Rice Production Worldwide; Springer International Publishing AG: Cham, Switzerland, 2017; p. 547. [Google Scholar]
- Fan, C.; Xing, Y.; Man, H.; Lu, T.; Han, B.; Xu, C.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nature Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Shomura, A.; Izawa, T.; Ebana, K.; Ebitani, T.; Kanegae, H.; Konishi, S.; Yano, M. Deletion in a gene associated with grain size increased yields during rice domestication. Nature Genet. 2008, 40, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J.; et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nature Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nature Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, K.; Hirotsu, N.; Madoka, Y.; Murakami, N.; Hara, N.; Onodera, H.; Kashiwagi, T.; Ujiie, K.; Shimizu, B.; Onishi, A.; et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nature Genet. 2013, 45, 707–711. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Kurata, N.; Wei, X.; Wang, Z.X.; Wang, A.; Zhao, Q.; Zhao, Y.; Liu, K.; Lu, H.; Li, W.; et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Japan Ministry of Agriculture, Forestry and Fisheries. TPP related information. Available online: http://www.maff.go.jp/j/kanbo/tpp/ (accessed on 19 September 2019).
- Japan Meteorological Agency. Heisei 30 July 7 heavy rain and floods. Available online: https://www.data.jma.go.jp/obd/stats/data/bosai/report/2018/20180713/20180713.html (accessed on 19 September 2019).
- Japan Meteorological Agency. 2018 (Heisei 30) Typhoons (Quick Report). Available online: https://www.jma.go.jp/jma/press/1812/21f/typhoon2018.pdf (accessed on 19 September 2019).
- Japan Ministry of Agriculture, Forestry and Fisheries. Damage situation by Heisei 30 July 7 heavy rain and floods. Available online: http://www.maff.go.jp/j/saigai/ooame/20180628.html (accessed on 19 September 2019).
- Japan Meteorological Agency. Abnormal Weather Risk Map. Available online: http://www.data.jma.go.jp/cpdinfo/riskmap/heavyrain.html (accessed on 19 September 2019).
- Japan Meteorological Agency. Long-term changes in the number and frequency of short-term heavy rains observed at AMeDAS. Available online: http://www.jma.go.jp/jma/kishou/info/heavyraintrend.html (accessed on 19 September 2019).
- Tomita, M. Combining two semidwarfing genes d60 and sd1 for reduced height in ‘Minihikari’, a new rice germplasm in the ‘Koshihikari’ genetic background. Genet. Res. Camb. 2012, 94, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Hauksdóttir, H.; Troy, A.; Herschleb, J.; Kraft, E.; Callis, J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant. Physiol. 2005, 137, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.S.; Kim, Y.J.; Markkandan, K.; Koo, Y.J.; Song, J.T.; Seo, H.S. GW2 functions as an E3 Ubiquitin ligase for rice Expansin-Like 1. Int. J. Mol. Sci. 2018, 19, 1904. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Saito, K.; Yamazaki, K.; Mikami, T. Environmental response of yielding capacity in isogenic lines for grain size of rice. Japan. J. Breed. 1987, 37, 309–317. [Google Scholar] [CrossRef]
- Takano-Kai, N.; Jiang, H.; Kubo, T.; Sweency, M.; Matsumoto, T.; Kanamori, H.; Padhukasaharam, B.; Bustamante, C.; Yoshimura, A.; Doi, K.; et al. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 2009, 182, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Official Gazette No. 7080, 14 Aug. 2017, Ministry of Agriculture, Forestry and Fisheries. Plant varietal registration application No. 32364, “Koshihaikri Suruga Gg” 2017, Tokyo, Japan. Available online: http://www.hinshu2.maff.go.jp/vips/cmm/apCMM111.aspx?SHUTSUGAN_NO=32364&LANGUAGE=English (accessed on 14 August 2017).
- Official Gazette No. 7080, 14 Aug. 2017, Ministry of Agriculture, Forestry and Fisheries. Plant varietal registration application No. 32365, “Koshihikari Suruga d60Gg” 2017, Tokyo, Japan. Available online: http://www.hinshu2.maff.go.jp/vips/cmm/apCMM111.aspx?SHUTSUGAN_NO=32365&LANGUAGE=English (accessed on 14 August 2017).
- Mishra, R.; Joshi, R.K.; Zhao, K. Genome editing in rice: Recent advances, challenges, and future implications. Front Plant Sci. 2018, 9, 1361. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).