Abstract
For the viticulture of the future, it will be an essential prerequisite to manage grapevine diseases with fewer chemical inputs. The development and the deployment of novel mildew resistant varieties are considered one of the most promising strategies towards a sustainable viticulture. In this regard, a collection of 102 accessions derived from crossing Vitis hybrids with V. vinifera varieties was studied. In addition to the true-to-type analysis, an exhaustive genetic characterization was carried out at the 11 reliable mildew resistance (R) loci available in the literature to date. Our findings highlight the pyramiding of R-loci against downy mildew in 15.7% and against powdery mildew in 39.2% of the total accessions. The genetic analysis was coupled with a three-year evaluation of disease symptoms in an untreated field in order to assess the impact of the R-loci arrangement on the disease resistance degree at leaf and bunch level. Overall, our results strongly suggest that R-loci pyramiding does not necessarily mean to increase the overall disease resistance, but it guarantees the presence of further barriers in case of pathogens overcoming the first. Moreover, our survey allows the discovery of new mildew resistance sources useful for novel QTL identifications towards marker-assisted breeding.
1. Introduction
Plant diseases cause billions of dollars in lost harvest annually, and in some instances, these losses have severe consequences for humans. One of the most convenient, inexpensive and environmentally sound ways to control plant disease is to utilize disease-resistant varieties, and plant breeders make extensive use of classically defined R genes [1]. Recent work has revealed the structure of a number of plant R genes, and a striking degree of similarity among these genes has been observed. The majority of resistance genes encode proteins classified as NBS-LRR proteins because they contain a nucleotide binding site (NBS) domain and a leucine–rich repeat (LRR) domain, followed by Toll/Interleukin-1-receptors (TIR); Coiled coil (CC); Transmembrane domain (TrD); PEST aminoacid domain; Endocytosis cell signaling domain (ECS); Nuclear localization signal (NLS); WRKY amino acid domain; Helminthosporium carbonum (HC) toxin reductase enzyme. Some categories can be more frequently associated with specific resistances (e.g., fungi vs oomycetes) [2]. Many resistance genes occur in complex loci that contain multiple copies of closely-related gene sequences. The concept of haplotype is used to describe the precise complement of related genes occurring in a particular variant of a complex locus. By contrast, there are some simple loci that encode only a single known resistance allele [3].
In grapevine, up to now a list of 27 genomic regions is reported associated with downy mildew (DM) resistance and 13 related to powdery mildew (PM) resistance [4], hereafter referred as disease resistance (R) loci. So far the thorough information about responsible genes has been achieved through map-based cloning approaches in a few cases: Run1 [5,6], Rdv1 [7] and Rpv1 [8]. The genetic base of resistance to PM, DM and other grapevine diseases originates from Vitis species which are natural sources of resistance, mainly deriving from North America and more recently from the Far East [9]. Most of them confer varying levels of partial resistance, while others coming from Muscadinia rotundifolia and V. piasezkii confer total resistance in the genetic context where they are studied. The disease resistance breeding history in grapevine began in the late 19th century, when the import of grapevine material from America into Europe (primarily France) enabled the introduction of serious, previously absent grapevine pests and pathogens for which the native European vines (V. vinifera L.) had no resistance. In particular, the North American genotypes were used for the reason of being resistant to phylloxera and the first interspecific hybrids, mainly rootstocks, were bred to overcome the insect threat [10]. Starting in the second half of the 19th century, several attempts on combining different resistance traits of American grapevines (V. riparia, V. labrusca, V. aestivalis and V. berlandieri) with qualitative characteristics of European species were made, leading to the creation of interspecific resistant varieties. Besides the phylloxera plague, a serious invasion of fungal diseases contributed to the massive destruction of European vineyards and led to the increase of hybrids. Very important in the history of interspecific breeding are the genotypes coming from France called first-generation hybrids, usually meaning crossings between American species and cultivated French varieties, also called “direct producers”, i.e., grapes grown on own roots and used for wine making [10]. This kind of crossings was made in the first quarter of the 20th century mainly by the breeder Albert Seibel. The second-generation hybrids, basically crossings between first generation hybrids among themselves or with cultivated European varieties, came out later in the century and were performed by Bertille Seyve and Victor Villard. They contain a higher percentage of the vinifera genome, thereby increasing the quality of the wine [11]. More recently, marker-assisted selection in combination with several backcrossing with vinifera varieties led to the development of fungi resistant grapes carrying multiple disease resistance genes and a significant percentage (more than 85%) of vinifera in their pedigree [12,13].
Despite a European Council Regulation (No 1493) in 1999 [14] that enabled to produce “quality wines” only from varieties belonging to botanical species V. vinifera, resistant cultivars are frequently used in some northern European viticultural regions. Environmental, health and cost concerns are leading producers to reconsider hybrids or interspecific crossings. First and foremost is the worry about the need for intensive use of fungicides to control diseases. A 2003 report of the European Food Safety Authority (EFSA) estimated that viticulture uses 40% of the crop protection products in European agriculture. For this reason, the future trend is addressed to reduce pesticides as much as possible [15]. In this respect, the almost 100 years of breeding efforts for interspecific crossings cannot be considered as unnecessary. Eventually, the attitude is changing: In a 2009 revision [16], the European Union relaxed the regulations prohibiting hybrids. Although the highest classification (Protected Designation of Origin, PDO) requires V. vinifera varieties, hybrids can be used in the next level (called Protected Geographical Indication, PGI). Interspecific, disease resistant hybrids are generally referred to as PIWI (from German: pilzwiderstandsfähig, meaning “fungal disease resistant”) and they are now accepted as V. vinifera varieties in the most European Catalogues [17]. Nowadays “PIWI” is also the name of a producer group devoted to the “dissemination of fungus-resistant grape varieties” with 350 members from 17 European and North American countries, some of whom running private breeding programs [18]. Moreover, in order to sustain the resistant variety growth in the marketplace, producers will need to overcome the stigma still associated with hybrid-derived wines.
Nowadays, on the one hand, the situation seems very favourable for the genetic improvement for disease resistance in grapevines: Many resistance sources are available, and several resistance factors with various effects have already been discovered. On the other hand, we know that disease resistances are not necessarily stable traits, and the protection could be “quickly” overcome by a virulent strain of the pathogen [19]. The advent of molecular genetics and associated technology like marker-assisted selection has led to the emergence of a new field in plant breeding, gene pyramiding [20]. It is now widely accepted that pyramiding resistance genes could be an effective plan to control a large range of pathogen strains as well as combining various defence mechanisms, all valuable strategies to increase resistance durability. Unfortunately, durability—the preservation of disease resistance genes over time—also depends on environmental conditions and agronomic practices, which influence the development of pathogen populations. From a practical point of view, the sustainable management of resistance aims at reducing the selection pressure applied by the resistance genes on the pathogen populations thanks to potentially durable genetic constructions and resistance deployment strategies, including cultivation practices [21]. One of the missing links for further improvement in breeding is a phenotypic evaluation of the genetic resources in the same environmental conditions taking into account the presence of resistance loci pyramids in their genome.
The aim of this study was to fill this gap assessing DM and PM resistance symptoms in a set of 102 accessions grown in an untreated field located in Northern Italy. The three-year experiment allowed the evaluation of the genetic material’s response to DM and PM in field compared to the R-loci arrangement in every accession and could gather the effect of different R-loci, alone or in pyramids, on mildew disease outbreaks in the same environmental conditions.
2. Results and Discussion
2.1. Fingerprinting
The present collection was studied within the international, collaborative project VITISANA and is thus referred to as the VITISANA collection in this work. It comprises 102 accessions, 18 of which were confirmed as true-to-type (TTT) by DNA profiling and comparing profiles at the Vitis International Variety Catalogue (VIVC) database [9]. Twenty-five profiles corresponded to well-known accessions without genetic information available at VIVC database. For four accessions (‘Duna Gyöngye’, ‘Odysseus’, ‘Viktoria Gyöngye’ and ‘Zarya Severa’) the TTT was not validated since they showed another genetic profile according to the VIVC database. On a total of 55 accessions, including 37 progeny individuals and 18 breeding lines, TTT results could not be disclosed since they are part of selections by private breeders (data on pedigree not available) (Table S1). For this reason, to increase the genetic information, a dendrogram reporting the genetic distance among all studied accessions is depicted in Figure 1. The dendrogram showed five pair identities (‘3/23/08’ = ‘3/1/06’; ‘Zarya Severa’ = ‘GM6495-3’; ‘Viktoria gyöngye’ = ‘Duna Gyöngye’; ‘Lela’ = ‘Odysseus’, ‘IV045’ = ‘IV069’), revealing the presence of 97 unique genetic profiles (genotypes). Taking into consideration the origin of the genotypes, we noticed a cluster comprising most of the progeny individuals in the first node located at the lower part of Figure 1. We speculated that this group shared at least one parent, thus identified as the candidate MW1 or IV062 genotypes based on 9 (highly polymorphic and neutral) reference SSR data.
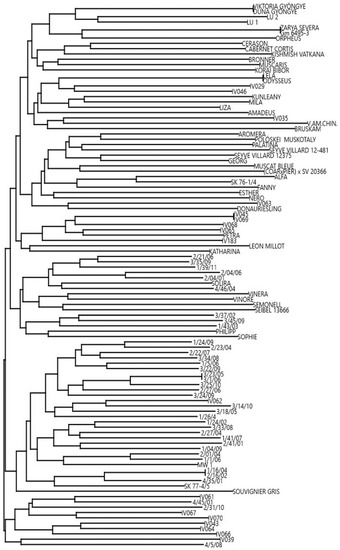
Figure 1.
Dendrogram showing the genetic distance based on cluster analysis (Neighbour Joining clustering; Euclidean similarity).
2.2. R-Loci Characterization
In an “all-vs-all” approach, all Vitis hybrids were examined at the 11 screenable and reliable R-loci highlighting the presence of eight R-loci (Rpv1, Rpv3, Rpv10 and Rpv12 for DM and Run1, Ren1, Ren3, Ren9 for PM) in a single (e.g., Ren1), combined (e.g., Rpv12 + Ren9) and stacked/pyramided (e.g., Rpv10 + Rpv3-3) status. The latter definition is in agreement with [22]. The Rpv3 locus represents a peculiar case where different resistant haplotypes were characterized [23] and for which we referred to a paired status (e.g., Rpv3-1 + Rpv3-2). The absence of both Run2.1/2.2 variants as well as Rpv14 and Ren2 underlined the respective lack of cultivars derived from M. rotundifolia and of V. cinerea accessions in the pedigree of the analyzed genotypes. Certain R-loci were not taken into consideration from the beginning since their donor was private and not exploitable to develop any accessions belonging to the VITISANA collection; this is the case of V. romanetii (Ren4, [24,25]) and V. piasezkii (Ren6 and Ren7, Rpv15 and Rpv16, [26]; Pap et al., in preparation). Not for all R-loci, an appropriate choice of markers could be retrieved from literature. Although we have taken into consideration the exhaustive overview of traits and original donor variety or species accession available at [9], we identified only for some R-loci associated with DM and PM resistance a defined set of associated SSR markers that were robust and exploitable for Marker-Assisted Parental Selection (MAPS) and the derived Marker-Assisted Seedling Selection (MASS) practice. Reasons for this are (i) the lack of clear information, especially in the oldest publications, (ii) large genomic intervals that would require additional fine mapping, and (iii) the absence of confirmed QTLs that would request marker validation in additional populations. In fact, moving from publication to application domain is challenging. Especially for fruit trees, most of the publication results derive from investments from funders with a strategic scientific mission, leading to rare emphasis on applied value in breeding programs.
As stable and co-dominant markers, SSRs are currently the marker system of choice for the Marker-Assisted Breeding (MAB) program in grapevine, where reliable, efficient and cost-effective molecular markers have to be available. This type of markers demonstrated to provide robust phenotype correlation with disease resistance (e.g., [22]) as well as other traits. Indeed, SSRs have some limitations, such as the need of an expensive equipment for allele sizing through capillary electrophoresis and high mutation rates compared to other DNA markers. The latter tendency generates size variation in DNA regions otherwise identical-by-descent and by their model of evolution, which vice versa produce identical electromorphs via independent mutational events—known as homoplasy—confounding the studies of genetic variation within and among populations (reviewed by [27]). From here comes the need of developing new flanking markers and to convert the original ones into less variable marker types, as the point mutation-based. To date, although few SNPs – also in terms of haplotype blocks – have been developed for grapevine Marker-Assisted Selection (MAS) applications (e.g., [28,29]), they are becoming more favoured as a marker system, since they are amenable to high-throughput genotyping platform [30]. Lately, to bridge the gap between marker development and MAS implementation, a novel practical strategy with a semi-automated pipeline, which incorporates trait associated SNP discovery, low-cost genotyping through amplicon sequencing and decision making, has been developed [31]. Unlike microsatellites and more similar to point mutations, each InDel is a unique and irreversible molecular event, which helps tagging more effectively a given haplotype. For this reason, [27] have very recently discovered InDel tags for the Rpv3-1 haplotype and proposed them as a significant improvement in terms of marker informative content, ease of allele scoring and MAS efficiency.
A list of the R-loci detected in each genotype of the VITISANA collection is shown in Table 1. Considering the incidence of each R-locus independently from its combination with other R-loci, a prevalence of Rpv3, Ren3 and Ren9 was observed. Regarding Rpv3-dependent resistance to DM, the Rpv3-1 haplotype was the most frequent (56.9%, deriving from ‘Seibel 4614′), while the Rpv3-2 (6.8%, conserved in the ‘Munson’ lineage) and the Rpv3-3 haplotype (3.9%, tracing back to ‘Noah’) were fewer. The Rpv3-1 haplotype was firstly identified in the German hybrid ‘Regent’ [32,33] and in the Hungarian hybrid ‘Bianca’ [34] through QTL mapping. The Rpv3-1 presence originates from ‘Seibel 6468′, the only offspring of the ancestor ‘Seibel 4614′ that disseminated extensively the haplotype in the germplasm repositories until now. ‘Seibel 6468′ participated in the generation of ‘Villard blanc’ (also known as ‘Seyve Villard 12-375′), one of the most deployed hybrids in grapevine breeding programs. The predominance of this haplotype definitely indicates its fixation during selection since it confers a superior resistance with an ETI (effector triggered immunity) associated necrosis perfectly capable to restrict the pathogen [27]. Rpv3-1 resistance depends on an inducible response specifically elicited by an avirulent strain of P. viticola and is a typical Hypertensive Response (HR), compatible with the cascade of events initiated by the products of NB-LRR and receptor like protein kinase genes, located within the Rpv3-1 locus [35]. Upon the comprehensive study reported by [23], two further wild relative Rpv3 haplotypes have been validated in segregating populations: Rpv3-2 has recently been confirmed by QTL mapping in ‘GF.GA-47-42′ × ‘Villard Blanc’ segregating population [36], while Rpv3-3 has been characterized in a ‘Merzling’ × ‘Teroldego’ progeny [37]. These two haplotypes are less represented in grapevine breeding selections [23] and therefore are novel and valid allelic variants, considering that Rpv3-1 was discovered to be ineffective against a specific P. viticola isolate [19,38].

Table 1.
Characterization of resistance (R) loci in each studied accession based on the “all-vs-all” approach: The occurrence of the R-locus is highlighted in red, while its absence is represented by an empty cell.
The second R-locus against DM well represented in the VITISANA collection was Rpv12 (17.3%), coming from ‘Zarya Severa’ and deriving from V. amurensis. The Amur grape is native to the cool climate of the Far East (Siberia, China, Korea and Japan) arousing the interest of breeders concerned to incorporate cold tolerance into V. vinifera (e.g., expedition by Vavilov in 1920–1940). Moreover, they noticed that some accessions were not significantly damaged by P. viticola under conditions highly conductive to DM. Soviet breeders (Michurin, Negrul and Potapenko) contributed to the introduction of these accessions into the Russian breeding program, and thanks to the networking with the Soviet bloc, Eastern European breeders shared Amur material that since 1960s was present in the Continental Europe [39]. Before Rpv12, another V. amurensis resistance gene, Rpv10 coming from ‘Severnyi’ [40], a full sibling of ‘Zarya Severa’, was discovered. In the VITISANA collection Rpv10 is present with a percentual of 7.8%. For both Rpv10 and Rpv12, only QTL mappings are available in the literature to date, while lacking gene expression or functionals studies. For both loci, the presence of large CC-NBS-LRR clusters has been detected along the reference genome [39,40].
Concerning the resistance to PM, Ren3 (50%) and Ren9 (49%) (both derived from ‘Regent’) were the most abundant, followed by a few genotypes with Run1 (8%, derived from M. rotundifolia) and only one genotypes with Ren1 (coming from V. vinifera cv. Kishmish vatkana). Run1 (and Run2), as an example, derive from M. rotundifolia. Although introduced in Europe in the late 19th century together with most of the other American Vitis species, it has not elicited any real interest in European growers, since all of the few cultivation attempts failed at that time [41]. Only a pseudo-backcross strategy succeeded in the introduction of the single locus in the V. vinifera genome [42], allowing to use it for resistance breeding programs. Also Ren1 from V. vinifera cv. Kishmish vatkana [43] was only recently utilized for pyramiding multiple resistances. Both Ren3 [32,33,44,45] and Ren9 [46] are located on chromosome 15 and are frequently inherited together. Being firstly discovered in ‘Regent’, the two R-loci originate from ‘Chambourcin’, one of the resistant varieties coming from French breeding efforts [23] and a popular parental line for breeders. This is probably the reason why, compared to the other PM resistance traits, these loci are quite widespread among the VITISANA genotypes. All the major PM responsive QTLs known as Run/Ren loci have been mapped in positions where various RGAs encoding TIR-NBS-LRR and CC-NBS-LRR type resistance proteins where mapped [47].
Interestingly, 12.7% and 33.3% of the genotypes were missing any of the analysed R-loci associated with DM and PM resistance, respectively. This suggests that these genotypes either are susceptible to one of the two diseases or represent new sources of resistance against DM or PM. The fact that most genotypes lack PM resistance demonstrates that breeding activities, concentrated in temperate-humid climates, are preferably focused on introducing resistance to DM.
Considering the total number of R-loci detected in each genotype, a molecular picture of the VITISANA collection is shown in Figure 2. Within the collection, 23.5% of the genotypes carried a single R-locus associated with DM or PM resistance, and the same percentage carried two loci. Further, our findings showed that pyramiding (stacking of at least two R-loci against the same disease) of Rpv loci occurred in 15.7% and of Run/Ren loci in 39.2% of the collection (data not shown). A particular case is represented by the paired status Rpv3-1 + Rpv3-2 detected in ‘MW1’ and ‘Katharina’ at the hypervariable Rpv3 locus. In addition, 40.2% contained three loci with the combination 2 Rpv loci + 1 Ren/Run locus or 1 Rpv locus + 2 Run/Ren loci. Only 4.9% contain four R-loci (2 Rpv + 2 Run/Ren loci) and therefore resulted pyramided for both diseases. In 7.9% of the genotypes, no Rpv and Run/Ren locus was found at all according to analyses with the markers able to reliably foresee resistance traits; these selections represent putative novel sources of disease resistance, prior the consideration of their phenotypic data. Taking into account the dendrogram (Figure 1) and considering the R-loci arrangement, we observed a group (from ‘Lela’ to ‘Liza’ in Figure 1) carrying Rpv12, followed by a cluster (from ‘IV045′ to ‘Petra’) with Rpv12 in some cases combined with Run1. We also identified a group from ‘IV067’ to ‘IV066’ characterized by Rpv1 + Rpv12 + Run1; these genotypes belong to the same breeding objectives of the private breeding platform InnoVitis based on closely related resistance sources (Tutzer E., personal communication). As expected, progeny individuals carrying Rpv3-1 + Ren3 + Ren9 clustered together.
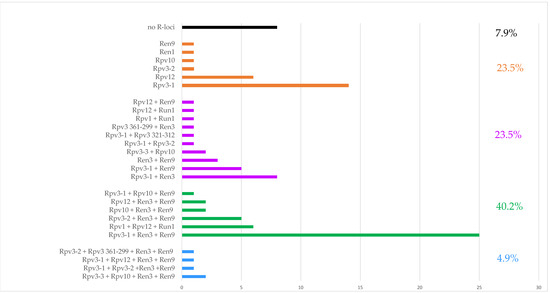
Figure 2.
Grouped bar chart showing the percentual of accessions with single resistance locus, combination of two, three and four resistance (R) loci and absence of any studied R-locus. Rpv: Resistance to Plasmopara viticola; Run: Resistance to Uncinula necator (from Muscadinia spp.); Ren: Resistance to Erysiphe necator (from Vitis spp.).
These results provided an overview on the genetic material present in the VITISANA collection. Putting them in relation with other data is challenging since few articles describe the presence of resistance QTLs for DM and PM in genotypes derived from breeding programs (e.g., [22,25,47,48]). The literature rather tends to concentrate on QTL studies and association analyses in order to deepen the knowledge on R-loci, as well as reported reviews on process [12] and dissertations on perspectives, especially in the post-genomics area [49,50]. On the contrary, data on MAB activities traditionally are not reported. In fact, as mentioned above about QTL validation, translating the research findings into application cases is mostly considered as not relevant and not required. In addition, the breeders’ mentality consists of considering the R-loci characterization as a simple, although advanced, tool in order to reach quickly the goals of yield and quality grape.
Worldwide schemes for pyramiding resistance QTLs are currently applied in breeding programs for wine grapes, table grapes and rootstocks to boost cultivar development via MAS, including early seedling selection and parental choice prior to crossing, all focused on QTLs with major effect and none on QTLs with minor effects. The shared idea was to combine resistance QTLs with complementary modes of action for breeding effective and potentially durable resistance, relying on the fact that the effects of resistance QTLs are often additive. In addition, another relevant factor to be taken into account is that, if the target QTL contains resistance genes and their homologs tightly linked to genes with large negative effects on other traits, these undesirable genes maybe transferred together with the target gene into the recipient line and result in the reduced performance of other traits (linkage drag) [20]. Finally, given the relatively recent history of pyramiding in grapevine, the last element to consider is the (future) genetic load. The average individual taken from a population with a low genetic load will generally, when grown in the same conditions, have more surviving offspring than the average individual from a population with a high genetic load. Deleterious mutation load is the main contributing factor to genetic load overall. Inbreeding increases homozygosity. In the short run, an increase in inbreeding increases the probability with which offspring get two copies of a recessive deleterious alleles, lowering fitness via inbreeding depression. However, in a species that habitually inbreeds, e.g., through self-fertilization, recessive deleterious alleles are purged [51]. Since grapevine breeding plans in case foresee one step of self-crossing, the possibility to purge deleterious alleles is remote. Another contributor to the genetic load overall is the recombination/segregation load. Combinations of alleles that have evolved to work well together may not work when recombined with a different suite of coevolved alleles, leading to outbreeding depression. Segregation load is the presence of underdominant heterozygotes (i.e., heterozygotes that are less fit than either homozygote). Recombination load arises through unfavourable combinations across multiple loci that appear when favourable linkage disequilibria are broken down. Recombination load can also arise by combining deleterious alleles subject to synergistic epistasis, i.e., whose damage in combination is greater than that predicted from considering them in isolation [52]. In the case of mainly outcrossed species as grapevine, these events can definitely occur.
2.3. DM and PM Resistance Evaluation in an Untreated Field
Even though the collection consists of 102 accessions, only 89 were available for field disease evaluations. The reason is attributable to low yields for 13 genotypes, impeding DM and PM bunch assessment in both periods during the first two years. Comparing the distribution of the genotypes according to standard scores defined by the International Organisation of Vine and Wine (OIV) and describing DM resistance level on leaves and bunches, for both organs we noticed a clear difference between the three years. In 2016, resistance levels on leaves and bunches ranged from low to mid degree, whereas in the two following years, the distributions significantly skewed towards a high (2017) and a moderate level of resistance (2018) (Figure 3A,B, Figure S1, Table S2). PM resistance was observed to be significantly lower in 2016, while increasing in 2017 and 2018, with 2016 remaining the most susceptible year in both disease observations.
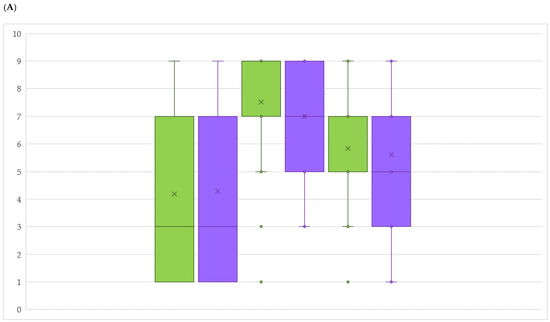
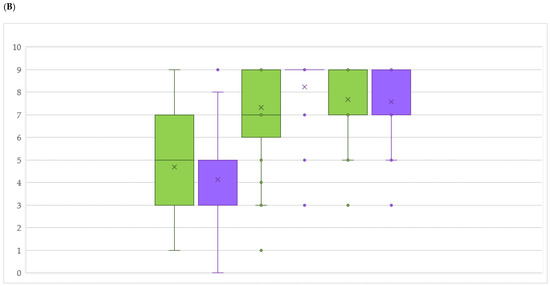
Figure 3.
Box plots comparing OIV scores of resistance level in years 2016, 2017 and 2018 for downy (A) and powdery (B) mildew on leaves (green) and bunches (violet). Per each year, the lowest score between evaluations was chosen.
Weather patterns revealed that 2016 was the rainiest year with a seasonal rainfall of 522.4 mm, whereas in 2017 and 2018 the rainfall in the same period was 491.4 mm and 381.2 mm, respectively (Figure S2). Interestingly, in 2016 the rainfall was particularly concentrated in April and May (sum of 183.4 mm), whereas it reached only 91 mm and 127 mm, respectively, in the two successive years. Indeed, the differences in DM resistance between the years can be attributed to the varying weather conditions, especially to rainfall [53]. Rain can initiate the process of oospore germination, breaking the dormancy (primary infection); in the successive periods, temperature and availability of water are fundamental to produce sporangia (secondary infection) [54]. Since the mean temperature registered during the three considered growing seasons were not so diverse, the different rainfall amount might explain the significant difference in the disease symptoms found both on leaves and grapes in 2016 (more severe) than in 2017 and 2018 (less severe).
Exploring PM resistance behaviour over years, in 2016 the rainfall was high between May and June, determining the timing of ascospore release, followed by a peak of humidity in June allowing fungus growth (Figure S2). Under these conditions the more susceptible conditions in 2016 can be explained compared to less humid periods in 2017 and 2018, that probably limit the development of the disease. As stated in [55], the ideal conditions for the growth of PM were the temperature from 20°C to 28°C with 80%–90% relative humidity. In 2017 and 2018, the mean temperature was higher with peaks near 35°C in July and August, in contrast to 2016 where average temperatures were lower (Figure S3). As reported by [38,56], high temperatures could inhibit spore germination and slow down the growth of the fungus, until the death of the spores when temperatures reached 40°C.
Pairwise correlation analyses reveal that, in a 3-year average, field resistance before veraison significantly correlates with resistance before harvest (p < 0.001, Figure S4). Especially for DM symptoms, correlation reached values of ρ = 0.746 and 0.824 for leaves and grapes, respectively. This correlation was slightly lower in PM symptoms with respectively ρ = 0.579 and 0.571.
To date, very few articles are available to be compared to the data presented here evaluated in such a range of genotypes. Some research dealt with the evaluation of hybrids in terms of performance (e.g., [57]) and most studies on QTL analysis of disease resistance against DM and PM resorted to use regularly leaf disc experiments (in vitro assay). For DM assessment in untreated fields, our study could be compared with the recent work of [58] where 28 promising hybrids were screened with a leaf discs assay and evaluated for foliar and cluster downy mildew resistance in an untreated field trial over three successive years. The common genotypes were only height, namely, ‘Aromera’, ‘Bronner,’ ‘Cabernet Cortis’, ‘Fanny’, ‘Leon Millot’, ‘Muscaris’, ‘Nero’ and ‘Pölöskei Muskotaly’. ‘Bronner’ resulted as the most resistant genotype at both leaves and cluster level in both works. Two additional resistant genotypes were ‘Muscaris’, that in the previous work appeared highly resistant as well, and ‘Leon Millot’. Surprisingly, ‘Leon Millot’ had a very high level of resistance in three successive years for both leaves and bunches, while in [58] the level of resistance was medium high for the leaves, and moderate for the grape cluster. Considering the remaining common genotypes, ‘Aromera’ shows a lower level of resistance in both organs, ‘Cabernet Cortis’ and ‘Fanny’ have higher levels of resistance in the leaves. In ‘Nero’ we noticed a lower level for leaves, but a higher level for bunches. Finally, ‘Pölöskei Muskotaly’ has shown the same mean behaviour for both. Another comparison for DM assessment using OIV 452 descriptor (leaves) in field exposed to natural pressure of pathogen was the work by [59]: The common genotypes were ‘Lela’, ‘Liza’, ‘Mila’, ‘Petra’, ‘Cerason’, ‘Seibel 13666′ and ‘Seyve Villard 12375′. ’Lela’, ‘Mila’, ‘Seibel 13666′ and ‘Seyve Villard 12375′ showed similar data with slight differences in the mean scores (OIV 452). Very different, on the contrary, were ‘Liza’ and ‘Petra’ that showed clearly higher values. Totally different was ‘Cerason’ that resulted susceptible, compared to a medium resistance in the Czech fields. In another work by [60], the only genotype in common was ‘Leon Millot’ where it showed once again a high level of field resistance, but the assessments were evaluated for several years carrying one or two fungicide treatments and the assessment of mildew damage was evaluated with a five grade scale taking into account leaves, shoots and berries. In [61], evaluations for DM and PM resistance were performed under field conditions in Hungary. The common genotypes were five, ‘Amadeus’, ‘Korai Bibor’, ‘Orpheus’, ‘Viktoria Gyöngye’ and ‘Duna Gyöngye’. Since these last two genotypes resulted not to be TTT in our work, they could not be compared. For DM resistance, the remaining genotypes displayed the same resistance levels for foliar (‘Korai Bibor’ and ‘Orpheus’) and bunches (‘Amadeus’ and ‘Orpheus’). In ‘Amadeus’ foliar resistance was detected at a lower degree, the same happened for bunches in ‘Korai Bibor’. For PM resistance, ‘Orpheus’ had similar result whereas ‘Korai Bibor’ showed definitely lower resistance levels in bunches whereas ‘Amadeus’ showed a higher resistance levels in both organs, especially in leaves.
2.4. Comparison between Mildew Resistance in Untreated Field and the Arrangement of Rpv or Run/Ren Loci
Merging the field resistance data with the presence of single or stacked R-loci, the contribution of a particular genetic arrangement to the level of resistance in the field could be analysed. We excluded resistance data and genetic arrangements represented by a single genotype, since they were not informative during ANOVA testing. Concerning DM resistance (Figure 4A), the single Rpv10 and Rpv12 showed a significantly higher degree of 3-year field resistance on leaves compared to genotypes with the single Rpv3-1. About bunches, this higher rank is confirmed only for the comparison between Rpv12 and Rpv3-1. The stacked Rpv1 + Rpv12 genotypes presented a significantly higher resistance than the single Rpv3-1 genotypes both at leaf and bunch level. Regarding bunches only, DM resistance was significantly higher in genotypes containing Rpv10 stacked with Rpv3-3 compared to genotypes with the single Rpv3-1 and Rpv3-2. No other R-loci (single or stacked) presented significant differences. Interestingly, genotypes without any detected Rpv locus revealed themselves not to be significantly more susceptible except for the stacked Rpv3-3 + Rpv10 genotypes at bunch level. Our findings highlight that as single players the resistance genes coming from V. amurensis (Rpv10 and Rpv12) guarantee a strong barrier against the pathogen, although they do not seem to benefit from stacking other studied Rpv loci. These findings are in contrast with previous results mainly on leaves and can be attributed to differences in the genetic backgrounds. In fact, as reported by [22], a higher resistance behaviour could be pointed out in the seedlings containing both resistance genes coming from the two parental lines ‘Regent’ (Rpv3) and ‘VHR 3082-1-42′ (Rpv1). In [39], the authors describe Rpv12 having an additive effect with Rpv3 to protect grapevines against natural infections. Also in [40], the F1 sub-population which contains the Rpv3 as well as the Rpv10 locus showed a significantly higher degree of resistance, indicating additive effects by pyramiding of R-loci.
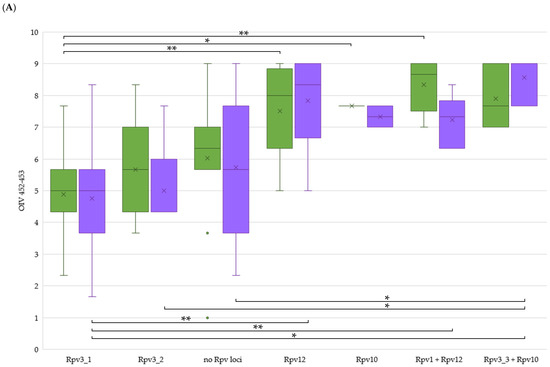

Figure 4.
Evaluation of field resistance to downy (A) and powdery (B) mildew on leaves (green) and bunches (violet) on clustered accessions according to the presence of resistance (R) loci. Per each accession, OIV scores were averaged by 3-years. Per each year, the lowest score between evaluations was chosen. Box plots show medians by middle line and means by x. *: p < 0.05; **: p < 0.01.
Regarding PM (Figure 4B), Run1 genotypes showed a significantly higher resistance compared to single Ren9 and stacked Ren3 + Ren9 genotypes at leaf level. On bunches, both Run1 and stacked Ren3 + Ren9 genotypes displayed a significantly higher resistance compared to the single Ren9 genotypes. In the case of the single Ren9 locus, bunch resistance levels were significantly decreased even compared to accessions without any studied R-locus, indicating a strong lack of resistance efficiency of this single locus. In fact, resistance QTL can sometimes only be detected under certain environmental conditions (soil, climate, pathogen population), or in specific genetic backgrounds. Thus, stable QTL are highly sought after for their applicability in breeding [62]. Our findings show that Run1 confers a very strong resistance which derived from Muscadinia compared to the other loci derived from Vitis spp. Unlike Rpv loci in our study, Ren loci comparison revealed that pyramiding (i.e., Ren3 + Ren9) allows the reinforcement of PM resistance at bunch level, the most delicate and relevant organ of the grapevine. In general, there is rather limited knowledge about the resistance mechanisms encoded in the various Ren/Run loci [63]. In the case of the flanked Ren3/Ren9 loci, they exhibit a hypersensitive response to E. necator evident at 5 days post inoculation. PM resistance of ‘Regent’ relies on a “post-invasion” mechanism that restricts pathogen development and finally impairs the formation of conidia [46].
While observing seven genotypes without any known R-loci (‘Amadeus’, ‘Bruskam’, ‘Leon Millot’, ‘Orpheus’, ‘IV035′ and ‘IV063′), two of them, ‘Bruskam’ and ‘Leon Millot’, exhibited a high level of OIV scores (7 < average OIV descriptor ≤ 9) both in leaves and clusters for DM and in clusters for PM, while the foliar resistance against PM was < 7. In addition, ‘Amadeus’ had a high resistance against PM and against DM only in bunches, while the foliar resistance against DM was < 7. For the remaining three genotypes (‘Orpheus’, ‘IV063′ and ‘Semonell’) the levels of the average OIV score were medium (5 < average OIV descriptor ≤ 7) on both organs for both mildews. ‘IV035′ shows a medium level, except for higher level in bunches against PM. If we consider the resistance to a single disease, ten genotypes were lacking Rpv and 34 were missing Run/Ren loci. Fifty percent of the genotypes without any screenable Rpv locus disclosed a ≥ 5 average OIV score, whereas the percentage of genotypes without feasible Run/Ren loci with a medium high resistance level (> 5 average OIV score) was even higher (94%). In general, genotypes showing no analyzed R-loci may indicate the possible presence of minor loci (in the case of low-mid phenotypic scores) or even the discovery of novel, not yet identified, R-loci (in case of high phenotypic scores). These genotypes should be considered as precious resources in the perspective of pyramiding towards durable resistance. Indeed, while MAS facilitates major resistance gene pyramiding, it appears inapplicable to capture small-effect loci. However, field observations suggest that putative minor factors are involved in the expression of weaker but significant effects that can enhance the protection conferred by major genes and improve even more the stability [27]. Thus, in parallel to pursuing QTL studies, the implementation of genome-wide association studies and genomic selection in grapevine breeding [64] will certainly bring new opportunities to combine both major R genes and quantitative resistance and thus construct new varieties with highly durable resistance. The durability of grapevine QTL pyramids is now under evaluation. Durable disease resistance is a complex phenomenon having no one genetic or molecular basis, and the success of an integrated strategy can be judged only in retrospect. Finally, we suggest that the combination of disease resistance genes or QTLs with other approaches for pathogen control (pesticide ad hoc application, farming practices) may be a relevant management strategy to slow down the evolution of virulent pathogen genotypes.
3. Materials and Methods
3.1. Genetic Material and DNA Isolation
The plant material consisted of 102 grapevine accessions divided into 65 breeding lines and 37 progeny individuals. The 65 breeding lines originated from breeding programs in various European institutions located in Germany, France, Austria, Hungary, Czech Republic, Russia and Switzerland. The remaining 37 genotypes were progeny individuals derived from different crosses made by a private breeder (InnoVitis, Marlengo (BZ), Italy) with the aim to introgress disease resistance traits into good quality V. vinifera backgrounds. The whole genetic material (Table S3)—in this study referred as VITISANA collection—was cultivated in an unsprayed vineyard located in a private winery in Marlengo (BZ, Italy) (N 46.670938, E 11.131313, 401 m a.s.l.). Each genotype was present at least in triplicate, managed since 2010 using a Guyot training system with a planting density of 2 m × 0.8 m in terraced fields.
Genomic DNA was extracted from young frozen leaves (0.08 g) using DNeasy Plant Kit (Qiagen, Hilden, Germany) in accordance with manufacturer’s instructions. After extraction, DNA was quantified using a NanoDrop 8000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and the DNA concentration was normalized to 10 ng/µL.
3.2. Trueness-to-Type Analysis
All the hybrids were genetically characterized with the nine reference SSRs, internationally approved for the genetic fingerprinting and the consequent identification of grapevine varieties according to the GenRes081 and GrapeGen06 EU project [65,66]. The amplifications were performed in a GeneAmp 9700 thermal cycler (Thermo Fisher Scientific, Waltham, MA, USA) in a 10 µL final volume using Qiagen Multiplex Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The nine SSR primer pairs were divided into three multiplex PCRs using different fluorescent dyes as reported in the Table S4. The following PCR profile was applied: Precycle 15 min at 95°C, 40 cycles of 40 sec denaturation at 95°C, 90 sec annealing at 55°C and 90 sec extension at 72°C, final extension of 30 min at 60°C. Capillary electrophoresis was carried out in an ABI 3130xl Genetic Analyzer (Life Technologies, Foster City, CA, USA) and the fragments (alleles) were sized with GeneMapper v4.0 in binning mode, using GeneScan 500LIZ size standard as an internal ladder (Life Technologies, Foster City, CA, USA). The trueness-to-type (TTT) was verified against the VIVC database [9].
3.3. R-Loci Analysis
Following the “all-vs-all” approach reported by [67,68], all Vitis hybrids were examined at the 11 actually screenable and reliable R-loci: Five associated with DM resistance (Rpv1, Rpv3, Rpv10, Rpv12, Rpv14) and six associated with PM resistance (Run1, Run2, Ren1, Ren2, Ren3 and Ren9). The symbol of the resistance genes, the name of the causal agents of the diseases (traits), the resistance-related markers, the alleles associated with the resistance and the genotypes of origin are reported in Table 2. PCR amplifications were carried out according to the protocols optimized by [67]. Herein absence of R-loci (no R-loci) stands solely for the analysed loci and leaves the possibility of known loci without exploitable markers or new /unknown loci.

Table 2.
Resistance (R) loci against Plasmopara viticola and Erysiphe necator with chromosome, used associated markers and their relative resistance alleles/haplotype, and used reference genotypes.
3.4. DM and PM Symptom Phenotyping in Field
Symptoms of DM and PM natural infections were assessed on the 102 accessions in untreated plots over three growing seasons (2016, 2017 and 2018). The scores were collected twice during each season, before veraison and before harvest. The OIV descriptors 452 and 453 were used for DM and OIV descriptors 455 and 456 for PM symptom assessments, on leaf laminas and clusters respectively. The organs were visually inspected by the same two trained evaluators and the scoring was carried out in the same two days for all hybrids. The maximum level of symptom expression (lowest OIV score) between the two times was considered within each season. Weather conditions including average rainfall, temperature and relative humidity were tracked from April to September in the local weather website (http://meteo.provincia.bz.it/dati-storici.asp, indicating Merano for the location). The weather station is located at about 2.0 km distance from the vineyard.
3.5. Statistical Analysis
Based on the 9 universal SSR markers, a genetic distance analysis was attempted by means of PAST v.3.14 software [69], applying Neighbour-joining with Euclidean distance. Statistical analyses on field resistance evaluations were performed using SPSS Statistics v24 (IBM). The year effect was analysed by pairwise comparing the mean values of the lowest assessments for each year with Friedman’s two-way Analysis of Variance (ANOVA) by ranks adjusted by Bonferroni correction (p < 0.001). Relationships between assessments before veraison and before harvest were calculated using Spearman’s bivariate correlation tests (p < 0.001) on a 3-year mean. One-Way ANOVA together with Post-Hoc Games–Howell test (p < 0.05) was used for pairwise comparisons of groups having different R-loci.
4. Conclusions
The analysis of all reliably applicable R-loci in the VITISANA collection—the so-called “all-vs-all” approach—turned out to be crucial to detect the presence of Rpv and Run/Ren loci in a definitive way, regardless of the supposed resistance donor(s) according to the historical pedigree information. The list of reliable markers is limited compared to the entire number of QTLs discovered and published until now on grapevine. For this reason, during selection activities, it is important to include also field evaluations in order to recover (mid)-resistant genotypes that would otherwise be discarded during sole marker analysis. In this regard, our survey allowed the discovery of several new sources of disease resistance useful for next coming QTL identifications, which are valuable and exploitable for MAB purposes. In addition, this study highlights Ren loci stacking to be effective on PM resistance especially at bunch level. At the same time, in the case of the presence of a prominent Rpv locus, pyramiding does not necessarily mean an increase of DM resistance level, but potentially provides a second barrier in case of pathogens overcoming the first one. As for traditional varieties, resistant varieties should be adapted to particular terroirs. Here there is still a huge lack of experience, which would enable possible adaptations to be identified in order to boost the ongoing diffusion of these novel varieties requested for sustainability issues.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/14/3526/s1.
Author Contributions
E.Z. contributed to the set-up of R-loci analysis, drafted the manuscript and contributed to the discussion of the results. (C.D.) Chiara Dolzani set up, optimized and performed the R-loci analysis. M.S. performed the phenotyping analyses. V.G. performed the TTT analysis. P.B. optimized the R-loci analyses and curated the bibliography. D.N. performed the R-loci analyses. G.B. and (C.D.) Cinzia Dorigatti contributed to the phenotyping analyses. R.V. contributed to the original idea and supported the project. T.L. performed statistical analyses, revised the manuscript and contributed to the discussion of the results. S.V. conceptualized as well as coordinated the project, drafted the manuscript and contributed to the discussion of the results. All authors read and approved the final manuscript.
Acknowledgments
The authors are grateful to Erhard Tutzer (Plonerhof Wine Growing Estate, Marlengo, IT) for providing grapevine genetic material, for the kindness of permitting the phenotyping assessments in his fields and, last but not least, his dedication and enthusiasm. The authors also thank Tiago C. Tomazetti for contributing to the PAST analysis and Hermann Stuppner for supporting the PhD fellowship of V.G. This work was financed by the EGTC “EUREGIO Tyrol, South Tyrol and Trentino” in the frame of the first call of the basic research fund: Project IPN 31 “VITISANA—Dissecting genetic traits in resistant grapevines.
Conflicts of Interest
All authors declared no conflicts of interest.
References
- Agrios, G. Plant Pathology, 3rd ed.; Academic Press: New York, NY, USA, 1988. [Google Scholar]
- Gururani, M.A.; Venkatesh, J.; Upadhyaya, C.P.; Nookaraju, A.; Pandey, S.K.; Park, S.W. Plant disease resistance genes: Current status and future directions. Physiol. Mol. Plant Pathol. 2012, 78, 51–65. [Google Scholar] [CrossRef]
- Dodds, P.N.; Lawrence, G.J.; Pryor, A.; Ellis, J.G. Genetic Analysis and Evolution of Plant Disease Resistance Genes. Mol. Plant Pathol. 2000, 4, 92–112. [Google Scholar] [CrossRef]
- Table of Loci for Traits in Grapevine Relevant for Breeding and Genetics. Available online: http://www.vivc.de/docs/dataonbreeding/20181001_Table of Loci for Traits in Grapevine.pdf (accessed on 30 October 2018).
- Pauquet, J.; Bouquet, A.; This, P.; Adam-Blondon, A.F. Establishment of a local map of AFLP markers around the powdery mildew resistance gene Run1 in grapevine and assessment of their usefulness for marker assisted selection. Theor. Appl. Genet. 2001, 103, 1201–1210. [Google Scholar] [CrossRef]
- Barker, C.L.; Donald, T.; Pauquet, J.; Ratnaparkhe, M.B.; Bouquet, A.; Adam-Blondon, A.F.; Thomas, M.R.; Dry, I. Genetic and physical mapping of the grapevine powdery mildew resistance gene, Run1, using a bacterial artificial chromosome library. Theor. Appl. Genet. 2005, 111, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hausmann, L.; Eibach, R.; Welter, L.J.; Töpfer, R.; Zyprian, E.M. A framework map from grapevine V3125 (Vitis vinifera ‘Schiava grossa’ × ’Riesling’) × rootstock cultivar “Börner” (Vitis riparia × Vitis cinerea) to localize genetic determinants of phylloxera root resistance. Theor. Appl. Genet. 2009, 119, 1039–1051. [Google Scholar] [CrossRef]
- Feechan, A.; Anderson, C.; Torregrosa, L.; Jermakow, A.; Mestre, P.; Wiedemann-Merdinoglu, S.; Merdinoglu, D.; Walker, A.R.; Cadle-Davidson, L.; Reisch, B.; et al. Genetic dissection of a TIR-NB-LRR locus from the wild North American grapevine species Muscadinia rotundifolia identifies paralogous genes conferring resistance to major fungal and oomycete pathogens in cultivated grapevine. Plant. J. 2013, 76, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Maul, E.; Töpfer, R. VIVC-Vitis International Variety Catalogue. Available online: http://www.vivc.de/ (accessed on 31 May 2019).
- Alleweldt, G.; Possingham, J.V. Progress in grapevine breeding. Theor. Appl. Genet. 1988, 75, 669–673. [Google Scholar] [CrossRef]
- Raddova, J.; Stefkova, A.; Radek, S.; Baranek, M. Genetic analysis of Vitis interspecific hybrids occuring in vineyards of the Czech Republic. Pak. J. Bot. 2016, 48, 681–688. [Google Scholar]
- Töpfer, R.; Hausmann, L.; Harst, M.; Maul, E.; Zyprian, E.; Eibach, R. New Horizons for Grapevine Breeding. Fruit Veg. Cereal Sci. Biotechnol. 2011, 5, 79–100. [Google Scholar]
- Pedneault, K.; Provost, C. Fungus resistant grape varieties as a suitable alternative for organic wine production: Benefits, limits, and challenges. Sci. Hortic. (Amsterdam). 2016, 208, 57–77. [Google Scholar] [CrossRef]
- Council Regulation (EC) No. 1493/1999 of 17 May 1999 on the Common Organization of the Market in Wine. Available online: https://eur-lex.europa.eu/eli/reg/1999/1493/2009-01-20 (accessed on 28 June 2019).
- European Food Safety Authority (EFSA) Annual Report 2003. Available online: https://www.efsa.europa.eu/sites/default/files/corporate_publications/files/ar03en.pdf (accessed on 28 June 2019).
- Council Regulation (EC) No 491/2009 of 25 May 2009 amending Regulation (EC) No 1234/2007 Establishing a Common Organisation of Agricultural Markets and on Specific Provisions for Certain Agricultural Products (Single CMO Regulation). Available online: https://eur-lex.europa.eu/eli/reg/2009/491/oj (accessed on 28 June 2019).
- Sivčev, B.V.; Sivčev, I.L.; Ranković Vasić, Z.Z. Natural process and use of natural matters in organic viticulture. J. Agric. Sci. 2010, 55, 195–215. [Google Scholar] [CrossRef][Green Version]
- PIWI International. Available online: https://www.piwi-international.de/de/ (accessed on 8 May 2019).
- Peressotti, E.; Wiedemann-Merdinoglu, S.; Delmotte, F.; Bellin, D.; Di Gaspero, G.; Testolin, R.; Merdinoglu, D.; Mestre, P. Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol. 2010, 10, 147. [Google Scholar] [CrossRef] [PubMed]
- Malav, A.K.; Indu; Chandrawat, K.S. Gene Pyramiding: An Overview. Int. J. Curr. Res. Biosci. Plant Biol. 2016, 3, 22–28. [Google Scholar] [CrossRef]
- Delmotte, F.; Bourguet, D.; Franck, P.; Guillemaud, T.; Reboud, X.; Vacher, C.; Walker, A.-S. Combining Selective Pressures to Enhance the Durability of Disease Resistance Genes. Front. Plant Sci. 2016, 7, 1916. [Google Scholar] [CrossRef]
- Eibach, R.; Zyprian, E.; Welter, L.; Töpfer, R. The use of molecular markers for pyramiding resistance genes in grapevine breeding. J. Grapevine Res. 2007, 46, 120–124. [Google Scholar]
- Di Gaspero, G.; Copetti, D.; Coleman, C.; Castellarin, S.D.; Eibach, R.; Kozma, P.; Lacombe, T.; Gambetta, G.; Zvyagin, A.; Cindrić, P.; et al. Selective sweep at the Rpv3 locus during grapevine breeding for downy mildew resistance. Theor. Appl. Genet. 2012, 124, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Mahanil, S.; Ramming, D.; Cadle-Davidson, M.; Owens, C.; Garris, A.; Myles, S.; Cadle-Davidson, L. Development of marker sets useful in the early selection of Ren4 powdery mildew resistance and seedlessness for table and raisin grape breeding. Theor. Appl. Genet. 2012, 124, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Riaz, S.; Tenscher, A.C.; Ramming, D.W.; Walker, M.A. Using a limited mapping strategy to identify major QTLs for resistance to grapevine powdery mildew (Erysiphe necator) and their use in marker-assisted breeding. Theor. Appl. Genet. 2011, 122, 1059–1073. [Google Scholar] [CrossRef]
- Pap, D.; Riaz, S.; Dry, I.B.; Jermakow, A.; Tenscher, A.C.; Cantu, D.; Oláh, R.; Walker, M.A. Identification of two novel powdery mildew resistance loci, Ren6 and Ren7, from the wild Chinese grape species Vitis piasezkii. BMC Plant Biol. 2016, 16, 170. [Google Scholar] [CrossRef] [PubMed]
- Foria, S.; Magris, G.; Morgante, M.; Di Gaspero, G. The genetic background modulates the intensity of Rpv3-dependent downy mildew resistance in grapevine. Plant Breed. 2018, 137, 220–228. [Google Scholar] [CrossRef]
- Barba, P.; Cadle-Davidson, L.; Harriman, J.; Glaubitz, J.C.; Brooks, S.; Hyma, K.; Reisch, B. Grapevine powdery mildew resistance and susceptibility loci identified on a high-resolution SNP map. Theor. Appl. Genet. 2014, 127, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Zyprian, E.; Šimon, S.; Schwander, F.; Töpfer, R. Efficiency of Single Nucleotide Polymorphisms to improve a genetic map of complex pedigree grapevines. Vitis-J. Grapevine Res. 2015, 54, 29–32. [Google Scholar]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP markers and their impact on plant breeding. Int. J. Plant Genom. 2012, 2012, 728398. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Fresnedo-Ramírez, J.; Wang, M.; Cote, L.; Schweitzer, P.; Barba, P.; Takacs, E.M.; Clark, M.; Luby, J.; Manns, D.C.; et al. A next-generation marker genotyping platform (AmpSeq) in heterozygous crops: A case study for marker-assisted selection in grapevine. Hortic. Res. 2016, 3, 16002. [Google Scholar] [CrossRef] [PubMed]
- Welter, L.J.; Göktürk-Baydar, N.; Akkurt, M.; Maul, E.; Eibach, R.; Töpfer, R.; Zyprian, E.M. Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L). Mol. Breed. 2007, 20, 359–374. [Google Scholar] [CrossRef]
- van Heerden, C.J.; Burger, P.; Vermeulen, A.; Prins, R. Detection of downy and powdery mildew resistance QTL in a ‘Regent’ × ‘RedGlobe’ population. Euphytica 2014, 200, 281–295. [Google Scholar] [CrossRef]
- Bellin, D.; Peressotti, E.; Merdinoglu, D.; Wiedemann-Merdinoglu, S.; Adam-Blondon, A.-F.; Cipriani, G.; Morgante, M.; Testolin, R.; Di Gaspero, G. Resistance to Plasmopara viticola in grapevine ‘Bianca’ is controlled by a major dominant gene causing localised necrosis at the infection site. Theor. Appl. Genet. 2009, 120, 163–176. [Google Scholar] [CrossRef]
- Casagrande, K.; Falginella, L.; Castellarin, S.D.; Testolin, R.; Di Gaspero, G. Defence responses in Rpv3-dependent resistance to grapevine downy mildew. Planta 2011, 234, 1097–1109. [Google Scholar] [CrossRef]
- Zyprian, E.; Ochßner, I.; Schwander, F.; Šimon, S.; Hausmann, L.; Bonow-Rex, M.; Moreno-Sanz, P.; Grando, M.S.; Wiedemann-Merdinoglu, S.; Merdinoglu, D.; et al. Quantitative trait loci affecting pathogen resistance and ripening of grapevines. Mol. Genet. Genom. 2016, 291, 1573–1594. [Google Scholar] [CrossRef]
- Vezzulli, S.; Malacarne, G.; Masuero, D.; Vecchione, A.; Dolzani, C.; Goremykin, V.; Mehari, Z.H.; Banchi, E.; Velasco, R.; Stefanini, M.; et al. The Rpv3-3 Haplotype and Stilbenoid Induction Mediate Downy Mildew Resistance in a Grapevine Interspecific Population. Front. Plant Sci. 2019, 10, 234. [Google Scholar] [CrossRef]
- Delmotte, F.; Mestre, P.; Schneider, C.; Kassemeyer, H.-H.; Kozma, P.; Richart-Cervera, S.; Rouxel, M.; Delière, L. Rapid and multiregional adaptation to host partial resistance in a plant pathogenic oomycete: Evidence from European populations of Plasmopara viticola, the causal agent of grapevine downy mildew. Infect. Genet. Evol. 2014, 27, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Venuti, S.; Copetti, D.; Foria, S.; Falginella, L.; Hoffmann, S.; Bellin, D.; Cindrić, P.; Kozma, P.; Scalabrin, S.; Morgante, M.; et al. Historical Introgression of the Downy Mildew Resistance Gene Rpv12 from the Asian Species Vitis amurensis into Grapevine Varieties. PLoS ONE 2013, 8, e61228. [Google Scholar] [CrossRef] [PubMed]
- Schwander, F.; Eibach, R.; Fechter, I.; Hausmann, L.; Zyprian, E.; Töpfer, R. Rpv10: A new locus from the Asian Vitis gene pool for pyramiding downy mildew resistance loci in grapevine. Theor. Appl. Genet. 2012, 124, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Bouquet, A. Contribution á L’étude de L’espèce Muscadinia Rotundifolia (Michx) Small et de Ses Hybrides Avec Vitis Vinifera L.-Applications en Sélection. Dissertation. Ph.D. Thesis, University of Bordeaux, Bordeaux, France, 1983. [Google Scholar]
- Bouquet, A. Introduction dans l’espèce Vitis vinifera L. d’un caractère de résistance à l’oidium (Uncinula necator Schw. Burr.) issu de l’espèce Muscadinia rotundifolia (Michx.) Small. Vignevini 1986, 12, 141–146. [Google Scholar]
- Kozma, P.; Kiss, E.; Hoffmann, S.; Galbács, Z.; Dula, T. Using the powdery mildew resistant Muscadinia rotundifolia and Vitis vinifera ‘Kishmish vatkana’ for breeding new cultivars. Acta Hortic. 2009, 827, 559–564. [Google Scholar] [CrossRef]
- Fischer, B.M.; Salakhutdinov, I.; Akkurt, M.; Eibach, R.; Edwards, K.J.; Töpfer, R.; Zyprian, E.M. Quantitative trait locus analysis of fungal disease resistance factors on a molecular map of grapevine. Theor. Appl. Genet. 2004, 108, 501–515. [Google Scholar] [CrossRef]
- Akkurt, M.; Welter, L.; Maul, E.; Töpfer, R.; Zyprian, E. Development of SCAR markers linked to powdery mildew (Uncinula necator) resistance in grapevine (Vitis vinifera L. and Vitis sp.). Mol. Breed. 2007, 19, 103–111. [Google Scholar] [CrossRef]
- Zendler, D.; Schneider, P.; Töpfer, R.; Zyprian, E. Fine mapping of Ren3 reveals two loci mediating hypersensitive response against Erysiphe necator in grapevine. Euphytica 2017, 213, 68. [Google Scholar] [CrossRef]
- Goyal, N.; Bhatia, G.; Sharma, S.; Garewal, N.; Upadhyay, A.; Upadhyay, S.K.; Singh, K. Genome-wide characterization revealed role of NBS-LRR genes during powdery mildew infection in Vitis vinifera. Genomics 2019, in press. [Google Scholar] [CrossRef]
- Katula-Debreceni, D.; Lencsés, A.K.; Szőke, A.; Veres, A.; Hoffmann, S.; Kozma, P.; Kovács, L.G.; Heszky, L.; Kiss, E. Marker-assisted selection for two dominant powdery mildew resistance genes introgressed into a hybrid grape population. Sci. Hortic. (Amsterdam) 2010, 126, 448–453. [Google Scholar] [CrossRef]
- Di Gaspero, G.; Cattonaro, F. Application of genomics to grapevine improvement. Aust. J. Grape Wine Res. 2010, 16, 122–130. [Google Scholar] [CrossRef]
- Merdinoglu, D.; Schneider, C.; Prado, E.; Wiedemann-Merdinoglu, S.; Mestre, P. Breeding for durable resistance to downy and powdery mildew in grapevine. OENO One 2018, 52, 203–209. [Google Scholar] [CrossRef]
- Klekowski, E. Genetic load and its causes in long-lived plants. Trees 1988, 2, 195–203. [Google Scholar] [CrossRef]
- King, J.L. The gene interaction component of the genetic load. Genetics 1966, 53, 403–413. [Google Scholar] [PubMed]
- Savary, S.; Delbac, L.; Rochas, A.; Taisant, G.; Willocquet, L. Analysis of Nonlinear Relationships in Dual Epidemics, and its Application to the Management of Grapevine Downy and Powdery Mildews. Phytopathology 2009, 99, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Rossi, V.; Caffi, T.; Giosuè, S.; Bugiani, R. A mechanistic model simulating primary infections of downy mildew in grapevine. Ecol. Model. 2008, 212, 480–491. [Google Scholar] [CrossRef]
- Boso, S.; Gago, P.; Santiago, J.L.; Martínez, M.C. Variation in Sensitivity of Different Grapevine Genotypes to Erysiphe necator Growing under Unfavourable Climatic Conditions. S. Afr. J. Enol. Vitic. 2018, 39, 100–105. [Google Scholar] [CrossRef]
- Keller, M.; Rogiers, S.Y.; Schultz, H.R. Nitrogen and ultraviolet radiation modify grapevines’ susceptibility to powdery mildew Cold hardiness in grapevines View project. Vitis 2003, 42, 87–94. [Google Scholar]
- Pacifico, D.; Gaiotti, F.; Giusti, M.; Tomasi, D. Performance of interspecific grapevine varieties in north-east Italy. Agric. Sci. 2013, 4, 91–101. [Google Scholar] [CrossRef]
- Vezzulli, S.; Vecchione, A.; Stefanini, M.; Zulini, L. Downy mildew resistance evaluation in 28 grapevine hybrids promising for breeding programs in Trentino region (Italy). Eur. J. Plant Pathol. 2018, 150, 485–495. [Google Scholar] [CrossRef]
- Pavloušek, P. Evaluation of foliar resistance of grapevine genetic resources to downy mildew (Plasmopara viticola). Acta Univ. Agric. Silvic. Mendel. Brun. 2012, 60, 191–198. [Google Scholar] [CrossRef]
- Lisek, J. Yielding and healthiness of selected grape cultivars for processing in central Poland. J. Fruit Ornam. Plant Res. 2010, 18, 265–272. [Google Scholar]
- Krizsics Cziskász, A.; Kozma, P. Characterization of fungus resistant grape varieties and candidate varieties in Pécs area. Acta Hortic. 2003, 763–765. [Google Scholar] [CrossRef]
- Pilet-Nayel, M.-L.; Moury, B.; Caffier, V.; Montarry, J.; Kerlan, M.-C.; Fournet, S.; Durel, C.-E.; Delourme, R. Quantitative Resistance to Plant Pathogens in Pyramiding Strategies for Durable Crop Protection. Front. Plant Sci. 2017, 8, 1838. [Google Scholar] [CrossRef]
- Qiu, W.; Feechan, A.; Dry, I. Current understanding of grapevine defense mechanisms against the biotrophic fungus (Erysiphe necator), the causal agent of powdery mildew disease. Hortic. Res. 2015, 2, 15020. [Google Scholar] [CrossRef]
- Fodor, A.; Segura, V.; Denis, M.; Neuenschwander, S.; Fournier-Level, A.; Chatelet, P.; Homa, F.A.A.; Lacombe, T.; This, P.; Le Cunff, L. Genome-Wide Prediction Methods in Highly Diverse and Heterozygous Species: Proof-of-Concept through Simulation in Grapevine. PLoS ONE 2014, 9, e110436. [Google Scholar] [CrossRef]
- This, P.; Jung, A.; Boccacci, P.; Borrego, J.; Botta, R.; Costantini, L.; Crespan, M.; Dangl, G.S.; Eisenheld, C.; Ferreira-Monteiro, F.; et al. Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theor. Appl. Genet. 2004, 109, 1448–1458. [Google Scholar] [CrossRef]
- Maul, E.; Sudharma, K.N.; Kecke, S.; Marx, G.; Müller, C.; Audeguin, L.; Boselli, M.; Boursiquot, J.M.; Bucchetti, B.; Cabello, F.; et al. The European Vitis Database (www.eu-vitis.de): A technical innovation through an online uploading and interactive modification system. Vitis 2012, 51, 79–85. [Google Scholar]
- Migliaro, D.; Peressotti, E.; Dolzani, C.; Bettinelli, P.; Banchi, E.; Riaz, S.; Cappellin, L.; Zini, E.; Walker, M.A.; Reisch, B.I.; et al. Unravelling the knot of grapevine hybrid kinship: A 50 SSR- and 12 R-loci-based genetic diversity analysis. in preparation.
- Vezzulli, S.; Dolzani, C.; Migliaro, D.; Banchi, E.; Stedile, T.; Zatelli, A.; Dallaserra, M.; Clementi, S.; Dorigatti, C.; Velasco, R.; et al. The FEM grapevine breeding program for downy and powdery mildew resistances: Towards a green viticulture. In Proceedings of the XII International Conference on Grapevine Breeding and Genetics, Bordeaux, France, 15–20 July 2018; Volume 2018, p. 37. [Google Scholar]
- Hammer, D.A.T.; Ryan, P.D.; Hammer, Ø.; Harper, D.A.T. Past: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).