Abstract
The EXO70 gene family is involved in different biological processes in plants, ranging from plant polar growth to plant immunity. To date, analysis of the EXO70 gene family has been limited in Triticeae species, e.g., hexaploidy Triticum aestivum and its ancestral/related species. By in silico analysis of multiple Triticeae sequence databases, a total of 200 EXO70 members were identified. By homologue cloning approaches, 15 full-length cDNA of EXO70s were cloned from diploid Haynaldia villosa. Phylogenetic relationship analysis of 215 EXO70 members classified them into three groups (EXO70.1, EXO70.2, and EXO70.3) and nine subgroups (EXO70A to EXO70I). The distribution of most EXO70 genes among different species/sub-genomes were collinear, implying their orthologous relationship. The EXO70A subgroup has the most introns (at least five introns), while the remaining seven subgroups have only one intron on average. The expression profiling of EXO70 genes from wheat revealed that 40 wheat EXO70 genes were expressed in at least one tissue (leaf, stem, or root), of which 25 wheat EXO70 genes were in response to at least one biotic stress (stripe rust or powdery mildew) or abiotic stress (drought or heat). Subcellular localization analysis showed that ten EXO70-V proteins had distinct plasma membrane localization, EXO70I1-V showed a distinctive spotted pattern on the membrane. The 15 EXO70-V genes were differentially expressed in three tissue. Apart from EXO70D2-V, the remaining EXO70-V genes were in response to at least one stress (flg22, chitin, powdery mildew, drought, NaCl, heat, or cold) or phytohormones (salicylic acid, methyl jasmonate, ethephon, or abscisic acid) and hydrogen peroxide treatments. This research provides a genome-wide glimpse of the Triticeae EXO70 gene family and those up- or downregulated genes require further validation of their biological roles in response to biotic/abiotic stresses.
1. Introduction
The exocyst complex is an evolutionarily conserved octameric tethering factor, which mediates the fusion of post-Golgi secretory vesicle with the plasma membrane (PM) and plays a major role in exocytosis [1,2]. EXO70 is a key member of the exocyst complex and has been found to be widely present in yeast, mammals and plants [3]. In yeast and mammals, the EXO70 only has a single copy, while plants have multiple copies of EXO70 genes [4] ranging from 21 to 47 EXO70 members in potatoes (Symphytum tuberosum), Arabidopsis, Populus trichocarpa and rice [1,5,6]. The EXO70s of land plants possibly originated from three ancient EXO70 genes and thus can be divided into three groups EXO70.1, EXO70.2 and EXO70.3. They have been further duplicated independently in the moss, lycophyte and angiosperm lineages, and in the subsequent lineage-specific multiplications which are represented by nine subgroups (EXO70A-EXO70I) [4,7,8,9].
The function of EXO70 has been extensively studied in yeast, and mammals [3,10,11,12,13,14,15]. In plants, EXO70s have been proven to play diverse roles in regulating plant growth and coping with adverse biotic/abiotic stresses. In Arabidopsis, EXO70A1 has been implicated in a wide range of developmental processes, including the differentiation of tracheary elements [16,17], the development of seed coat, root hair and stigmatic papillae [18], the recycling of the auxin efflux carrier proteins (PIN1 and PIN2) [19] and the formation of the Casparian strip [20]. EXO70C1 and EXO70C2 regulated the polarized growth and maturation of the pollen tube [21,22]. EXO70H4 regulates trichome cell wall maturation by mediating the secretion and accumulation of callose and silica [23,24]. The rice OsEXO70A1 is necessary for vascular bundle differentiation and assimilation of mineral nutrients [5]. The legumes EXO70J7, EXO70J8 and EXO70J9 are members of an atypical subgroup of EXO70 proteins (EXO70J) that regulate leaf senescence and nodule formation [25]. In Nicotiana benthamiana, silencing all the paralogue genes in subgroups EXO70A (six), C (three), D (four) and G (six) resulted in a smaller leaf phenotype [6].
Evidence has accumulated for the critical role of EXO70s in plant-pathogen interactions or responses to abiotic stresses. In N. benthamiana, the silencing of two EXO70B paralogues led to increased susceptibility to Phytophthora infestans [6]. Three of the 23 members of the Arabidopsis EXO70 gene family (EXO70B1, EXO70B2 and EXO70H1) have been proven to be involved in plant immunity [26]. AtEXO70B1 and AtEXO70B2 belong to the same subgroup and are both involved in plant immunity, of which AtEXO70B1 a negative regulator, while AtEXO70B2 is a positive regulator. AtEXO70B1 underwent autophagic transport, and the loss-of-function of exo70B1 led to reduced numbers of internalized autophagosomes, accumulation of salicylic acid (SA), and finally, ectopic hypersensitive responses and enhanced resistance to several pathogens. AtEXO70B1’s regulation of disease resistance, either by interacting with TIR-NBS2, a truncated version of the classical nucleotide binding (NB) domain and a leucine-rich repeat (LRR)-containing (NLR) intracellular immune receptor-like protein [27,28,29], or by interacting with RIN4, a well-known regulator of pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) [29]. AtEXO70B2 regulated innate immunity via interacting with a negative PTI regulator, AtPUB22, which mediated the ubiquitination and degradation of AtEXO70B2 and contributed to PTI. The exo70B2 mutants showed aberrant papillae with halos and were susceptible to different PAMPs and pathogens [30,31]. AtEXO70B1 and AtEXO70B2 also contribute to the abiotic stress response; both were positive regulators of stomatal movement. The response to mannitol (drought) treatments is in either an abscisic acid (ABA)-dependent or -independent manner [32,33]. AtEXO70H1 is a homolog of AtEXO70B2 and is also involved in plant immunity [31]. Three of the 47 EXO70 members of rice (OsEXO70E1, OsEXO70F2 and OsEXO70F3) were reported to participate in plant immunity [5]. OsEXO70E1 is attributed to planthopper resistance by interacting with a broad resistance protein, Bph6. Interaction of the two proteins increased exocytosis and blocked the feeding of a planthopper by cell wall thickening at the infection sites [34]. The importance of EXO70 in plant immunity was also shown by the fact that some of the EXO70s were targets of the secreted effectors of the plant pathogen. Both OsEXO70F2 and OsEXO70F3 were targets of the Magnaporthe oryzae effector AVR-Pii, and OsEXO70F3 was proven to play an important role in Pii-dependent resistance by interacting with AVR-Pii [35].
Accumulated evidence has shown that a large number of EXO70s exist in plants; however, only a few have had their biological roles elucidated [5,35,36]. Due to the huge genome size and complexity [37], the knowledge of the EXO70 gene family from the Triticeae species is rather limited. In the last five years, the genome sequences of wheat and its ancestor species have been released, which makes genome-wide identification of a gene family in the Triticeae species feasible [38,39,40,41,42,43].
Haynaldia villosa L. (2n = 2x = 14, VV) is a diploid wild relative of wheat. Previous studies showed that H. villosa is a valuable genetic resource harboring many elite traits, such as resistance to several wheat diseases and tolerance to abiotic stresses [44,45,46]. In the present study, different members of the EXO70 gene family are identified by browsing the released genome sequences of the Triticea species. Specific primer pairs are designed, EXO70s are cloned from H. villosa and their potential functions are elucidated by expression profiles based on in silico analysis and quantitative RT-PCR (qRT-PCR). The obtained results would help us to understand the evolution and diversification of the EXO70s among Triticeae species and their potential roles in plant immunity and responses to abiotic stresses.
2. Results
2.1. Identification and Phylogenetic Relationship Analysis of the EXO70 Gene Family in Triticeae Species
In total, 200 EXO70 genes were identified from the public database of five Triticeae species. Among them, there were 26 each from T. urartu, Ae. Tauschii and H. vulgare; 47 from T. dicoccoides; and 75 from common wheat (T. aestivum), respectively. Fifteen EXO70s from H. villosa were obtained by homology cloning (Figure 1a). The evolutionary relationship of the above 215 Triticeae EXO70s, along with 22 from Brachypodium distachyon, 41 from rice and 23 from Arabidopsis, were phylogenetically analyzed (Figure 1b, Table S1). These EXO70s were divided into three major groups, EXO70.1, EXO70.2 and EXO70.3, which were further assigned to nine subgroups, from EXO70A to EXO70I, according to a phylogenetic tree (Figure 1b). The EXO70A subgroup belongs to the group EXO70.1, in which 43 (14.28%) EXO70s were included; the EXO70B, C, D, E, F, H, and I subgroups belong to the group EXO70.2, in which 229 (76.08%) EXO70s were included; the EXO70G subgroup belongs to the group EXO70.3, in which 29 (9.63%) EXO70s were included. The EXO70I subgroup has the most members (74, 24.58%), followed by EXO70F (48, 15.95%) and EXO70A (43, 14.28%) (Figure 1c). Based on the subgroups and genome allocation, the wheat EXO70s were designated [7,47]. For example, the EXO70B1 from T. dicoccoides located on chromosome 1A was assigned TdEXO70B1-1A. In Arabidopsis, the EXO70I subgroup is missing, whereas the EXO70I subgroup in Triticeae species appeared to be the most divergent. However, our analysis led to a new insight: the EXO70I subgroup belongs to EXO70.2, rather than EXO70.3 from other species [36] (Figure 1b,c).
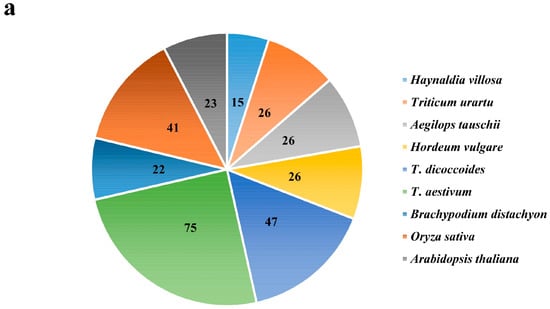
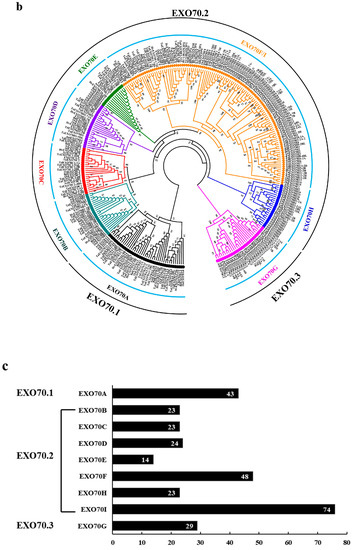
Figure 1.
The number and phylogenetic relationships of EXO70 family genes from T. aestivum, T. urartu, Ae. tauschii, T. dicoccoides, H. vulgare, A. thaliana, Oryza sativa, Brachypodium distachyon and H. villosa. (a) The total number of EXO70 gene family in nine species. (b) The phylogenetic tree of nine Triticeae species. Species abbreviations: Ta, Triticum aestivum; Tu, Triticum urartu; Aet, Aegilops tauschii; Td, Triticum dicoccoides; Hv, Hordeum vulgare; At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Os, Oryza sativa; -V, Haynaldia villosa. (c) The number of the EXO70 gene family from nine species in each of the subgroups. The horizontal/longitudinal coordinate axis represents the number of genes and different subgroups, respectively.
The 15 cloned EXO70s from H. villosa were designated as EXO70A1-V to EXO70I1-V according to the phylogenetic relationship to wheat EXO70s. They belong to nine subgroups, three each to EXO70A and F, two each to EXO70D and G and one each to EXO70B, C, E, H and I (Figure 1). Their CDS length ranges from 801 bp (EXO70H1-V) to 2007 bp (EXO70C1-V) and their isoelectric point varies from 4.52 (EXO70B1-V) to 10.19 (EXO70G1-V) (Table 1).

Table 1.
EXO70 genes cloned from H. villosa.
DNAMan was used to explore the amino acid sequence feature of H. villosa EXO70s. The pfam03081 domain at the C-terminus was relatively conservative, shown by the fact that all of the EXO70s had the same amino acid in the 11 sites (in the red rectangle in Figure 2a). R programming language was used to visualize the sequence similarities. The same subgroups shared more common sequences, e.g., EXO70D1-V and EXO70D2-V shared about 80% similarity, while different subgroups had low sequence similarities, e.g., EXO70H1-V and EXO70I1-V only had 21.46% similarity (Figure 2b).
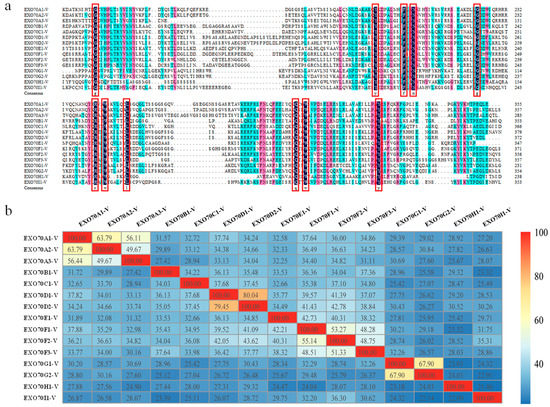
Figure 2.
The amino acid sequence feature analysis of EXO70 gene in H. villosa. (a) The amino acid sequence of pfam03081 domain for each EXO70 was selected for multiple sequence alignment analysis by DNAMan. The 11 identical amino acids were indicated in red frame. The three colors of black, red and blue represent the level of similarity of amino acids, from high to low. (b) Sequence similarity analysis using the R programming language. The color scale bar represents sequence similarity between different genes. Red and yellow indicate that the sequence similarity was greater than 80% and 60%, respectively. Blue indicates that the sequence similarity was less than 40%.
2.2. The Number of EXO70s in Genomes of Triticeae Species
The diploid species B. distachyon and Arabidopsis have 22 and 23 EXO70s, respectively, while diploid rice has 41, nearly twice as many. We identified 22 EXO70s each in A and D of common wheat and B of the tetraploid wheat, 25 in A of tetraploid wheat, 26 in D of Ae. Tauschii and H of barley and 29 EXO70s in B of common wheat. For the A genome, tetraploid has three more EXO70s than hexaploidy wheat; for the B genome, hexaploidy has seven more than tetraploid wheat; for the D genome, Ae. tauschii has four more EXO70s than hexaploidy wheat. In common wheat, the chromosomes 5A, 5B and 5D had the same number of EXO70s, belonging to the same gene type and having the same gene order. The chromosomes 7A, 7B and 7D also have the same number of EXO70s; however, their gene type and gene order were not completely the same. For example, TaEXO70D is only present on 7A and 7B, but not on 7D; TaEXO70G1 is only present on 7A and 7D, but not 7B. The gene number and order for EXO70s in the remaining five homoeologous were also different among corresponding homoeologous chromosomes, mainly due to their difference for subgroup EXO70I (Table 2 and Table S2).

Table 2.
Number of EXO70 from different species in each of the chromosomes.
The three analyzed diploid Triticea species, H. vulgare, T. uratu and Ae. tauschii, each had 26 EXO70s and had the same number as the EXO70B, C, D and H subgroups. They had the least EXO70Hs and the most EXO70Is. T. uratu only had one EXO70G, while the other two species had three. However, T. uratu had more EXO70I than the other two species (Table 3). The variation among different diploid species may be due either to the quality of the genome assembly or the duplication or deletion during evolution. The tetraploid T. dicoccoides and hexaploid T. aestivum have 47 and 75 EXO70s, which are about twice and three times the number in diploid species, respectively. In addition, the number of EXO70s from each subgroup is also exactly (EXO70C, H) or almost (EXO70A, B, D, E, F, I) 2 or 3 times that of the diploid species (Table 3). This indicated that, in polyploid wheat, the increased number of EXO70s is mostly due to the genome polyploidization.

Table 3.
Numbers of EXO70 paralogs encoded by the surveyed genomes in total and individual subgroups.
We only identified 15 EXO70s in diploid H. villosa, which is much less than the average number for other diploid species. H. villosa had the same number of subgroups D and H, but fewer members of subgroups B, C, E, F and I. This is due either to the lack of the sequence of H. villosa, or the divergence of H. villosa from other Triticea species (Table 3).
2.3. The Chromosomal Distribution of EXO70s in Triticeae Species
The identified 174 EXO70s from four Triticeae species (T. aestivum, Ae. tauschii, T. dicoccoides and H. vulgare) were assigned to corresponding chromosomes (including three on unknown chromosome). With the exception of wheat 6D, all chromosomes have at least one EXO70 gene (Figure 3). The EXO70s were not evenly distributed on difference chromosomes or in different homologous groups. In total, there were 9, 44, 27, 14, 19, 7 and 51 EXO70s in the homologous groups 1 to 7, with group 7 having the most EXO70s (29.31%), followed by group 2 (25.29%) (Table 2). The EXO70s in group 7 was dispersely distributed along the chromosome, while those in group 2 were clutered at the dital region of the long arm (Figure 3). We found that nine groups EXO70s were conversely present on the same homoeologous groups of the four species, such as the B1 in group1, F2/3 in group 2, E1 and F3/2 in group 3, C2/C1 in group 4, CI and D1 in group 5, F1 and F4 in group 7 (Table S2, Figure 3).
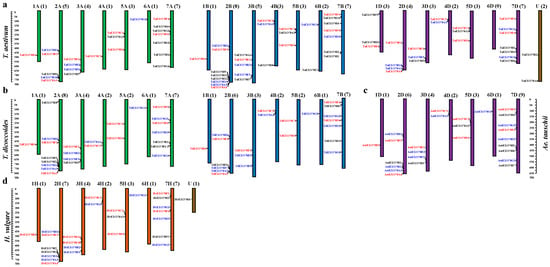
Figure 3.
The chromosomal distribution of the EXO70 gene family in four Triticeae species. (a–d) represent T. aestivum, T. dicoccoides, A. tauschii and H. vulgare, respectively. Chromosome numbers are indicated at the top of each bar and the number in parentheses corresponds to the number of EXO70 genes present on that chromosome. The name of each gene is to the left of each chromosome. Gene names labeled with red, blue, or black indicate that they are conserved in four species, missing in one due to incomplete data, or missing in more than two, respectively.
For homologous chromosomes from different genomes of wheat and its ancestral/related species, most of the corresponding EXO70 orthologs were present at the syntenic genome regions. For example, EXO70B1s were on the long chromosome arm of homoeologous group 1 and EXO70E1 on group 3 chromosomes (Figure 3). There are some exceptions. The EXO70G1 is present on all group 6 chromosomes except for sub-genome 6D of the hexaploidy wheat. The EXO70H1 is located on chromosomes 2A, 2B of common wheat and T. dicoccoides, 2D of Ae. tauschii and 2H of barley and on 1D of common wheat (Figure 3). We found that the EXO70C2 is on the 4BS of common wheat and T. dicoccoides (Figure 3a,b), on the 4DS of common wheat and Ae. tauschii (Figure 3a,d), but on the 4AL of common wheat and T. dicoccoides (Figure 3a,b). This also supports the presence of an inter-arm translocation of 4A during the evolution from diploid to tetraploid wheat [48,49].
2.4. The Diversification of Gene Structure of Triticeae EXO70s
The C-terminal PFam03081 domain, which may determine the function or structure of the proteins, is a specific characteristic of the EXO70 superfamily [47]. All the predicted 200 and 15 homologous cloned EXO70 proteins possessed such a domain; however, their amino acid sequence length is different for different EXO70s, varying from 103aa to 669aa and with an average length of 345aa (Table S1).
The exon–intron structure of the 200 Triticeae EXO70s was visualized by using the online Gene Structure Display Server and compared among different subgroups or within each individual subgroup. The exon-intron structures for most of the EXO70s within the same subgroups were relatively conserved among Triticeae species, was similar to in Arabidopsis or rice [7,36]. Compared with other subgroups, the subgroup EXO70A with 30 genes has the most introns on average, e.g., TuEXO70A3 has the most introns (20) and HvEXO70A4-2H has the fewest introns (five) (Figure 4a). The 170 EXO70 genes in the remaining eight subgroups only have about one intron on average. Among them, 83 genes (41.50%) are intronless, including eight EXO70Bs, seven EXO70Cs, nine EXO70Ds, one EXO70Es, 18 EXO70sF, 12 EXO70Gs, five EXO70Hs and 23 EXO70sI; 60 genes (30.00%) have one intron (e.g., AetEXO70B1 and TaEXO70C1-5BL), 19 genes (9.50%) have two introns (e.g., TuEXO70C2 and TaEXO70G2-6BS) and eight genes (4.00%) have three to six introns (e.g., AetEXO70I3 and TaEXO70D1-5AL). We also observed that genes that have a closer phylogenetic relationship in the same subgroup have a similar gene structure; however, within the same subgroup, some genes showed quite different gene structure. For instance, TaEXO70D1-5AL has six introns in EXO70D (Figure 4d) and the three copies of TaEXO70I5 and AetEXO70I3 have five introns, while the other genes in the same subgroup (EXO70D or EXO70I) only have one or two introns (Figure 4i). The most diversified gene structure of the EXO70A subgroup may be a clue that their diversified biological role is different from that of other subgroups.
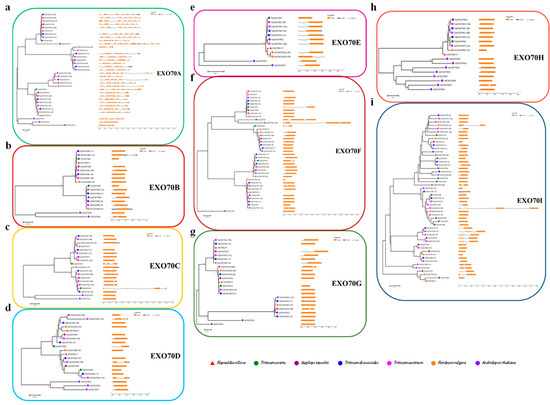
Figure 4.
Phylogenetic analysis and exon–intron structures of EXO70 gene family in common wheat and related Triticeae species. The phylogenetic analysis was performed using the sequences of the conserved domain of EXO70; proteins were aligned by ClustalW, constructed by MEGA6 using the N-J method, with 1000 bootstrap replicates; the branch length scale bar indicates the evolutionary distance. The left column identifies subgroups and is marked with different alternating background tones to make subgroup identification easier. Introns and exons are represented by black lines and colored boxes, respectively.
2.5. The Expression Pattern of TaEXO70 Genes
The expression patterns of different members in a gene family will help us predict their potential biological roles. To elucidate the potential roles of the identified EXO70s, their expression in different tissues or in responses to various biotic and abiotic stresses was investigated by in silico expression profiling or qRT-PCR analysis. The expression patterns of wheat TaEXO70s in different tissues (root, stem and leaf of seeding stage), under two biotic stresses (stripe rust pathogen CYR31 and powdery mildew pathogen E09) and two abiotic stresses (drought and heat) [50,51] were first investigated using the wheat RNA-seq data from the publicly available databases. The expression level was measured as tags per million (TPM). To facilitate the portraits of transcript abundance, we assume the expression was high if TPM ≥ 2.5; moderate if 2.5 > TPM ≥ 1.5; low if 1.5 >TPM > 0; and undetectable if TPM = 0. All 75 TaEXO70 genes exhibited significantly diverse expression patterns (Figure 5). Their expression can be classified into five groups. The group “e” includes 23 TaEXO70s (30.67%). Their transcript abundance was undetectable; among them 11 were from subgroup I and six were from subgroup C (Figure 5e). The group “d” includes 20 genes (26.67%). They had detectable but weak transcript abundance; among them six were from subgroup A and six were from subgroup I (Figure 5d). The group “c” has six genes (8.00%), whose expression is high in roots and stems, whereas it does not respond to the four stresses (Figure 5c). The group “b” has 15 genes (20.00%). These genes were found to be negligibly to moderately expressed in the three tissue types and in response to one or two stresses. Among them, nine were from the subgroup F (Figure 5b). The group “a” has ten genes (13.33%). Their expression was generally high, either in different tissues or in response to the four stresses and none of them were from the subgroup I (Figure 5a).
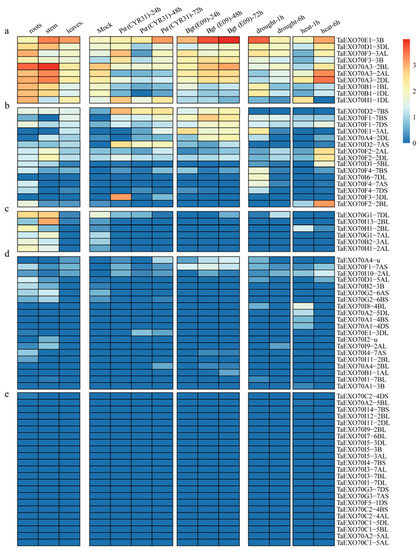
Figure 5.
Heat map of the expression profiling of wheat EXO70 genes in different tissues and under various stresses. The color scale bar represents the expression values of the genes. (a–e): Genes with different expression types. Abbreviations: Bgt, powdery mildew; Pst: Stripe rust.
Six genes were upregulated in response to both biotic and abiotic stresses. TaEXO70E1-3B was the only gene that was highly expressed in three tissues and upregulated in response to all four stresses. Its homologs on 3AL TaEXO70E1-3AL displayed moderate expression in three organs and were only responsive to Bgt inoculation and drought stress. The TaEXO70D1-5DL and TaEXO70F3-3B also displayed moderate expression in three organs and stresses. The TaEXO70H1-1DL expression was higher in root/leaf tissue and showed upregulation under both Pst infection and heat treatment. The TaEXO70B1-1BL/1DL expression was higher in root/stem tissue, and it was downregulated in response to Bgt and Pst inoculation but upregulated by drought treatment. TaEXO70A3-2AL/2BL/2DL had a similar expression pattern, being expressed in three tissues (stems > roots > leaves) and downregulated by Pst inoculation but upregulated by both drought and heat treatment. TaEXO70D2-7BS and TaEXO70G1-7DL were both only responsive to biotic stresses; however, they showed opposite patterns, with TaEXO70D2-7BS upregulated and TaEXO70G1-7DL downregulated. Twelve genes were only responsive to one of the four stresses; five were only upregulated in response to Bgt or Pst inoculation; and seven were only upregulated in response to drought or heat stress. Conservation roles were observed for all three homolog genes from different genomes, such as TaEXO70A3(2AL/2BL/2DL) and TaEXO70F2(2AL/2BL/2DL).
2.6. Differential Expression of 15 EXO70 Genes from H. villosa
The expression patterns of 15 EXO70-Vs in different tissues and in response to different stresses or treatments in H. villosa were investigated by qRT-PCR. Different expression patterns were observed for the analyzed genes (Figure 6). Six of the genes (EXO70D1-V, EXO70A2-V, EXO70F3-V, EXO70H1-V, EXO70I1-V and EXO70G2-V) were not differentially expressed in all three tissues. Four genes (EXO70B1-V, EXO70G1-V, EXO70A1-V, EXO70C1-V) showed more abundant transcript in stem; seven genes (EXO70E1-V, EXO70A3-V, EXO70B1-V, EXO70G1-V, EXO70F1-V, EXO70D2-V and EXO70F2-V) showed higher expression level in leaves (Figure 6a).
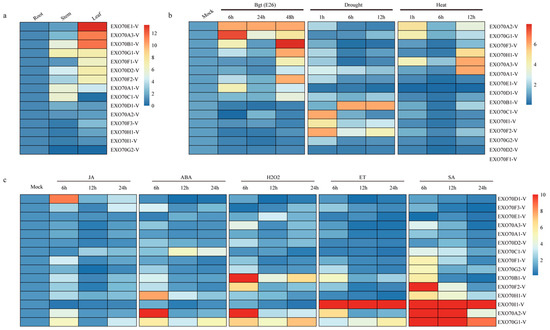
Figure 6.
Heat map of the expression profiling of wheat and H. villosa EXO70 genes in tissues and in response to biotic/abiotic stress, phytohormones and H2O2 treatments. (a) Tissue-specific expression pattern of 15 EXO70-V genes in H. villosa. (b) Expression levels of EXO70-V genes in biotic/abiotic stresses of H. villosa. (c) Expression profiling of EXO70-V genes in response to phytohormones and H2O2 treatments. The scale bar showing expression level of the genes. Abbreviations: Bgt, powdery mildew; SA, salicylic acid; MeJA, methyl jasmonate; ET, ethephon; ABA, abscisic acid; H2O2, hydrogen peroxide.
EXO70A2-V showed a similar expression level in all the three tissues; however, its expression was significantly increased in response to Bgt inoculation and treatments by chitin, flg22, heat stress, phytohormones, or H2O2. EXO70G1-V showed a similar expression level in the stems and leaves; its expression was also significantly increased by Bgt inoculation and treatment by chitin, flg22, four phytohormones, or H2O2. The expression of EXO70H1-V was strongly upregulated by Bgt inoculation and cold stress, and moderately upregulated by chitin, Flg22, ET and heat stress. EXO70I1-V only responded to abiotic stresses (drought, salt and cold) and ABA treatment. EXOG2-V was responsive to flg22 treatment and drought stress, and EXO70A3-V was only responsive to heat stress (Figure 6b,c). The divergence of expression patterns of different gene members indicated their clear-cut roles in the adaptation of H. villosa to various environments or stresses.
2.7. The Subcellular Localization of EXO70s from H. villosa
Knowledge of the subcellular localization of a plant protein can help us predict its potential role in the biological process. The subcellular localization of EXO70-Vs was investigated by transiently expressing the construct into leaves of Nicotiana tabacum via Agrobacterium method. Eleven EXO70-Vs generated fluorescence signals. Compared with the relatively even distribution of GFP signals in the cell (Figure 7a), the 11 confusion proteins had distinct localization patterns. EXO70A1, A3 and F1-V displayed weak signals on the plasma membrane (PM) (Figure 7b–d), while the PM signals for EXO70C1-V and EXO70D2-V were more intensive (Figure 7e,f). EXO70B1, E1 and F3-V displayed signals both in the PM and the nucleus (Figure 7g–i). EXO70D1-V and EXO70F2-V also produced signals in the PM; in addition, they also had small and discrete spot signals in the PM (Figure 7j,k). EXO70I1-V was the only one with no continuous PM localized signal; however, we observed discrete punctate signals along the PM (Figure 7l).
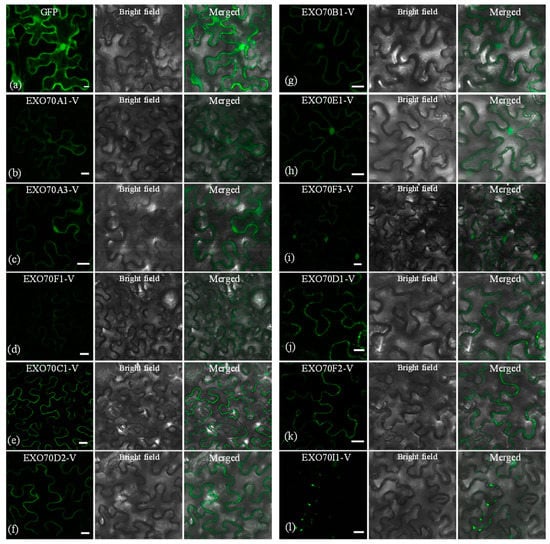
Figure 7.
Subcellular localization of H. villosa EXO70-GFP-Vs proteins. H. villosa EXO70-GFP-Vs proteins were transiently expressed in N. benthamiana leaves carried out with injection of Agrobacterium and examined by a confocal microscope. Green fluorescence was observed 48 h after infection. Bar = 20 μm. (a) The empty GFP vector was used as the control. The green channel shows that GFP signals were localized in the nucleus and cytoplasmic and plasma membranes. (b–l) The subcellular localization pattern of EXO70A1-V to EXO70I1-V, respectively.
3. Discussion
3.1. Evolutionary Relationship of the EXO70 Gene Family in Wheat and Its Relatives
The evolutionary relationships of the EXO70 gene family (between wheat, T. urartu, Ae. tauschii, T. dicoccoides, H. vulgare and H. villosa) have been speculated about based on the total number, classification, chromosomal distribution and structure. The surveyed diploid species of seven chromosome pairs except for H. villosa all possessed 26 EXO70 genes, which suggested this gene family appeared before the divergence among Triticum species [52]. Allohexaploid wheat originated from two hybridizations between three diploid progenitors approximately 2.5–4.5 million years ago [53]. The number of EXO70 genes in tetraploid and common wheat (a total of 47 and 75, respectively) is approximately twice and three times as many as diploids, implying they have undergone one and two rounds of polyploidization events [54]. Although the polyploidization event induced rapid and extensive genetic and epigenetic changes in the genome which were related to a large range of molecular and physiological adjustment [55] as well as a significant loss of gene family members upon domestication [53], by comparing with diploid species, the EXO70 gene family did not go through a wide range of expansion or diminution in tetraploid wheat and allohexaploid wheat. This deduction is also supported by their chromosomal location analysis, which showed that 73.1% of genes have good collinearity among wheat, Ae. tauschii, T. dicoccoides and H. vulgare, and most of the same type of orthologous genes maintain the relative order of their ancestral genes (Figure 3 and Figure 4). As for the small difference in the number of genes on individual subgroups among wheat and its relatives, this may be because of the quality of the genome assembling or the gene duplication of subgroup EXO70I.
Phylogenetic analysis showed that all three groups (EXO70.1, EXO70.2 and EXO70.3) and nine subgroups (EXO70A to EXO70I) are represented in each of the six Triticeae species. A similar gene structure was found in the same subgroup; subgroup EXO70A consists of multiple introns, while the other eight subgroups had fewer or were intronless. The variable intron numbers confirmed the classifications of the EXO70 genes. Additionally, EXO70 subgroups diversified before the divergence within polyploid wheat and related species during the evolutionary process of the EXO70 gene family; however, no new groups/subgroups have emerged.
EXO70I members were most represented in wheat and its five relatives (57), as well as in rice (16), but not in Arabidopsis [6,36]. EXO70I belongs to EXO70.2, not EXO70.3, which is different from what was found in previous studies [1,7,36]. This suggests that the EXO70I subgroup arose before the evolutionary divergence of rice from other Triticeae crops and disappeared during the evolution of Arabidopsis. The EXO70I subgroup underwent rapid divergence, producing a large number of members; this event can probably be explained by unequal cross-over or segmental chromosomal duplication [56,57]. During long-term natural selection, numerous EXO70 genes diverged and evolved in order to respond to various conditions. The study of Arabidopsis showed that the duplicated gene loss process is non-random; those involved in DNA repair are more likely to be lost, while genes involved in signal transduction and transcription have been preferentially retained [56]. Therefore, the function of EXO70I subgroup in Arabidopsis was inclined to responses to DNA repair, and in grass species may participate in signal transduction. Research on a larger range of species is needed to figure out whether the EXO70I branch is unique to monocotyledons.
3.2. Diversification of Subcellular Localization Pattern of the EXO70 in H. villosa
Protein subcellular localization analysis provided important clues to their specialized biological functions [58]. The diversification of EXO70-V subcellular localization patterns implies functional differentiation. Except for EXO70I1-V, all EXO70 genes showed plasma membrane (PM) signals. The EXO70D1/F2-V-GFP locates to the PM merged with some small, discrete punctate. At the same time, EXO70I1-V-GFP only gave rise to smaller fluorescent discrete punctate along with PM, which are similar to AtEXO70E2 in Arabidopsis protoplasts, which was a maker of a novel double-membraned structure termed EXPO (exocyst-positive organelles). AtEXO70E2 was involved in unconventional protein secretion for cytosolic proteins that lack a signal peptide, because of its ability to recruit several other exocyst complex subunits [8,9,59,60]. Therefore, EXO70D1, F2 and I1-V might have the ability to recruit different partners, then form various complexes to execute different biological functions. AtEXO70A1 was distributed in different patterns in different systems’ cytosol; it showed up in the nucleus and numerous small punctate structures in the BY-2 cell [36], at the apex of growing tobacco pollen tubes [61] and is strongly present in the cell plate [62]. In the study, EXO70A1/A3-V showed a weak PM signal, while EXO70A2-V was characterized by mis-localizations. This was probably because AtEXO70A1 took part in a different vesicular transport process. Moreover, despite both AtEXO70B2 and AtEXO70H1 participating in the interaction between plants and pathogens, the signal of EXO70B2-GFP was mainly found in the cytoplasm, while EXO70H1-GFP was in vesicle-like structures in Nicotiana benthamiana leaf [31]. In our analysis, EXO70B1-V was present in the PM and nucleus (Figure 7g). It is likely that they went through different action sites to take part in the process of disease resistance. An exocyst is a tethering factor that mediates secretory vesicles to the plasma membrane before SNARE-mediated fusion [63,64]. EXPOs deliver cytosolic proteins to the cell surface [65,66] and therefore all of those were related to PM. The results explained why most genes had the PM location pattern.
3.3. Function Conserve or Differentiation of the EXO70 Gene Family in Common Wheat and H. villosa
An orthologous gene is one that diverged after evolution to give rise to different species; this gene generally maintains a similar function to that of the ancestral gene that it evolved from [67]. In our study, some EXO70 orthologous genes from H. villosa exhibited a similar expression pattern to common wheat. For example, EXO70A3-V and TaEXO70A3 showed a high expression level under heat stress; EXO70B1-V was preferred to TaEXO70B1-3AL, which was induced by Bgt at a late stage (48 h), but not by drought and heat stress; EXO70E1-V and TaEXO70E1-3B were upregulated by Bgt treatment; the expression of EXO70H1-V and TaEXO70H1-1DL was increased in response to heat (Figure 5 and Figure 6). Therefore, it is reasonable to presume that some EXO70 genes from H. villosa may have a similar function to the corresponding EXO70 genes from common wheat. In plants, AtEXO70B1, AtEXO70B2, AtEXO70H1 and OsEXO70E1 are known for their roles in innate immunity [6,27,28,29,30,31,34]. In the literature, genes such as TaEXO70B1/B2 (with their homologous alleles), EXO70B1-V, TaEXO70E1-3B, EXO70E1-V, TaEXO70H1-1DL and EXO70H1-V were induced by Pst/Bgt treatment. Thus, we hypothesize that those genes also play an important role in plant defense responses and it is worth conducting further study to prove their function.
The long-term evolutionary fate of paralogous genes will still be determined by functions, with the genes that appeared to be sub-functionalized or neo-functionalized probably having higher rates of gene birth because of the increased adaptability. In contrast, the functional redundancy gene is unlikely to be stably maintained in the genome [55,57,68,69]. Paralogous/orthologous genes may diverge in expression to achieve more complex control of the same genetic network, balancing the relationship between internal growth and external environmental stimuli so that they “pay” the least and get the most [70]. In Arabidopsis, EXO70C1/C2 were involved in pollen development and mainly localized pollen related tissue [21,22]. In common wheat, six TaEXO70C genes had an undetectable expression level in roots/stem/leaves (Figure 5a), but EXO70C1-V showed a moderate expression level in the stem and responded to drought and ABA treatment (Figure 6). Studies have shown that ABA has contributed to osmotic stress tolerance by regulating stomatal aperture and guard cells [71]. Therefore, EXO70C1-V perhaps plays an important role in drought tolerance. In rice, OsExo70F3 interacts with AVR-Pii and plays a crucial role in triggered immunity [35]. In our research, including the copy number, 13 TaEXO70F members were identified and EXO70F1-V, EXO70F2-V and EXO70F3-V were cloned from H. villosa. Expression studies have revealed that 11/13 common wheat varieties were induced by stress treatment (Figure 5), and three EXO70-V genes showed clearly diverse expression patterns, of which EXO70F1-V was only induced by SA, EXO70F2-V in response to drought, H2O2 and SA at an early stage, and EXO70F3-V in response to Bgt treatment at a late stage (48 h) (Figure 6). Research shows that both SA and H2O2 function as a key regulator against pathogens and stress tolerance [72,73,74]. Thus, we can infer the function of EXO70F genes not only in plant defense responses but also in abiotic stress.
In N. benthamiana, EXO70D and EXO70G mainly affect the size of the leaf [6]. In wheat, TaEXO70D2-7BS and TaEXO70G1/G2 (with their homeoalleles) were induced by Pst and Bgt treatments. EXO70D1-V was upregulated by Bgt and MeJA treatments, EXO70G1-V had an increased expression level under phytohormones and H2O2 treatments, while EXO70G2-V was induced by drought and SA. MeJA is important for regulating the growth of plants and promotes plant resistance of various stresses [75]. This might suggest that EXO70D1-V plays an important role in plant growth and defense responses EXO70G are multifunctional. Of 22 EXO70I genes from common wheat, only five genes had distinct inducible expression. For instance, TaEXO70I6-7DL and TaEXO70I8-4BL were upregulated by drought and heat, respectively. EXO70I1-V was induced by ET and SA and maintained a high expression level. ET and SA regulate many diverse metabolic and developmental processes in plants, such as seed germination, abiotic stress response and pathogen defense [76,77]. Thus, we hypothesize that EXO70I1-V might play a vital role in growth or against multiple stresses. EXO70 genes provide diverse expression patterns in different tissues and stresses, implying that EXO70 genes may play an essential role in plant adaptation to a complicated and changeable environment.
4. Materials and Methods
4.1. Plant Materials
H. villosa (genome VV, accession no. 91C43), from the Cambridge Botanical Garden, Cambridge, UK, was used for gene cloning and expression analysis. Powdery mildew susceptible variety Sumai 3 was used for propagation of fresh spores of powdery mildew isolate E26. Nicotiana benthamiana plants were used for subcellular localization analysis. All the materials were grown in a greenhouse under a 14 h light/10 h dark cycle at 24 °C/18 °C, with 70% relative air humidity.
4.2. Plant Treatments
The seedlings of H. villosa were grown in liquid or soil until the three-leaf stage. For heat shock or drought stress treatment, the plants were transferred to 42 °C conditions, or dipped into 20% PEG 6000 and leaves were sampled at 0, 1, 6, or 12 h after treatment. For powdery mildew treatment, the plant was inoculated with pathogen isolate E26 and the leaf tissues were sampled at 0, 24, 48 and 72 h after inoculation. For phytohormones and H2O2 treatments, the plants were sprayed with 5 mmol salicylic acid (SA), 0.1 mmol methyl jasmonate (MeJA), 0.1 mmol ethephon (ET), 0.2 mmol abscisic acid (ABA) and 7 mmol hydrogen peroxide (H2O2), respectively and all leaf tissues were collected at 0, 6, 12, or 24 h after spraying [78]. All the samples were rapidly frozen in liquid nitrogen, then stored in an ultra-freezer (−80 °C) before use.
4.3. RNA Isolation and Real-Time PCR Analysis
Total RNA was extracted using a Trizol Reagent kit (Invitrogen, CA, USA) according to the manufacturer’s instructions and analyzed by gel electrophoresis. The first-strand cDNA was synthesized with random oligonucleotides using the HiScript® II Reverse Transcriptase system (Vazyme, Nanjing, China). qRT-PCR was carried out in a total volume of 20 μL containing 2 μL of cDNA, 0.4 μL gene-specific primers (10 μM), 10 μL SYBR Green Mix and 7.2 μL of RNase free ddH2O, using the Roche LightCycler480 Real-time System (Roche, Basel, Swiss Confederation). The expression was represented in the form of relative fold change using the 2−ΔΔCT method [79]. Differentially expressed genes between each two samples pair were defined as two-fold up-regulated or two-fold down-regulated genes. Primers used for qRT-PCR are designed by Primer3 (Table S3). Three biological replications were performed. Heat map analysis of the expression data was performed using heat map drawing software MeV (version No. 4.7, Institute for Genomic Research, MD, USA)
4.4. Identification of EXO70 Gene Families in Wheat and Related Triticeae Species
We searched for the keywords ‘exocyst subunit exo70 family protein’ in the annotated proteins database of Hordeum vulgare (HH, 2n = 2x = 14, accession No. FJWB02000000) and obtained entries containing gene ID and protein sequences [42]. Then we performed a Conserved Domains (CD) search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and reserved the protein sequences that contain the typical pfam03081 domain [47].
The identified EXO70 protein sequences of H. vulgare were used as query sequences to blast (E-value ≤ 10−10) against protein database of the other species, including Triticum urartu (AA, 2n = 2x = 14, accession No. NMPL02000000) [39], T. aestivum (AABBDD, 2n = 6x = 42, accession No. NMPL02000000) [80], Brachypodium distachyon (BdBd, 2n = 2x = 14, accession No. ADDN03000000) [81], Oryza sativa Japonica (2n = 2x = 24, accession No. AP008207–AP008218) [82], T. dicoccoides (AABB, 2n = 4x = 28, accession No. FXXJ01000000) [43] and Aegilops tauschii (DD, 2n = 2x = 14, accession No. AOCO02000000) [83]. After removing the redundant gene sequences for each the species, the alignment hits were validated by performing a CD search as described above.
4.5. Cloning and Protein Sequences Analysis of EXO70 Genes from Haynaldia villosa
According to the sequences obtained from the database of H. vulgare, primers (Table S3) for cloning the full-length cDNA of the EXO70 gene from Haynaldia villosa were designed with online software Primer3 designing tool (v. 0.4.0, University of California, USA) [84] (Table S3). Mixed root, stem and leaf tissue cDNA of H. villosa served as a template for the isolation. This was performed at 95 °C for 30 s, followed by 35 cycles of 95 °C for 15 s, 58 °C for 15 s or 30 s and 72 °C for 3 min and then by 5 min at 72 °C in Phanta Max Super-Fidelity DNA polymerase (Vazyme, Nanjing, China). Before subcloning into their destination vectors, the PCR-amplified cDNA products were first cloned into the pTOPO-Blunt Vector (Aidlab, Beijing, China) as per the manufacturer’s instructions. Multiple sequence alignments were conducted using DNAMan (Lynnon Corporation, Quebec, QC, Canada) software. The sequences similarity was visualized using the R programming language.
4.6. Subcellular Localization Assay
The ORFs of EXO70-V genes (without stop codon) were amplified from the pTOPO-Blunt Vector, then inserted into the pCambia1305-GFP vector, which contains a green fluorescent protein (GFP) reporter gene driven by the CaMV 35S promoter, using homologous cloning technology as per the manufacturer’s instructions (Vazyme, Nanjing, China) (Table S3). Then it was introduced into Agrobacterium tumefaciens (strain GV3101) bacteria by a freeze–thaw procedure and grown in Luria-Bertani (LB) medium at 28 °C for 2 or 3 d.
Agrobacterium tumefaciens (strain GV3101) bacteria containing fusion constructed were grown in Luria-Bertani (LB) medium with both rifampicin and kanamycin (0.05 µg/mL) at 28 °C overnight. The bacterial cells were centrifuged and resuspended in an infiltration solution (10 mM MES pH 5.6, 0.1 mM Acetosyringone, 10 mM MgCl2) to a final OD600 = 1.5. Bacterial suspensions were infiltrated into five- to six-week growing stage leaves of N. benthamiana by depressing the plunger of a 1-mL disposable needleless syringe into the abaxial side of leaves [85,86]. The fluorescence signals were observed 48–60 h after injection and images were captured using a confocal laser scanning microscope (LSM780; Carl Zeiss, Jena, Germany) according to the methods described by Wang et al. [87].
4.7. Phylogenetic Analysis of EXO70 Gene Family
Multiple sequence alignment was conducted by ClustalW which was integrated in Mega v6.0 [88]. Phylogenetic analysis was performed through online software PhyML 3.0 [89] using maximum-likelihood method with default parameter [90]. EXO70 proteins are rather diverse at their N-terminal and could not be aligned reliably, so we only used the conserved domain proteins to construct the phylogenetic tree and removed five genes where the length of the domain is fewer than 200 amino acids for further analysis.
4.8. Chromosomal Distribution and Exon-Intron Structure Analysis
Chromosomal information of predicted EXO70 genes from each species was obtained after using cDNA sequences as a query sequence blasted to the genomic sequence to determine their chromosomal locations. Then we drew their locations onto the physical map of each chromosome using MapInspect tool (http://mapinspect.software.informer.com/).
The gff3 files of each species was downloaded from the Ensembl Plants FTP server (http://plants.ensembl.org/index.html) on 20 January 2018 for exon–intron structure analysis; the image of the exon–intron structure was obtained using the online Gene Structure Display Server (last accessed date on 20 January 2018) with the gff3 files for each species [91]. The corresponding evolutionary tree were constructed by Mega v6.0 [88], all sequences were aligned by ClustalW using the default parameters [92,93], used the Neighbor-Joining method with the pairwise deletion option, Poisson correction and bootstrap analysis conducted with 1000 replicates [26,94]
4.9. RNA-seq Expression Analysis
Publicly available RNA-seq data were retrieved from the expVIP [50] were used to analyze the expression pattern of predicted wheat EXO70 genes in different tissues and stresses. The tissue-specific expression data were compiled from three wheat tissues (leaf, stem, root) collected from Chinese Spring at seeding development. The biotic stress expression data included two diseases (stripe rust and powdery mildew pathogen) and were collected from disease-resistant wheat varieties N9134 (at 7 days seedling stage). The abiotic stress expression data, including drought and heat treatments, were collected from heat-resistant wheat cultivar TAM107 (at 7 days seedling stage). The relative expression of each TaEXO70 gene in different tissues and stresses was presented as a heat map, which was constructed by the heat map drawing software MeV (version No. 4.7, Institute for Genomic Research, MD, USA).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/20/1/60/s1.
Author Contributions
X.W., J.X., H.W. and J.Z. conceived and designed the study; J.Z., X.Z., W.W. and J.X. analyzed the data; H.Z., J.L. and M.L. collected the plant materials; J.Z. and X.Z. performed the experiments; J.Z., X.W. and J.X. wrote the manuscript. All authors reviewed and edited the manuscript.
Funding
This work was supported by the National Key Research and Development program (2016YFD0101004, 2016YFD0102001-004), the National Science Foundation of China (Nos. 31471490, 31661143005, 31290213), the Important National Science & Technology Specific Projects in Transgenic Research (2014ZX0800907B), the Chinese High Tech Program of China (No. 2011AA1001), the Program of Introducing Talents of Discipline to Universities (No. B08025), the 333 Talent Project of Jiangsu Province, the Shanghai Agriculture Applied Technology Development Program, China Grant (No. Z201502) and Development of Key Agricultural Varieties in Jiangsu Province (PZCZ201706).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ABA | abscisic acid |
| DNA | deoxyribonucleic acid |
| EXPO | exocyst-positive organelle |
| ET | ethephon |
| GFP | green fluorescent protein |
| H2O2 | hydrogen peroxide |
| MeJA | methyl jasmonate |
| PM | plasma membrane |
| Pm | powdery mildew |
| qRT-PCR | quantitative real time polymerase chain reaction |
| RNA | ribonucleic acid |
| SNARE | soluble NSF attachment protein receptor |
| SA | salicylic acid |
References
- Cvrckova, F.; Grunt, M.; Bezvoda, R.; Hala, M.; Kulich, I.; Rawat, A.; Zarsky, V. Evolution of the land plant exocyst complexes. Front. Plant Sci. 2012, 3, 159. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Guo, W. The exocyst complex in polarized exocytosis. Curr. Opin. Cell Biol. 2009, 21, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Wang, Y.; Yao, X.; Xu, Z.; An, L.; Yin, M. The role of Exo70 in vascular smooth muscle cell migration. Cell. Mol. Biol. Lett. 2016, 21, 20. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Elias, M. The exocyst complex in plants. Cell Biol. Int. 2003, 27, 199–201. [Google Scholar] [CrossRef]
- Tu, B.; Hu, L.; Chen, W.; Li, T.; Hu, B.; Zheng, L.; Lv, Z.; You, S.; Wang, Y.; Ma, B.; et al. Disruption of OsEXO70A1 Causes Irregular Vascular Bundles and Perturbs Mineral Nutrient Assimilation in Rice. Sci. Rep. 2015, 5, 18609. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Overdijk, E.J.R.; Berg, J.A.; Govers, F.; Bouwmeester, K. Solanaceous exocyst subunits are involved in immunity to diverse plant pathogens. J. Exp. Bot. 2018, 69, 655–666. [Google Scholar] [CrossRef]
- Synek, L.; Schlager, N.; Elias, M.; Quentin, M.; Hauser, M.T.; Zarsky, V. AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. Plant J. 2006, 48, 54–72. [Google Scholar] [CrossRef]
- Wang, J.; Ding, Y.; Wang, J.; Hillmer, S.; Miao, Y.; Lo, S.W.; Wang, X.; Robinson, D.G.; Jiang, L. EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell 2010, 22, 4009–4030. [Google Scholar] [CrossRef]
- Ding, Y.; Wang, J.; Chun Lai, J.H.; Ling Chan, V.H.; Wang, X.; Cai, Y.; Tan, X.; Bao, Y.; Xia, J.; Robinson, D.G.; et al. Exo70E2 is essential for exocyst subunit recruitment and EXPO formation in both plants and animals. Mol. Biol. Cell 2014, 25, 412–426. [Google Scholar] [CrossRef]
- Dellago, H.; Loscher, M.; Ajuh, P.; Ryder, U.; Kaisermayer, C.; Grillari-Voglauer, R.; Fortschegger, K.; Gross, S.; Gstraunthaler, A.; Borth, N.; et al. Exo70, a subunit of the exocyst complex, interacts with SNEV(hPrp19/hPso4) and is involved in pre-mRNA splicing. Biochem. J. 2011, 438, 81–91. [Google Scholar] [CrossRef]
- Ren, J.; Guo, W. ERK1/2 regulate exocytosis through direct phosphorylation of the exocyst component Exo70. Dev. Cell 2012, 22, 967–978. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, J.; Yang, C.; Capraro, B.R.; Baumgart, T.; Bradley, R.P.; Ramakrishnan, N.; Xu, X.; Radhakrishnan, R.; Svitkina, T.; et al. Exo70 generates membrane curvature for morphogenesis and cell migration. Dev. Cell 2013, 26, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.; Zhang, J.; Zhang, Y.; Hsu, S.C.; Zhou, D.; Guo, W. Exo70 interacts with the Arp2/3 complex and regulates cell migration. Nat. Cell Biol. 2006, 8, 1383–1388. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yue, P.; Artym, V.V.; Mueller, S.C.; Guo, W. The role of the exocyst in matrix metalloproteinase secretion and actin dynamics during tumor cell invadopodia formation. Mol. Biol. Cell 2009, 20, 3763–3771. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Zheng, K.; Lv, X.; Hou, J.; Xu, L.; Zhao, Y.; Song, F.; Fan, Y.; Cao, H.; Zhang, W.; et al. Exo70 is an independent prognostic factor in colon cancer. Sci. Rep. 2017, 7, 5039. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, M.; Yu, D.; Ren, S.; Sun, S.; Liu, L.; Ketelaar, T.; Emons, A.M.; Liu, C.M. EXO70A1-mediated vesicle trafficking is critical for tracheary element development in Arabidopsis. Plant Cell 2013, 25, 1774–1786. [Google Scholar] [CrossRef] [PubMed]
- Vukasinovic, N.; Oda, Y.; Pejchar, P.; Synek, L.; Pecenkova, T.; Rawat, A.; Sekeres, J.; Potocky, M.; Zarsky, V. Microtubule-dependent targeting of the exocyst complex is necessary for xylem development in Arabidopsis. New Phytol. 2017, 213, 1052–1067. [Google Scholar] [CrossRef]
- Kulich, I.; Cole, R.; Drdova, E.; Cvrckova, F.; Soukup, A.; Fowler, J.; Zarsky, V. Arabidopsis exocyst subunits SEC8 and EXO70A1 and exocyst interactor ROH1 are involved in the localized deposition of seed coat pectin. New Phytol. 2010, 188, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Drdova, E.J.; Synek, L.; Pecenkova, T.; Hala, M.; Kulich, I.; Fowler, J.E.; Murphy, A.S.; Zarsky, V. The exocyst complex contributes to PIN auxin efflux carrier recycling and polar auxin transport in Arabidopsis. Plant J. 2013, 73, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Kalmbach, L.; Hematy, K.; De Bellis, D.; Barberon, M.; Fujita, S.; Ursache, R.; Daraspe, J.; Geldner, N. Transient cell-specific EXO70A1 activity in the CASP domain and Casparian strip localization. Nat. Plants 2017, 3, 17058. [Google Scholar] [CrossRef]
- Lai, K.S. Analysis of EXO70C2 expression revealed its specific association with late stages of pollen development. Plant Cell Tissue Organ Cult. 2015, 124, 209–215. [Google Scholar] [CrossRef]
- Synek, L.; Vukasinovic, N.; Kulich, I.; Hala, M.; Aldorfova, K.; Fendrych, M.; Zarsky, V. EXO70C2 Is a Key Regulatory Factor for Optimal Tip Growth of Pollen. Plant Physiol. 2017, 174, 223–240. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, M.; Jiang, L.; Liu, X.; Zhao, J.; Yan, S.; Yang, S.; Ren, H.; Liu, R.; Zhang, X. Transcriptome profiling reveals roles of meristem regulators and polarity genes during fruit trichome development in cucumber (Cucumis sativus L.). J. Exp. Bot. 2014, 65, 4943–4958. [Google Scholar] [CrossRef] [PubMed]
- Kulich, I.; Vojtikova, Z.; Glanc, M.; Ortmannova, J.; Rasmann, S.; Zarsky, V. Cell wall maturation of Arabidopsis trichomes is dependent on exocyst subunit EXO70H4 and involves callose deposition. Plant Physiol. 2015, 168, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, P.; Yang, Y.; Chi, Y.; Fan, B.; Chen, Z. Expression and Functional Analysis of a Novel Group of Legume-specific WRKY and Exo70 Protein Variants from Soybean. Sci. Rep. 2016, 6, 32090. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhou, Y.; Chi, Y.; Fan, B.; Chen, Z. Characterization of Soybean WRKY Gene Family and Identification of Soybean WRKY Genes that Promote Resistance to Soybean Cyst Nematode. Sci. Rep. 2017, 7, 17804. [Google Scholar] [CrossRef] [PubMed]
- Kulich, I.; Pecenkova, T.; Sekeres, J.; Smetana, O.; Fendrych, M.; Foissner, I.; Hoftberger, M.; Zarsky, V. Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic 2013, 14, 1155–1165. [Google Scholar] [CrossRef]
- Stegmann, M.; Anderson, R.G.; Westphal, L.; Rosahl, S.; McDowell, J.M.; Trujillo, M. The exocyst subunit Exo70B1 is involved in the immune response of Arabidopsis thaliana to different pathogens and cell death. Plant Signal. Behav. 2013, 8, e27421. [Google Scholar] [CrossRef]
- Zhao, T.; Rui, L.; Li, J.; Nishimura, M.T.; Vogel, J.P.; Liu, N.; Liu, S.; Zhao, Y.; Dangl, J.L.; Tang, D. A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet. 2015, 11, e1004945. [Google Scholar] [CrossRef]
- Stegmann, M.; Anderson, R.G.; Ichimura, K.; Pecenkova, T.; Reuter, P.; Zarsky, V.; McDowell, J.M.; Shirasu, K.; Trujillo, M. The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 2012, 24, 4703–4716. [Google Scholar] [CrossRef]
- Pecenkova, T.; Hala, M.; Kulich, I.; Kocourkova, D.; Drdova, E.; Fendrych, M.; Toupalova, H.; Zarsky, V. The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant-pathogen interaction. J. Exp. Bot. 2011, 62, 2107–2116. [Google Scholar] [CrossRef] [PubMed]
- Seo, D.H.; Ahn, M.Y.; Park, K.Y.; Kim, E.Y.; Kim, W.T. The N-Terminal UND Motif of the Arabidopsis U-Box E3 Ligase PUB18 Is Critical for the Negative Regulation of ABA-Mediated Stomatal Movement and Determines Its Ubiquitination Specificity for Exocyst Subunit Exo70B1. Plant Cell 2016, 28, 2952–2973. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Jeon, B.W.; Kim, S.Y.; Hwang, J.U.; Lee, Y. The ROP2-RIC7 pathway negatively regulates light-induced stomatal opening by inhibiting exocyst subunit Exo70B1 in Arabidopsis. New Phytol. 2016, 209, 624–635. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Xu, C.; Wu, D.; Zhao, Y.; Qiu, Y.; Wang, X.; Ouyang, Y.; Cai, B.; Liu, X.; Jing, S.; et al. Bph6 encodes an exocyst-localized protein and confers broad resistance to planthoppers in rice. Nat. Genet. 2018, 50297–50306. [Google Scholar] [CrossRef] [PubMed]
- Fujisaki, K.; Abe, Y.; Ito, A.; Saitoh, H.; Yoshida, K.; Kanzaki, H.; Kanzaki, E.; Utsushi, H.; Yamashita, T.; Kamoun, S.; et al. Rice Exo70 interacts with a fungal effector, AVR-Pii, and is required for AVR-Pii-triggered immunity. Plant J. 2015, 83, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Chong, Y.T.; Gidda, S.K.; Sanford, C.; Parkinson, J.; Mullen, R.T.; Goring, D.R. Characterization of the Arabidopsis thaliana exocyst complex gene families by phylogenetic, expression profiling, and subcellular localization studies. New Phytol. 2010, 185, 401–419. [Google Scholar] [CrossRef]
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; Jakobsen, K.S.; Wulff, B.B.; Steuernagel, B.; Mayer, K.F.; Olsen, O.A. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345, 1250092. [Google Scholar] [CrossRef]
- Zimin, A.V.; Puiu, D.; Hall, R.; Kingan, S.; Clavijo, B.J.; Salzberg, S.L. The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum. GigaScience 2017, 6, gix097. [Google Scholar] [CrossRef]
- Ling, H.Q.; Ma, B.; Shi, X.; Liu, H.; Dong, L.; Sun, H.; Cao, Y.; Gao, Q.; Zheng, S.; Li, Y.; et al. Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 2018, 557, 424–428. [Google Scholar] [CrossRef]
- Bettgenhaeuser, J.; Krattinger, S.G. Rapid gene cloning in cereals. Theor. Appl. Genet. 2018. [Google Scholar] [CrossRef]
- Luo, M.C.; Gu, Y.Q.; Puiu, D.; Wang, H.; Twardziok, S.O.; Deal, K.R.; Huo, N.; Zhu, T.; Wang, L.; Wang, Y.; et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 2017, 551, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Qian, C.; Cao, A.; Li, Y.; Jiang, Z.; Li, M.; Jin, X.; Hu, J.; Zhang, Y.; Wang, X.; et al. The Hv-SGT1 gene from Haynaldia villosa contributes to resistances towards both biotrophic and hemi-biotrophic pathogens in common wheat (Triticum aestivum L.). PLoS ONE 2013, 8, e72571. [Google Scholar] [CrossRef]
- Cao, A.; Xing, L.; Wang, X.; Yang, X.; Wang, W.; Sun, Y.; Qian, C.; Ni, J.; Chen, Y.; Liu, D.; et al. Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. USA 2011, 108, 7727–7732. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Hu, P.; Liu, J.; Witek, K.; Zhou, S.; Xu, J.; Zhou, W.; Gao, L.; Huang, Z.; Zhang, R.; et al. Pm21 from Haynaldia villosa Encodes a CC-NBS-LRR that Confers Powdery Mildew Resistance in Wheat. Mol. Plant 2018, 11, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; van Os, G.M.; Ren, S.; Yu, D.; Ketelaar, T.; Emons, A.M.; Liu, C.M. Expression and functional analyses of EXO70 genes in Arabidopsis implicate their roles in regulating cell type-specific exocytosis. Plant Physiol. 2010, 154, 1819–1830. [Google Scholar] [CrossRef] [PubMed]
- Miftahudin; Ross, K.; Ma, X.F.; Mahmoud, A.A.; Layton, J.; Milla, M.A.; Chikmawati, T.; Ramalingam, J.; Feril, O.; Pathan, M.S.; et al. Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics 2004, 168, 651–663. [Google Scholar] [CrossRef]
- Hao, M.; Luo, J.; Zhang, L.; Yuan, Z.; Zheng, Y.; Zhang, H.; Liu, D. In situ hybridization analysis indicates that 4AL-5AL-7BS translocation preceded subspecies differentiation of Triticum turgidum. Genome 2013, 56, 303–305. [Google Scholar] [CrossRef]
- Borrill, P.; Ramirez-Gonzalez, R.; Uauy, C. expVIP: A Customizable RNA-seq Data Analysis and Visualization Platform. Plant Physiol. 2016, 170, 2172–2186. [Google Scholar] [CrossRef]
- Pearce, S.; Vazquez-Gross, H.; Herin, S.Y.; Hane, D.; Wang, Y.; Gu, Y.Q.; Dubcovsky, J. WheatExp: An RNA-seq expression database for polyploid wheat. BMC Plant Biol. 2015, 15, 299. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gao, S.; Tang, Y.; Li, L.; Zhang, F.; Feng, B.; Fang, Z.; Ma, L.; Zhao, C. Genome-wide identification and evolutionary analyses of bZIP transcription factors in wheat and its relatives and expression profiles of anther development related TabZIP genes. BMC Genom. 2015, 16, 976. [Google Scholar] [CrossRef] [PubMed]
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D.; et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.; Levy, A.A. Allopolyploidy--a shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 2005, 109, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.; Levy, A.A. Genome evolution in allopolyploid wheat—A revolutionary reprogramming followed by gradual changes. J. Genet. Genom. 2009, 36, 511–518. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef]
- Sperschneider, J.; Catanzariti, A.M.; DeBoer, K.; Petre, B.; Gardiner, D.M.; Singh, K.B.; Dodds, P.N.; Taylor, J.M. LOCALIZER: Subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 2017, 7, 44598. [Google Scholar] [CrossRef]
- Lin, Y.S.; Ding, Y.; Wang, J.; Shen, J.B.; Kung, C.H.; Zhuang, X.H.; Cui, Y.; Yin, Z.; Xia, Y.J.; Lin, H.X.; et al. Exocyst-Positive Organelles and Autophagosomes Are Distinct Organelles in Plants. Plant Physiol. 2015, 169, 1917–1932. [Google Scholar]
- Robinson, D.G.; Ding, Y.; Jiang, L. Unconventional protein secretion in plants: A critical assessment. Protoplasma 2016, 253, 31–43. [Google Scholar] [CrossRef]
- Hala, M.; Cole, R.; Synek, L.; Drdova, E.; Kulich, I.; Pecenkova, T.; Hochholdinger, F.; Cvrckova, F.; Fowler, J.; Zarsky, V. Exocyst complex functions in plant development. Comp. Biochem. Phys. A 2008, 150, S188–S189. [Google Scholar] [CrossRef]
- Fendrych, M.; Synek, L.; Pecenkova, T.; Toupalova, H.; Cole, R.; Drdova, E.; Nebesarova, J.; Sedinova, M.; Hala, M.; Fowler, J.E.; et al. The Arabidopsis exocyst complex is involved in cytokinesis and cell plate maturation. Plant Cell 2010, 22, 3053–3065. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Guo, W. The Exocyst at a Glance. J. Cell Sci. 2015, 128, 2957–2964. [Google Scholar] [CrossRef] [PubMed]
- Lurick, A.; Kummel, D.; Ungermann, C. Multisubunit tethers in membrane fusion. Curr. Biol. 2018, 28, R417–R420. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Yang, Y.; Li, G.; Wang, F.; Fan, B.; Chen, Z. Identification and characterization of a novel group of legume-specific, Golgi apparatus-localized WRKY and Exo70 proteins from soybean. J. Exp. Bot. 2015, 66, 3055–3070. [Google Scholar] [CrossRef] [PubMed]
- Ebine, K.; Ueda, T. Roles of membrane trafficking in plant cell wall dynamics. Front. Plant Sci. 2015, 6, 878. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005, 39, 309–338. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Roulin, A.; Auer, P.L.; Libault, M.; Schlueter, J.; Farmer, A.; May, G.; Stacey, G.; Doerge, R.W.; Jackson, S.A. The fate of duplicated genes in a polyploid plant genome. Plant J. 2013, 73, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Schnable, J.C. Functional Divergence Between Subgenomes and Gene Pairs After Whole Genome Duplications. Mol. Plant. 2018, 11, 388–397. [Google Scholar] [CrossRef]
- Fernando, V.C.D.; Schroeder, D.F. Role of ABA in Arabidopsis Salt, Drought, and Desiccation Tolerance. In Abiotic and Biotic Stress in Plants—Recent Advances and Future Perspectives; IntechOpen: London, UK, 2016; Chapter 22. [Google Scholar]
- Kumar, D. Salicylic acid signaling in disease resistance. Plant Sci. 2014, 228, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Maity, A.; Sharma, J.; Sarkar, A.; More, A.K.; Pal, R.K.; Nagane, V.P.; Maity, A. Salicylic acid mediated multi-pronged strategy to combat bacterial blight disease (Xanthomonas axonopodis pv. punicae) in pomegranate. Eur. J. Plant Pathol. 2018, 150, 923–937. [Google Scholar] [CrossRef]
- You, J.; Chan, Z. ROS Regulation During Abiotic Stress Responses in Crop Plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Howe, G.A. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009, 21, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed]
- Bleecker, A.B.; Kende, H. Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 2000, 16, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, Y.; Fei, F.; Wang, Z.; Wang, W.; Cao, A.; Liu, Y.; Han, S.; Xing, L.; Wang, H.; et al. E3 ubiquitin ligase gene CMPG1-V from Haynaldia villosa L. contributes to powdery mildew resistance in common wheat (Triticum aestivum L.). Plant J. 2015, 84, 154–168. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Mayer, K.F.X.; Rogers, J.; Doležel, J.; Pozniak, C.; Eversole, K.; Feuillet, C.; Gill, B.; Friebe, B.; Lukaszewski, A.J.; Sourdille, P. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [Google Scholar]
- International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar] [CrossRef]
- Fujisawa, M.; Baba, T.; Nagamura, Y.; Nagasaki, H.; Waki, K.; Vuong, H.; Matsumoto, T.; Wu, J.Z.; Kanamori, H.; Katayose, Y. The map-based sequence of the rice genome. Nature 2005, 436, 793–800. [Google Scholar]
- Zhao, G.; Zou, C.; Li, K.; Wang, K.; Li, T.; Gao, L.; Zhang, X.; Wang, H.; Yang, Z.; Liu, X. The Aegilops tauschii genome reveals multiple impacts of transposons. Nat. Plants 2017, 3, 946–955. [Google Scholar] [CrossRef] [PubMed]
- Andreas, U.; Ioana, C.; Triinu, K.; Jian, Y.; Brant, C.F.; Maido, R.; Steven, G.R. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar]
- Chen, H.; Zou, Y.; Shang, Y.; Lin, H.; Wang, Y.; Cai, R.; Tang, X.; Zhou, J.M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008, 146, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Tan, L.; van Hooren, M.; Tan, X.; Liu, F.; Li, Y.; Zhao, Y.; Li, B.; Rui, Q.; Munnik, T.; et al. Arabidopsis EXO70A1 recruits Patellin3 to the cell membrane independent of its role as an exocyst subunit. J. Integr. Plant Biol. 2017, 59, 851–865. [Google Scholar] [CrossRef]
- Wang, Z.; Cheng, J.; Fan, A.; Zhao, J.; Yu, Z.; Li, Y.; Zhang, H.; Xiao, J.; Muhammad, F.; Wang, H.; et al. LecRK-V, an L-type lectin receptor kinase in Haynaldia villosa, plays positive role in resistance to wheat powdery mildew. Plant Biotechnol. J. 2018, 16, 50–62. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yue, H.; Feng, K.; Deng, P.; Song, W.; Nie, X. Genome-wide identification, phylogeny and expressional profiles of mitogen activated protein kinase kinase kinase (MAPKKK) gene family in bread wheat (Triticum aestivum L.). BMC Genom. 2016, 17, 668. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).