Abstract
Barnacles of the genus Neoverruca are abundant near deep-sea hydrothermal vents of the northwestern Pacific Ocean, and are useful for understanding processes of population formation and maintenance of deep-sea vent faunas. Using next-generation sequencing, we isolated 12 polymorphic microsatellite loci from Neoverruca sp., collected in the Okinawa Trough. These microsatellite loci revealed 2–19 alleles per locus. The expected and observed heterozygosities ranged from 0.286 to 1.000 and 0.349 to 0.935, respectively. Cross-species amplification showed that 9 of the 12 loci were successfully amplified for Neoverruca brachylepadoformis in the Mariana Trough. A pairwise FST value calculated using nine loci showed significant genetic differentiation between the two species. Consequently, the microsatellite markers we developed will be useful for further population genetic studies to elucidate genetic diversity, differentiation, classification, and evolutionary processes in the genus Neoverruca.
1. Introduction
Larval dispersal in ecosystems such as deep-sea hydrothermal vents is essential for maintenance of benthic populations and for establishment of new colonies. For vent-endemic benthos, planktonic gametes or larvae provide the only means of migration between hydrothermal vents [1]. However, for most vent-endemic species, migration and dispersal patterns among vent fields have not been well studied. Vent-endemic barnacles belonging to the family Neoverrucidae (Cirripedia: Thoracica) [2] consist of two genera, Neoverruca [2] and Imbricaverruca [3]. The former is abundant in hydrothermal vent fields of the northwestern Pacific [4,5]. Though the genus Neoverruca has no fossil record, the first appearance of close, but extinct ancestors is in the Mesozoic Era [2,6]. Hence, the genus Neoverruca is a very primitive barnacle. Presently, the genus Neoverruca contains only one described species, Neoverruca brachylepadoformis. Based on morphology, individuals in the Okinawa Trough and Izu-Ogasawara Arc are distinct from N. brachylepadoformis in the Mariana Trough, located south of the Izu-Ogasawara Arc [4,5]. Watanabe et al. [5] classified specimens from the Okinawa Trough and Izu-Ogasawara Arc as a new, but unnamed species, designated Neoverruca sp., and investigated population structure, focusing on mitochondrial cytochrome oxidase c subunit I (COI). They discovered significant genetic differentiation between the two regions. Further population genetic analysis of this species using high-resolution genetic markers will be needed for detailed estimation of genetic diversity and structure. Therefore, microsatellite markers have been developed for further population genetic studies of Neoverruca sp. We isolated and characterized novel polymorphic microsatellite loci for Neoverruca sp. using next-generation sequencing and investigated cross-species amplification for N. brachylepadoformis collected from the Mariana Trough. Furthermore, we estimated the extent of genetic differentiation between Neoverruca sp. and N. brachylepadoformis using developed microsatellite loci, since Watanabe et al. [5] previously showed their distinctness using molecular phylogenetic analysis based upon mitochondrial COI.
2. Results and Discussion
We obtained 291,118,462 bp (956,987 read pairs) of raw sequence data from genomic DNA of Neoverruca sp., and assembled each read pair. We used assembled 736,240 sequences longer than 100 bp (122,338,901 bp total, average 166 bp) for simple sequence repeat identification. Sequence repeats were used for microsatellite detection and primer design. Ninety-six primer pairs were designed for screening amplifiable microsatellite loci (3 mer repeats: 60 loci, 4 mer: 17, 5 mer: 11, 6 mer: 8). Twelve of these loci were successfully amplified and subsequently analyzed in Neoverruca sp., although Nsp_11 and Nsp_80 were not amplified in two and one individual(s), respectively. The number of alleles per locus ranged from 2 to 19 (Table 1). Values of observed and expected heterozygosities ranged from 0.286 to 1.000 and 0.349 to 0.935, respectively. Out of 12 loci, two loci, Nsp_09 and Nsp_11, showed significant deviation from Hardy-Weinberg equilibrium, and these two showed the existence of null alleles (99% confidence level). Significant linkage disequilibrium was detected in the combination of Nsp_21-Nsp_37 (p < 0.05).
Of the 12 loci isolated and characterized from Neoverruca sp., nine were successfully amplified in N. brachylepadoformis. Three loci, including Nsp_11 and Nsp_23, which showed high polymorphism in Neoverruca sp., were hardly amplified in N. brachylepadoformis. The number of individual N. brachylepadoformis that showed successful amplification at these nine loci was 14 to 19 (out of 19) (Table 2). These nine loci were also polymorphic in that species, with 3 to 9 alleles per locus (Table 2). Values of observed and expected heterozygosities ranged from 0.063 to 0.786 and 0.174 to 0.855, respectively. In N. brachylepadoformis, two loci, Nsp_21 and Nsp_52, showed significant deviation from Hardy-Weinberg equilibrium, but seven other loci did not. A pairwise FST value calculated using nine common loci indicated significant genetic differentiation between the two species (FST = 0.292, p = 0.001).
In this study, the number of useful loci and the significance of the pairwise FST implied significant differentiation between Neoverruca sp. and N. brachylepadoformis. This differentiation is consistent with divergent patterns in molecular phylogenetic trees previously constructed for the COI haplotype [5]. Using nuclear and multiple microsatellite loci, this study robustly demonstrated the genetic differentiation between these two species at the population level.
Further population genetic studies using these markers will enable us to elucidate the level of present and historical dispersal and recruitment between these areas, i.e., between the Okinawa Trough and the Izu-Ogasawara Arc, which influences the formation and maintenance of populations within these species. Furthermore, these markers are expected to help elucidate genetic diversity, differentiation, classification, and evolutionary processes in genus Neoverruca. They may even be useful in clarifying the taxonomic diversification concerning this genus.

Table 1.
Characteristics of the 12 developed polymorphic microsatellite loci in 15 individuals of Neoverruca sp. Columns show the locus name, the primer sequence, the repeat motif, the size range of amplification products, including the U19 sequence, numbers of alleles (NA), observed (HO) and expected (HE) heterozygosities, and the index of deviation from Hardy-Weinberg equilibrium (FIS), and GenBank accession number.
| Locus | Repeat Motif | Primer Sequence (5'–3') | Size Range (bp) | NA | HO | HE | FIS | Accession No. |
|---|---|---|---|---|---|---|---|---|
| Nsp_09 | (CTTT)3CTTC(TTCC)10TCCCTTCA(TTCC)4 | F: AGGAGGCTTTCATGTTTTCC | 128–274 | 11 | 0.533 | 0.833 | 0.360 ** | AB971583 |
| R: U19-AAATGCGTGAGAGTGAAAGG | ||||||||
| Nsp_11 †† | (GTGA)12 | F: CACTCCTTTCGCGATTCC | 298–498 | 19 | 0.692 | 0.935 | 0.259 * | AB971584 |
| R: U19-CTACCAGGTGGAGCGTGC | ||||||||
| Nsp_21 | (ACACG)11 | F: TGAAGCAAGCAATGATAAGC | 122–174 | 8 | 0.667 | 0.833 | 0.200 | AB971585 |
| R: U19-TGTTGCGTCGTGTCG | ||||||||
| Nsp_23 | (GCAC)20 | F: AACCGGGTTACCCAAAGG | 183–373 | 16 | 0.867 | 0.913 | 0.051 | AB971586 |
| R: U19-TGTGCTGACGGATGTGC | ||||||||
| Nsp_37 | (AGA)11 | F: U19-CACCCGAGACTTCGATGC | 158–167 | 3 | 0.467 | 0.504 | 0.075 | AB971587 |
| R: TGGGATGAATAAGAGCTGCC | ||||||||
| Nsp_52 | (ATC)10 | F: CTATACTGGTCGATGCGCC | 125–128 | 2 | 0.733 | 0.500 | −0.467 | AB971588 |
| R: U19-CCACTTTTGGAGTGCATGG | ||||||||
| Nsp_60 | (TCG)10 | F: GGATCCGTGTCCCTTATGC | 184–202 | 5 | 0.400 | 0.556 | 0.280 | AB971589 |
| R: U19-TAACTTCAGGGCGCTTCG | ||||||||
| Nsp_68 | (GTA)7 | F: U19-CTCGTGGGAACCCATCC | 93–112 | 6 | 1.000 | 0.758 | 0.320 | AB971590 |
| R: TCTAAACTCGCGCAAGCC | ||||||||
| Nsp_70 | (CAC)7(AAC)2 | F: U19-CCTCAGTCTGCACACCC | 102–114 | 5 | 0.400 | 0.564 | 0.291 | AB971591 |
| R: TGGAGGCGATGAAGATGG | ||||||||
| Nsp_73 | (AGC)3AAC(AGC)3(AAC)2AGCAAC(AGC)6(AAC)7(AGC)3 | F: ATGTGGGTCGGTCTCAGC | 122–137 | 6 | 0.733 | 0.704 | −0.041 | AB971592 |
| R: U19-TGCATTTGATGTTGCTGC | ||||||||
| Nsp_80 † | (CTA)7 | F: U19-TCTGGAACCGGTCTCACC | 128–134 | 3 | 0.286 | 0.349 | 0.182 | AB971593 |
| R: AATAATCCAGAGCGGGACG | ||||||||
| Nsp_81 | (ACC)7 | F: U19-CGCATAATGACAAACGC | 178–187 | 4 | 0.467 | 0.496 | 0.058 | AB971594 |
| R: CACTGAACATGCAAGCCG |
† Individual(s) that did not show amplification of specific loci († 1 individual, †† 2 individuals); * Significant deviation from Hardy-Weinberg equilibrium (* p < 0.05, ** p < 0.01).

Table 2.
Cross-species amplification for N. brachylepadoformis using nine loci developed from Neoverruca sp. Columns show the locus name, the number of individuals in which these loci were successfully amplified, the size range of amplification products, including the U19 sequence, numbers of alleles (NA), observed (HO) and expected (HE) heterozygosities, and the index of deviation from Hardy-Weinberg equilibrium (FIS).
| Locus | Succeed | Size Range (bp) | NA | HO | HE | FIS |
|---|---|---|---|---|---|---|
| Nsp_09 | 18/19 | 144–177 | 7 | 0.556 | 0.682 | 0.186 |
| Nsp_21 | 16/19 | 122–137 | 3 | 0.063 | 0.174 | 0.640 ** |
| Nsp_37 | 19/19 | 161–220 | 9 | 0.737 | 0.855 | 0.138 |
| Nsp_52 | 16/19 | 119–134 | 5 | 0.438 | 0.604 | 0.275 *** |
| Nsp_60 | 18/19 | 160–196 | 4 | 0.278 | 0.335 | 0.171 |
| Nsp_68 | 14/19 | 93–118 | 8 | 0.786 | 0.747 | −0.051 |
| Nsp_70 | 18/19 | 108–117 | 4 | 0.667 | 0.500 | −0.333 |
| Nsp_73 | 18/19 | 110–125 | 6 | 0.611 | 0.645 | 0.053 |
| Nsp_81 | 19/19 | 178–187 | 3 | 0.316 | 0.476 | 0.337 |
* Significant deviation from Hardy-Weinberg equilibrium (** p < 0.01, *** p < 0.001).
3. Experimental Section
For isolation of whole genomic DNA, Neoverruca sp. was collected at the Iheya Depression (27°32.994'N/126°58.158'E, 1402 m depth) in the Okinawa Trough during cruise of R/V “Kaiyo” (Cruise No. KY11-02 Leg. 2), using the remotely operated vehicle (ROV) “Hyper-Dolphin” (Dive No. HPD#1245). Specimens were preserved in ethanol, and genomic DNA was isolated using proteinase K and phenol-chloroform extraction. DNA was further purified using ethanol precipitation and a QIAquick PCR purification kit (Qiagen, Hilden, Germany). Extracted DNA was sequenced as 300 bp paired end reads using a MiSeq sequencer (Illumina, San Diego, CA, USA), according to manufacturer’s instructions. Sequencing adapters were trimmed with fastq-mcf in ea-utils version 1.1.2-537 [7] and sequences of all read pairs were assembled using fastq-join in ea-utils [7]. Then assembled sequences longer than 100 bp were selected. Detection of simple sequence repeats and PCR primer design in each assembled sequence were performed with PAL_FINDER version 0.02.04 [8]. Then we removed redundancies in the assembled sequences in which PAL_FINDER detected simple sequence repeats and designed primers using CDHIT-EST [9]. In order to select microsatellite loci that may be highly variable, we selected primer pairs amplifying longer repeat stretches (thresholds: 3 mer is 5 repeats or more, 4 to 6 mers are 10 or more, respectively).
To characterize microsatellite loci, we screened 15 individuals of Neoverruca sp. collected at the Iheya North field (27°47.196'N/126°53.862'E, 990 m depth and 27°47.226'N/126°53.832'E, 977 m depth, Figure 1) in the Okinawa Trough during cruise of the R/V “Natsushima” (Cruise No. NT00-08), using the submersible, “Shinkai 2000” (Dive No. 2K#1192 and 2K#1194). Specimens collected were preserved in ethanol, and genomic DNA was extracted using a DNeasy Blood & Tissue Kit (Qiagen). To assess amplification and evaluate polymorphism of the designed primer sets, we performed PCR as follows. The reaction mixture (5 µL) contained <20 ng of template genomic DNA, AmpliTaq Gold 360 Master Mix (Applied Biosystems, Foster City, CA, USA), and three primers for each locus: a non-tailed forward primer (0.5 µM), a reverse primer with a U19 (5'-GGTTTTCCCAGTCACGACG-3') tail (0.5 µM), and a U19 primer (0.5 µM) fluorescently labeled with FAM, VIC, NED, or PET, based on the method of Schuelke [10]. PCR amplification was performed under the following conditions: 95 °C for 5 min; followed by 35 cycles at 95 °C for 30 s, 54 °C for 30 s, and 72 °C for 1 min, with a final extension at 72 °C for 5 min. Amplified PCR products were analyzed using an automated capillary-based DNA sequencer (ABI 3130xl Genetic Analyzer, Applied Biosystems) and GeneMapper version 3.7 (Applied Biosystems). For successfully amplified microsatellite loci for Neoverruca sp., cross-species amplification was further examined in N. brachylepadoformis (19 individuals) with aforementioned procedures and conditions. These specimens were collected at the Alice Springs field (18°12.599'N/144°42.431'E, 3640 m depth, Figure 1) in the Mariana Trough during a cruise of the R/V “Atlantis II” using the submersible “Alvin”.
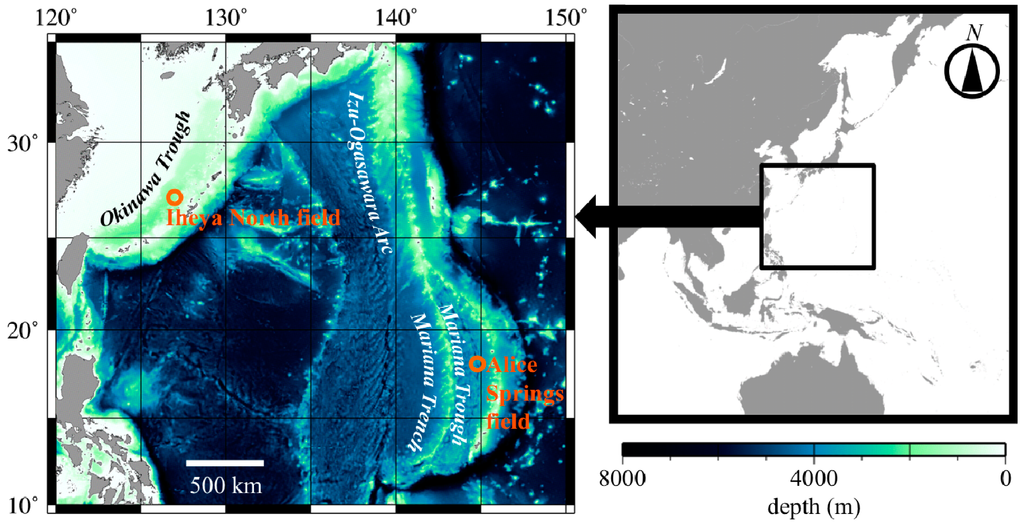
Figure 1.
Sampling sites for Neoverruca. Iheya North field (for Neoverruca sp.) is located in the Okinawa Trough and the Alice Springs field (N. brachylepadoformis) is in the Mariana Trough.
For successfully amplified microsatellite loci, the number of alleles, expected and observed heterozygosity, and deviation index from Hardy-Weinberg equilibrium (FIS) were calculated with GenAlEx version 6.5 [11]. The software Micro-Checker version 2.2.3 [12] performed 10,000 randomizations to check for null alleles at each locus at the 95% and 99% confidence levels. For all successful loci, linkage disequilibrium was estimated with Genepop version 4.2 [13,14,15] under the following Markov chain parameters: 10,000 dememorizations, 1000 batches, 10,000 iterations per batch. For evaluation of outcomes in cross-species amplification, the number of successful microsatellite loci was counted in N. brachylepadoformis. In addition, the number of alleles, expected and observed heterozygosity, and deviation index from Hardy-Weinberg equilibrium were also calculated in N. brachylepadoformis. To estimate the level of genetic differentiation between populations of Neoverruca sp. and N. brachylepadoformis, pairwise FST was calculated using GenAlEx focusing cross-species amplifiable loci. The test for statistical significance was based on 999 random permutations.
4. Conclusions
Twelve microsatellite loci developed in this study were successfully amplified and should have utility for further population genetics of Neoverruca sp. to estimate the processes of population formation and maintenance. In addition, nine of the primers can amplify microsatellites in N. brachylepadoformis. These markers are expected to help elucidate genetic diversity, differentiation, classification, and evolutionary processes in the genus Neoverruca.
Acknowledgments
We are grateful to Suguru Ohta for providing specimens of Neoverruca brachylepadoformis, and Hiroshi Miyake, the chief scientist of KY11-02 Leg. 2 cruise, captains and crews of R/V “Kaiyo” and “Natsushima”, and operation teams of the submersibles “Shinkai 2000” and ROV “Hyper-Dolphin” for collecting the specimens. We thank Steven D. Aird editing the manuscript. This work was financially supported by The Canon Foundation and Okinawa Institute of Science and Technology Graduate University.
Author Contributions
Nakajima, Watanabe, and Mitarai conceived and designed the experiments; Watanabe and Inagaki collected and treated specimens. Nakajima, Shinzato and Khalturina conducted the experiments; Satoh contributed to the experimental tools; Nakajima and Shinzato analyzed the data; Nakajima wrote the paper; Watanabe and Mitarai advised and helped for editing the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Adams, D.K.; Arellano, S.M.; Govenar, B. Larval dispersal: Vent life in the water column. Oceanography 2012, 25, 256–268. [Google Scholar] [CrossRef]
- Newman, W.A.; Hessler, R.R. A new abyssal hydrothermal verrucomorpha (Cirripedia; Sessilia): The most primitive living sessile barnacle. Trans. San Diego Soc. Nat. Hist. 1989, 21, 256–273. [Google Scholar]
- Newman, W.A. A new genus and species of barnacle (Cirripedia, Verrucomorpha) associated with vents of the Lau Back-Arc Basin: Its gross morphology, inferred first juvenile stage and affinities. Zoosystema 2000, 22, 71–84. [Google Scholar]
- Watanabe, H. Dispersal and evolution in chemoautosynthesis-based communities in the western Pacific: Verrucomorphs as test species for evolutionary studies on hydrothermal vent-endemic animals. Jpn. J. Benthol. 2003, 58, 44–49. [Google Scholar]
- Watanabe, H.; Tsuchida, S.; Fujikura, K.; Yamamoto, H.; Inagaki, F.; Kyo, M.; Kojima, S. Population history associated with hydrothermal vent activity inferred from genetic structure of neoverrucid barnacles around Japan. Mar. Ecol. Prog. Ser. 2005, 288, 233–240. [Google Scholar] [CrossRef]
- Newman, W.A. Juvenile ontogeny and metamorphosis in the most primitive living sessile barnacle, Neoverruca, from abyssal hydrothermal springs. Bull. Mar. Sci. 1989, 45, 467–477. [Google Scholar]
- Aronesty, E. Ea-Utils: “Command-Line Tools for Processing Biological Sequencing Data”. 2011. Available online: http://code.google.com/p/ea-utils (accessed on 19 November 2013).
- Castoe, T.A.; Poole, A.W.; de Koning, A.P.; Jones, K.L.; Tomback, D.F.; Oyler-Mccance, S.J.; Fike, J.A.; Lance, S.L.; Streicher, J.W.; Smith, E.N.; et al. Rapid microsatellite identification from Illumina paired-end genomic sequencing in two birds and a snake. PLoS One 2012, 7, e30953. [Google Scholar]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. Genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Micro-Checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. Genepop (version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar]
- Rousset, F. Genepop’007: A complete reimplementation of the Genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Genepop on the web. Available online: http://genepop.curtin.edu.au/ (accessed on 5 April 2014).
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).