Abstract
The traditional Mongolian medicine Erdun-Uril is a conventional combination of 29 herbs commonly used for the treatment of cerebrovascular ailments. It has the effects of reducing inflammation, counteracting oxidative stress, and averting strokes caused by persistent cerebral hypoperfusion. Prior research on Erdun-Uril has predominantly concentrated on its pharmacodynamics and mechanism of action; however, there has been a lack of systematic and comprehensive investigation into its chemical constituents. Therefore, it is crucial to establish an efficient and rapid method for evaluating the chemical constituents of Erdun-Uril. In this study, Erdun-Uril was investigated using UHPLC-Q-Exactive Orbitrap MS combined with parallel reaction monitoring for the first time. Eventually, a total of 237 compounds, including 76 flavonoids, 68 phenolic compounds, 19 alkaloids, 7 amino acids, etc., were identified based on the chromatographic retention time, bibliography data, MS/MS2 information, neutral loss fragments (NLFs), and diagnostic fragment ions (DFIs). And of these, 225 were reported for the first time in this study. This new discovery of these complex components would provide a reliable theoretical basis for the development of pharmacodynamics and quality standards of the Mongolian medicine Erdun-Uril.
1. Introduction
Erdun-Uril, also known as the Zhenbao pill or Sobud Uril, meaning “precious and miraculous pill”, originated from ancient Mongolian medicine books, and is recorded in the Mongolian medical classics Manag Mudigpo Yewa (Secret Supplement) and Mongolian Medical Jinkui [1,2]. It mostly consists of 29 herbs, including Glycyrrhiza uralensis Fisch, Aquilaria spp., Dalbergia odorifera T. Chen, Gardenia jasminoides J. Ellis, Carthamus tinctorius L., Piper longum L., Syringa oblata Lindl., and Lygodium japonicum (Thunb.) S., etc. [2,3]. Previous investigations [3,4,5,6] have shown that Erdun-Uril has many pharmacological effects, including antioxidant properties, vasodilation, and modulation of blood lipid levels, which have been widely used for the treatment of cerebrovascular ailments in clinical applications, such as cerebral hemorrhage, stroke, epilepsy, Alzheimer’s disease, and Parkinson’s disease [3,4,5,6,7]. Among them, Gardenia jasminoides Ellis, Cassiae Semen, and Terminalia chebula Retz. have all been reported to have lipid-regulating effects [8], whereas Gardenia jasminoides Ellis, along with drugs such as Syringa oblata Lindl. and Dalbergiae Odoriferae, has been shown to have antiplatelet coagulation and thrombosis-reducing effects [9], and Radix Aucklandiae and Inula helenium L. have been reported to have anti-atherosclerotic effects. Both Carthamus tinctorius L. and Glycyrrhiza uralensis Fisch. can synergistically improve energy metabolism as well as anti-inflammatory and analgesic effects in rats with cold coagulation and blood stasis [10]. At the same time, the currently available drug safety evaluation shows that the relevant biochemical indexes are still within the reasonable range when ten times the clinical dose of Erdun-Uril is given to rats, which indicates that the drug has a favorable safety profile [11]. However, compared with the research of pharmacological effects and clinical applications, there are few reports on the chemical composition of Erdun-Uril. Therefore, it is worth establishing a rapid and comprehensive analytical strategy to characterize its chemical composition.
UHPLC-Q-Exactive Orbitrap MS technology has high-throughput scanning and multiple detection capabilities and has been widely used to analyze complex systems [12,13,14]. The complete MS/dd MS2 mode typically acquires sample data by first detecting the primary precursor ion, then performing secondary mass spectrometry triggered by the top three higher intensities. However, the constraints of the analytical process make it challenging to extract and analyze minor components [15,16,17]. Recent research shows that the parallel reaction monitoring (PRM) mode enhances sensitivity compared to the full MS scan mode. [18,19,20]. Therefore, the aim of our study was to develop an analytical method based on UHPLC Q-Exactive Orbitrap MS technology combined with parallel reaction monitoring, enabling diagnostic fragment ions (DFIs) and neutral loss fragments (NLFs) to identify the major chemicals in Erdun-Uril.
During this procedure, a total of 237 components were detected using UHPLC-Q-Exactive Orbitrap mass spectrometry (Thermo Fisher Scientific Co., Ltd., Waltham, MA, USA), of which 225 were first reported in Erdun-Uril. These findings might assist us in conducting quality control of Erdun-Uril in future clinical trials and provide foundations for crucial research on the pharmacodynamic components of Erdun-Uril.
2. Results and Discussion
2.1. Analytical Strategy
The aim of this study was to methodically identify the chemical constituents of Erdun-Uril. Consequently, a highly effective approach utilizing UHPLC-Q-Exactive Orbitrap MS combined with parallel reaction monitoring (PRM) was developed. Initially, the chemicals in Erdun-Uril were extracted and enriched by applying ultrasonic extraction with 70% methanol. Furthermore, the sample was injected to the UHPLC-Q-Exactive Orbitrap mass spectrometer in order to acquire high-resolution MS data for Erdun-Uril. Additionally, the UHPLC-Q-Exactive Orbitrap MS paired with PRM scanning was used to gather the MS2 data for the trace components in Erdun-Uril. Finally, the potential chemicals were determined by comparing them with standards, summary DFIs, and neutral loss, as well as by referencing the literature.
2.2. Establishment of Diagnostic Fragment Ions (DFIs) and Neutral Loss Fragments (NLFs)
DFIs and NLFs are both well-acknowledged screening methods that are used to identify the complex traditional Chinese medicine system composition based on the accurate ion mass and specific fragment information provided by the high-resolution mass spectrum and standard reference [21,22,23]. In contrast to the traditional way of identifying compounds derived from the literature, this approach can deduce the specific structure of a compound based on mass spectrometry information, which is suitable for identifying the multiple compounds contained in Erdun-Uril. As an example, flavonoids possess 2-phenylchromanone as their parent structure, which is also subdivided into flavonols, flavonoid glycosides, and isoflavonoids based on the different substituent groups on the parent nucleus. Therefore, the cleavage fragments of 2-phenylchromanone can be used as the DFIs of flavonoids, and the fragment ions with different substituents can be used as the basis for the identification of compounds. Some fragment ions are detected by the NLF method in the form of a fixed mass difference between the fragments; NLFs are also valuable for the identification of compounds.
In this study, the fragment ion patterns of flavonols, dihydroflavonoids, and phenolic acids were investigated using LC-MS/MS. The fragmentation pathway of flavonols and dihydroflavonoids is shown in Figure 1A–C. The same fragmentations were identified as 178.9974 (C8H3O5), 151.0025 (C7H3O4), 121.0281 (C7H5O2), and 107.0124 (C6H3O2), which could be regarded as the DFIs of flavonols. In addition, the fragments 135.0075 (C7H4O3) and 119.0489 (C8H7O) were displayed in the fragmentation pathway as the DFIs of dihydroflavonoids. Likewise, phenolic acids contain phenolic hydroxyl and carboxyl groups and easily lose H2O, CO2, and CO neutral groups during cracking. This was confirmed by the fragment ions obtained from the secondary mass spectra of several phenolic acid standard controls, all of which showed neutral loss and generated [M − H2O], [M − CO], and [M − CO2] fragments during cleavage.
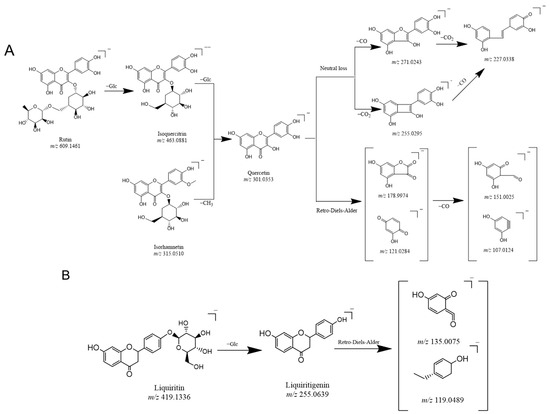
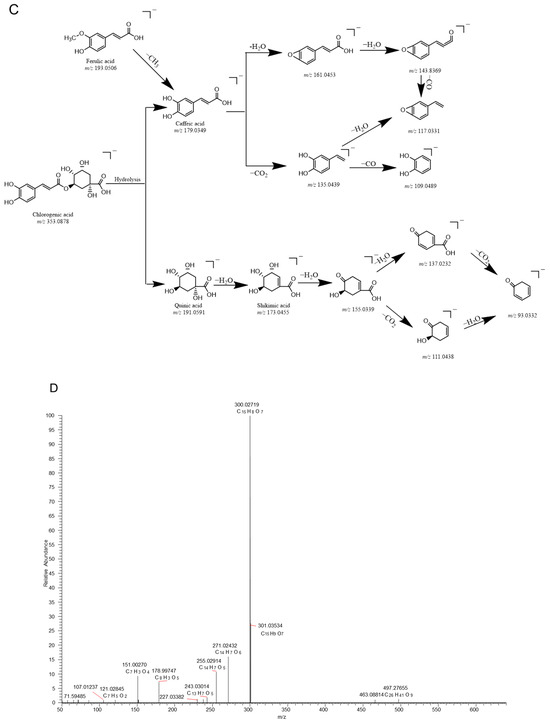
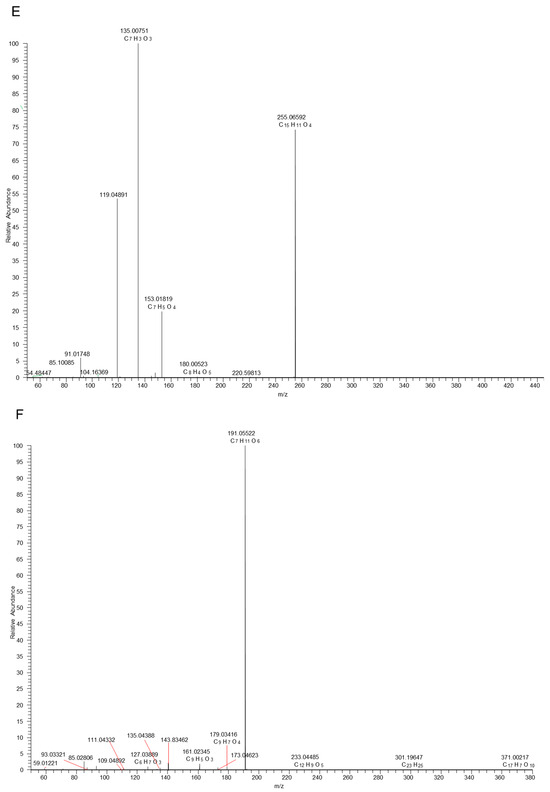
Figure 1.
Proposed selected fragmentation pattern of components identified in Erdun-Uril flavonols (A), dihydroflavonoids (B), phenolic acids (C), and mass spectra of standards: Rutin (D), liquiritin (E), and chlorogenic acid (F).
2.3. Characterization of the Chemical Constitution in Erdun-Uril
The table lists all the compounds detected in Erdun-Uril by UHPLC-Q-Exactive Orbitrap MS (Table 1 and Table S1). As the result, a total of 237 compounds (225 first reported), including 76 flavonoids, 68 phenolic compounds, 19 alkaloids, 7 amino acids, etc., were identified based on the diagnostic fragment ions, retention time, and MS2 database (mzVault, mzCloud, and literature). Figure 2 and Figure 3 show high-resolution ion chromatograms extracted from the Erdun-Uril extract in positive and negative ion modes, respectively.

Table 1.
Identification results of the chemical composition of Erdun-Uril.
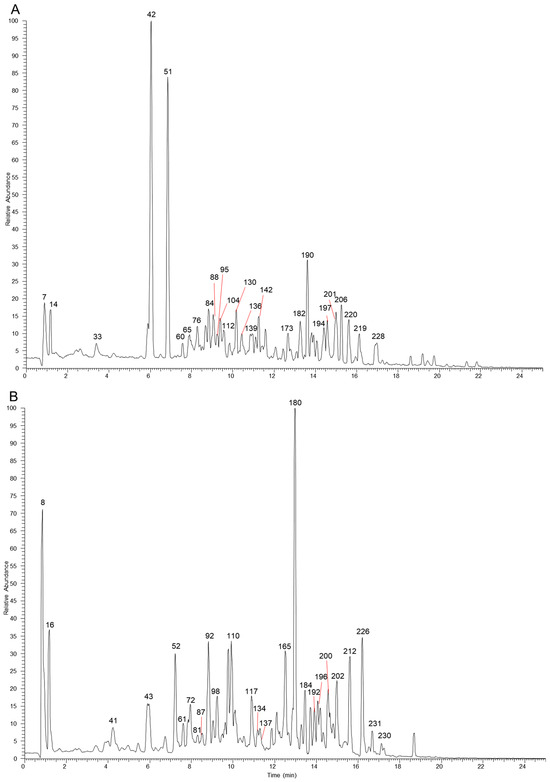
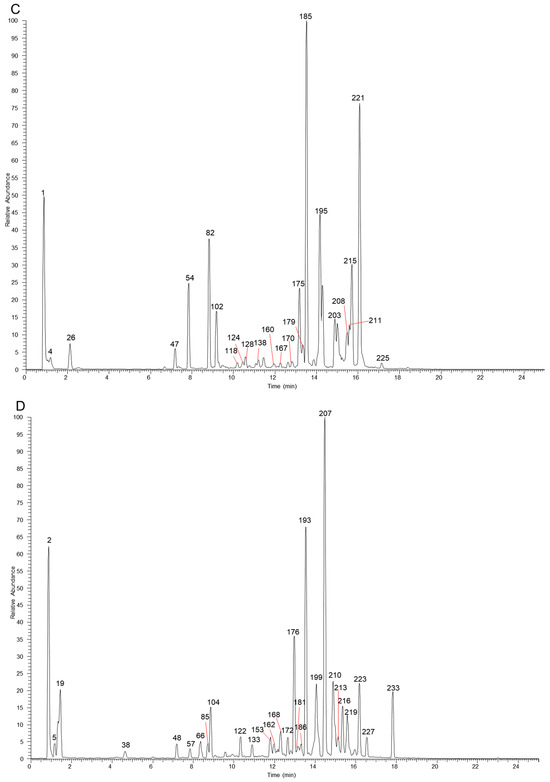
Figure 2.
The high-resolution extracted ion chromatograms (EIC) in 10 ppm for multiple compounds in Erdun-Uril in positive mode. (A) m/z 124.0393, 165.0546, 127.0389, 355.1023, 291.0863, 249.1485, 565.1551, 193.0495, 573.1578, 595.1657, 371.1125, 493.0976, 301.0706, 303.0499, 507.1133, 271.06, 287.055, 355.1176, 345.1696, 455.3519, 367.1176, 265.1223, 325.1434, 165.091, 236.2008, 279.159; (B) m/z 130.0498, 268.104, 163.0389, 347.17, 225.0757, 153.0546, 183.0651, 479.082, 419.1336, 463.0871, 317.0291, 551.1759, 269.0444, 537.233, 855.4008, 179.0338, 135.0804, 353.1383, 355.154, 163.0753, 372.1805, 312.1594, 314.175, 293.1747, 300.2897, 325.141; (C) m/z 118.0862, 152.0566, 136.0617, 166.0862, 177.0546, 167.0704, 209.0808, 319.1176, 179.0702, 431.1336, 285.0757, 219.1743, 248.1281, 369.1332, 318.1335, 274.1437, 203.1794, 311.1277, 193.0859, 251.1066, 233.1536, 231.1379, 165.091, 330.2063, 340.1907, 342.2063, 179.1066, 252.2321; (D) m/z 116.0706, 182.0811, 132.1019, 114.0913, 551.1963, 118.0651, 169.1223, 257.0808, 147.044, 337.0837, 258.1488, 823.411, 272.1281, 267.1015, 391.2842, 327.159, 288.1594, 397.1586, 281.1172, 195.1015, 328.1907, 224.2008, 233.1536, 221.1899, 209.1172, 344.222, 384.2533.
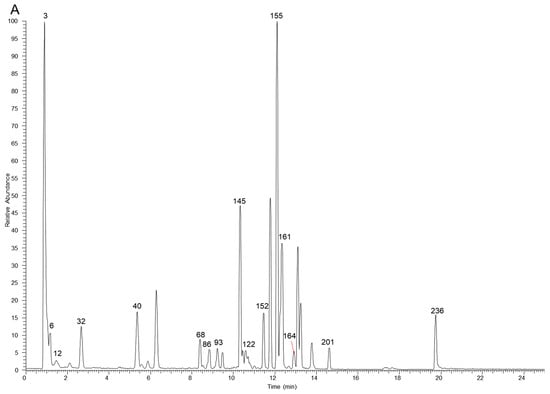
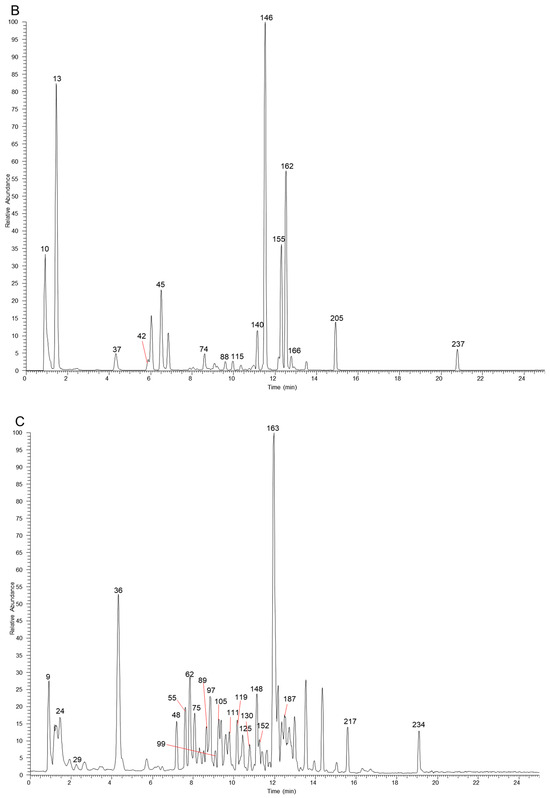
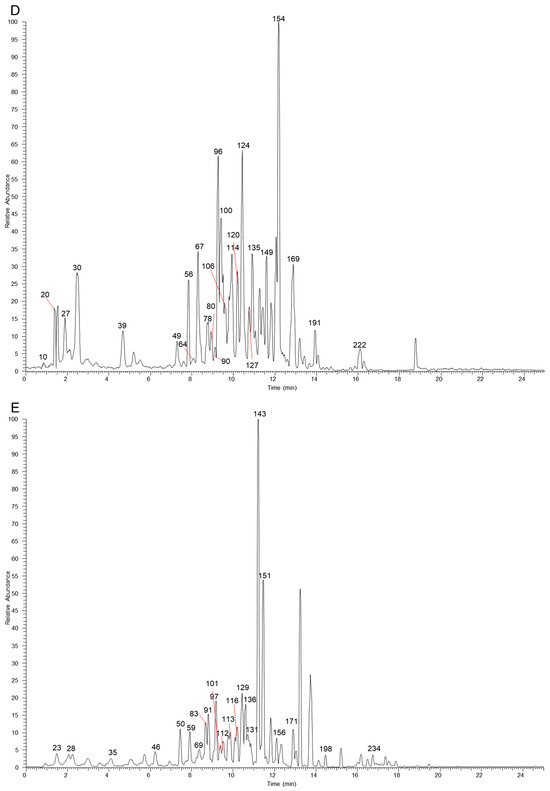
Figure 3.
The high-resolution extracted ion chromatograms (EIC) in 10 ppm for multiple compounds in Erdun-Uril in negative mode. (A) m/z 173.0455, 355.0306, 153.0193, 183.0298, 197.0455, 515.1194, 283.0611, 267.0662, 503.3378, 329.103, 279.2329, 191.0561, 255.0662, 323.1288, 179.0349, 463.0881; (B) m/z 133.0142, 499.1668, 169.0142, 137.0244, 353.0878, 611.1617, 300.9989, 593.1511, 559.1457, 271.0611, 514.2843, 329.0666, 821.3965, 299.0924, 357.1707, 355.155; (C) m/z 331.067, 373.114, 167.0349, 403.1245, 549.1824, 387.1296, 163.04, 183.1026, 449.1089, 447.0932, 269.0455, 137.0244, 419.0983, 285.0404, 477.1038, 313.0717, 269.0819, 329.2333, 239.0713, 343.0823, 325.1445, 227.2016, 633.0733, 463.0881; (D) m/z 125.0244, 369.0463, 391.1245, 405.1402, 785.0842, 225.0768, 447.09328, 287.0561, 521.1664, 431.0983, 447.0932, 565.1926, 491.0831, 315.0146, 491.1194, 253.0506, 301.0353, 263.1288, 225.1132, 329.0666, 299.0561, 651.2658, 337.1445, 277.2173; (E) m/z 483.078, 315.0721, 285.0615, 291.0146, 595.1668, 635.0889, 151.04, 317.103, 623.1617, 599.0678, 565.1562, 167.0349, 301.0717, 287.0561, 285.0768, 345.0979, 573.2341, 271.0975, 283.0611, 335.0924, 469.3323, 315.051, 609.1461, 193.0506.
2.3.1. Identification of Flavonoids of Erdun-Uril
Flavonoids are a large class of secondary metabolites widely distributed in plants [24]. It is well known that the common structural features of most flavonoids are based on diphenylpropane A (C6-C3-C6) as the basic skeleton, where the two aromatic rings A and B are linked by a three-carbon bond [25]. Based on a comparison with fragment ion details in the literature and reference standards, we detected a total of 76 flavonoids in this study (compounds 45, 50, 64–65, 67, 77, 79–81, 83–92, 94–95, 99–100, 104, 107, 109, 111, 113, 116, 118–119, 121–126, 128–131, 139–140, 142–144, 146–147, 149–150, 153–155, 162, 165–166, 172–173, 176–177, 179, 183, 187, 190, 193, 195, 197–198, 200–201). According to the connection position of the B ring of flavonoids (C-2 or C-3 position), the degree of oxidation of the central 3-carbon chain and whether the 3-carbon chain is linked, etc., the above flavonoid compounds are mainly divided into five categories: flavonoid glycoside, flavonols, dihydroflavonoids, isoflavones, and chalcones.
Compounds 64, 77, 79, 80, 86, 87, 84, 121, 124, 131, 139, 140, 150, and 201 were identified as orientin, rutin, isoquercitrin, vitexin, hyperoside, liquiritin, isoliquiritin, kaempferol, quercetin, isorhamnetin, genistein, naringenin, liquiritigenin, and glabridin, respectively, by comparing the retention times and MS2 data with the reference standards.
Compounds 85 and 174 possessed the same quasi-molecular ion and characteristic fragment ions as isoliquiritigenin. Thus, they were characterized as being isoliquiritigenin isomers. Likewise, compounds 126, 90, 109, 144, and 99 were deemed to be the isomers of quercetin, orientin, liquiritin, isorhamnetin, and genistein, respectively.
Compounds 111 and 147 were found at 9.82 and 11.59 min and yielded the same quasi-molecular ion [M + H]+ at m/z 301.0706 (−1.36 ppm, C16H12O6); their main fragment ions [M + H]+ were at m/z 257.0436, attributed to the neutral loss of the CO2 (44 Da), and at m/z 213.0537, attributed to the neutral loss of two CO2 (88 Da). Combined with the literature report [12,26], they were tentatively characterized as tectorigenin and its isomer. Similarly, compounds 173 and 188, 167 and 183, 184 and 196, and 84 and 113 were characterized as glycyrrhiza isoflavanone [27], glycycoumarin [28], gancaonin I, isoliquiritin apioside [28], and their isomers.
Compound 100 yielded deprotonated molecular ions [M − H]− at m/z 491.0831 (−0.46 ppm, C22H20O13), and the main fragment ions [M − H]− were at m/z 345.0809, attributed to the neutral loss of the rhamnose moiety (146 Da), and at m/z 300.0272 and 255.0295, were attributed to the loss of one and two COOH groups, respectively. Compound 100 had a better correspondence in both positive and negative ion modes; based on its retention time, fragmentation ions, and combined with the previous studies [29], it was confirmed as isorhamnetin 3-glucuronide. Additionally, compounds 119, 122, 124, 150, and 200 all had better correspondence in both positive and negative ion modes. According to a previous study [30,31,32], reference standards, and DFIs, they were tentatively characterized as luteolin, liquiritigenin, quercetin, isoliquiritigenin, and glabridin. Compounds 83 and 95 produced deprotonated molecular ions [M + H]+ at m/z 479.0820 (−1.43 ppm, C21H18O13) and 462.0792 (−0.94 ppm, C21H17O12). Their main fragment ions [M + H]+ were at m/z 272.0727 and 255.0797, attributed to the loss of the glucuronide, and m/z 113.0233 (C5H5O3) was their common fragment. Then, they were tentatively characterized as quercetin 3-O-beta-d-glucuronide and luteolin-7-O-beta-d-glucuronide.
Compounds 83, 87, and 142 yielded quasi-molecular ions [M − H]− at m/z 317.1030 (−1.27 ppm, C17H18O6), 449.1089 (−0.66 ppm, C21H22O11), and 345.0979 (−0.74 ppm, C18H18O7), which initially generated 151.0310 (C7H6O2) and 178.9976 (C8H4O5) by retro Diels–Alder (RDA) rearrangement. According to previous reports [33,34,35] and DFIs, they were tentatively characterized as 3′-hydroxy-8-methoxyvestitol, carthamidin-5-glucoside, and 5,7-dihydroxy-2′,3′,4′-trimethoxyisoflavanone.
Compounds 116, 120, 129, 155, 162, 165, and 197 yielded deprotonated molecular ions [M − H]− at m/z 301.0717 (−0.57 ppm, C16H14O6), 253.0506 (−1.63 ppm, C15H10O4), 285.0768 (−0.66 ppm, C16H14O5), 271.0975 (−1.63 ppm, C16H16O4), 239.0713 (−2.29 ppm, C15H12O3), 299.0924 (−1.13 ppm, C17H16O5), and 335.0924 (−0.08 ppm, C20H16O5), which initially generated 135.0079 (C7H3O3) by retro Diels–Alder (RDA) rearrangement. Thus, they were assigned as homoeriodictyol [36], soyside, sakuranetin, vestitol, 2′,4′-dihydroxychalcone, methylnissolin, and glabrone by searching in the databases such as the chemical structure database (ChemSpider) and MS2 database (mzVault and mzCloud) and DFIs.
2.3.2. Identification of Phenolic Acids of Erdun-Uril
A total of 68 individual phenolic acids constituents were putatively identified in Erdun-Uril using UHPLC-Q-Exactive Orbitrap MS. Seven compounds, such as shikimic acid (6), chlorogenic acid (42), caffeic acid (44), gallic acid (21), ellagic acid (74), quinic acid (3), and ferulic acid (112) were identified as having a pseudomolecular ion [M − H]− at m/z 173.0455 (−1.43 ppm, C7H10O5), 353.0878 (−0.19 ppm, C16H18O9), 179.0349 (−5.49 ppm, C9H8O4), 169.0142 (1.84 ppm, C7H6O5), 300.9989 (−1.30 ppm, C14H6O8), 191.0561 (−4.51 ppm, C7H12O6), and 193.0506 (−4.47 ppm, C10H10O4), respectively. These were unambiguously identified by comparing their accurate mass information and chromatography retention times with reference standards in negative ionization mode. Compounds 62, 70, and 71 were detected at 8.02, 8.44, and 8.50 min, respectively, and they possessed the same quasi-molecular ion [M − H]− at m/z 151.0400 and the main characteristic fragment ions [M − H]− at m/z 133.0153 (C8H6O2) and 107.0122 (C7H8O). In addition, compound 61 was found at 7.95 min, shared the same formula and similar fragment as compound 62, etc., and was observed at positive scan mode. Hence, they were identified as being 4-Hydroxyphenylacetic acid isomers. Compounds 233 and 236 were eluted at 15.26 and 15.36 min with the same deprotonated ions [M + H]+ at m/z 165.0910, which initially generated 137.0593 (C9H12O) and 119.0356 (C9H10) by neutral loss CO and H2O, respectively. Thus, they were identified as eugenol isomers. Peaks 98 and 106 were detected at 9.38 and 9.61 min, possessed the same quasi-molecular ions [M − H]− at m/z 315.0146, and the typical daughter ions at m/z 300.9984, obtained by the loss of the CH3 group, which further gave rise to the product ions at m/z 271.0463 and 227.0486 by sequential loss of a H2O group. In addition, m/z 300.9984 could be considered as a fragment of ellagic acid. Thus, compounds 98 and 106 were identified as derivatives of ellagic acid, and they were tentatively characterized as 3-O-methylellagic acid isomers. Likewise, compound 68 produced the main fragment at m/z 169.0131 and exhibited a similar cleavage pathway as gallic acid. Thus, it can be preliminary characterized as syringic acid. Compounds 12, 37, 40, and 46 were eluted at 1.15, 4.32, 5.36, and 6.91 min with deprotonated ions [M − H]− at m/z 355.0306 (C14H12O11), 137.0244 (C7H6O3), 183.0298 (C8H8O5), and 291.0146 (C13H8O8). According to the previous report [37,38] and DFIs, they were tentatively characterized as chebulagic acid, p-hydroxybenzoic acid, methyl gallate, and brevifolincarboxylic acid, respectively.
2.3.3. Identification of Amino Acids of Erdun-Uril
Compounds 1, 2, 5, 8, 19, and 26 were observed at 0.85, 0.87, 0.91, 0.93, 1.34, and 2.12 min, respectively, and produced deprotonated ions [M + H]+ at m/z 118.0862 (C5H11NO2), 116.0706 (C5H9NO2), 182.0811 (C9H11NO3), 130.0498 (C5H7NO3), 132.1019 (C6H13NO2), and 166.0862 (C9H11NO2). Comparing the MS2 fragment ions with data from the bibliography [39,40,41,42], compounds 1, 2, 5, 8, 19, and 26 were tentatively identified as valine, proline, tyrosine, L-Pyroglutamic acid, isoleucine, and phenylalanine, respectively.
3. Materials and Methods
3.1. Chemicals and Reagents
Thermo Fisher Scientific Co., Ltd. (Fair Lawn, NJ, USA) manufactured the Dionex UltiMate 3000 Ultra Performance Liquid Chromatograph and the Q-Exactive-Orbitrap-MS. Guangzhou Watsons Food & Beverage Co., Ltd. (Guangzhou, China) provided the purified water. Kunshan Ultrasonic Instrument Co., Ltd. (Kunshan, China) manufactured the KQ-300DE sonication instrument. Aladdin Industrial Corporation (Shanghai, China) provided the other analytical grade solvents used in the experiment. Chengdu Plant Standard Pure Biotechnology Co., Ltd. (Chengdu, China) provided the chemical reference standards for chlorogenic acid, orientin, isoquercitrin, vitexin, kaempferol, isorhamnetin, caffeic acid, glabridin, and naringenin. Cheng Du Herbpurify Co., Ltd. (Chengdu, China) provided the reference standards for gallic acid and corilagin. Chengdu Aifa Biotechnology Co., Ltd., located in Chengdu, China, supplied ellagic acid, rutin, isoliquiritin, quercetin, genistein, and liquiritigenin. We acquired quinic acid, hyperoside, liquiritin, and ferulic acid from various sources, including Shanghai Yuanye Bio-Technology Co., Ltd. (Shanghai, China), the National Institutes for Food and Drug Control, Sigma Aldrich (Shanghai) Trading Co., Ltd. (Shanghai, China), and Shandong West Asia Chemical Technology Co., Ltd. (Linyi, China), respectively. All reference standards had purities exceeding 98% as determined by HPLC-UV analysis.
3.2. Preparation of Specimen and Standard Solution
Erdun-Uril (211102) was acquired from Inner Mongolia Kulun Mongolian Pharmaceutical Co., Ltd. (Tongliao, China)
Prior to sample preparation, we powdered the Erdun-Uril substance and then passed it through a No. 40 mesh sieve. We treated the Erdun-Uril powder (2 g) with sonication (300 W, 40 kHz) in 10 mL of 70% (v/v) methanol for 1 h at room temperature (16–24 °C). The final product was then filtered and dried using rotary evaporation and nitrogen blowing. We then dissolved the solution again using methanol and centrifuged it at a speed of 12,000 rpm for 15 min to obtain the supernatant. For analysis using UHPLC-Q-Exactive Orbitrap MS, a total of 2 μL of material was prepared.
Standard solutions were prepared in methanol at a concentration of 1 mg/mL. These reference standard solutions were further diluted to obtain working solutions and then were stored at 4℃ before analysis.
3.3. Instruments and Conditions
In order to obtain a better chromatographic peak shape and separation resolution, various factors were set in the detection and identification process, including a column (Agilent Eclipse Plus C8 2.1 × 100 mm, 1.8 μm), column temperature (40 °C), and the mobile phase gradient.
Each LC-MS analysis was exercised on a Q-Exactive Focus Orbitrap MS connected to a Thermo Scientific Dionex Ultimate 3000 RS (Thermo Fisher Scientific Co., Ltd., Waltham, MA, USA) through an ESI source. Chromatographic separation was performed at 40 °C using an Agilent Eclipse Plus C8 (2.1 × 100 mm, 1.8 μm). The mobile phase was composed of 0.1% formic acid (A) and acetonitrile (B), and the flow rate was 0.3 mL/min. The following gradient was used: 0–2 min, 95–92%A; 2–5 min, 92–90%A; 5–12 min, 90–45%A; 12–18 min, 45–15%A; 18–23 min, 15–10%A; 23–25 min, 10–5% A; 25–30 min, 5–95%A.
All data were obtained in alternating positive and negative ion scanning mode using the following tuning method. In terms of the mass spectrometry conditions, the spray voltage was 3.5 kV in positive ion mode and 3.0 kV in negative ion mode. The spray voltage was 3.0 kV; the sheath gas and auxiliary gas pressure were 30 arb and 10 arb; the capillary temperature was 320 °C; the S-lens RF level was 50. In the mass range of m/z 100–1500, a high-resolution mass spectrum was obtained at a resolution of 70,000, which was detected by the Orbitrap analyzer. MS2 data at a resolution of 17,500 were obtained by data-dependent MS2 scanning or parallel reaction monitoring (PRM) mode. Nitrogen (purity ≥ 99.999%) served as the collision gas, which generated the fragment ions, and the energy level was set as a normalized collision energy of 30%.
3.4. Data Processing and Analysis
Xcalibur software version 4.2 (Thermo Fisher Scientific, San Jose, CA, USA) was used to obtain all high-resolution data, including the full-scan MS and MS2 data. Peaks detected with intensities over 10,000 were selected for identification. The chemical formulas for all parent and fragment ions of the selected peaks were calculated from the accurate mass using a formula predictor by setting the parameters as follows: C [0–90], H [0–90], O [0–90], and N [0–10]. The mass tolerance of MS and MS2 was within 10 ppm.
4. Conclusions
In this study, an efficient strategy based on UHPLC-Q-Exactive Orbitrap MS was established to detect chemical components in Erdun-Uril. A total of 237 constituents were identified, of which 225 were first reported in Erdun-Uril, including 76 flavonoids, 68 phenolic acids, 19 alkaloids, 7 amino acids, etc. These were detected and identified based on their chromatographic retention, MS and MS2, and bibliography data. These results are very useful for understanding the bioactive compounds of Erdun-Uril and their utilization. Overall, the results lay the foundation for in-depth research on the pharmacodynamic material basis of Erdun-Uril.
Supplementary Materials
The following Supporting Information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules29184349/s1; Table S1: Retention times and mass spectral data of Erdun-Uril.
Author Contributions
Y.H. and K.L.: investigation writing and draft preparation; S.Y. and B.Y.: performed experiments and analyzed the data; Z.C. and L.F.: data processing and curation; L.S. and B.G.: conceptualization; H.L. and W.C.: writing—review and editing, conceptualization, supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of Inner Mongolia Autonomous Region (2024LHMS08017) and the Flower Bud Program Project of the Baotou Medical College (HLJH202401).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data will be provided upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gaowa, S.; Bao, N.; Da, M.; Qiburi, Q.; Ganbold, T.; Chen, L.; Altangerel, A.; Temuqile, T.; Baigude, H. Traditional Mongolian Medicine Eerdun Wurile Improves Stroke Recovery through Regulation of Gene Expression in Rat Brain. J. Ethnopharmacol. 2018, 222, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Committee National Pharmacopoeia. Drug Standard of Ministry of Public Health of the Peoples Republic of China; Mongolian Medicines Fascicle; People’s Medical Publishing House: Beijing, China, 1998. [Google Scholar]
- Zhao, G.; Ren, Y.; Han, C.; Kong, L.; Jia, Y. Mechanism of Mongolian Medicine Erden-Uril on Osteoarthritis in Rats. Chin. J. Tissue Eng. Res. 2024, 28, 1193–1199. [Google Scholar]
- Ai, L. Clinical Efficacy of Mongolian Medicine Erdun Wurile and Shusha-7 in the Treatment of Depression. Master’s Thesis, Inner Mongolia Medical University, Hohhot, China, 2023. [Google Scholar]
- Si, Q.; Renmandoula, N. Research Progress of Mongolian Medicine Eerdun Wurile. J. Med. Pharm. Chin. Minor. 2024, 30, 41–45. [Google Scholar]
- Li, X.; Lin, Y.; Zhang, Y.; Chen, X.; Zhao, R.; Zhu, W.; Xie, Y.; Xie, W.; Bade, R.; Jiang, S.; et al. Study on the Mechanism of Mongolian Medicine Erdun- Wurile on Parkinson’s Disease Based on Network Pharmacology and Molecular Docking. J. Baotou Med. Coll. 2024, 40, 1–7. [Google Scholar]
- Bao, L.; Song, F.; Du, G.; Bao, S.; Saiyin, B.; Xilin, Q.; Zhang, W.; Tian, J.; Wuhan, Q. The Progress of Traditional Mongolian Medicine Erden-Uril. J. Chin. J. Ethnomed. Ethnopharm. 2019, 28, 53–56. [Google Scholar]
- Chen, G. Progress of Research on the Drug Composition and Pharmacological Effects of Gardenia Jasminoides. Spec. Econ. Anim. Plant 2022, 25, 20–22+32. [Google Scholar]
- Liu, Q.; Zhao, S.; Wang, X.; Hu, Z.; Chen, J. Overview of the Research on the Characteristics of the Eldon-Uriel Formula of Mongolian Medicine and the Treatment of Atherosclerosis. Her. Med. 2024, 43, 561–567. [Google Scholar]
- Dong, J.; Mao, J.; Long, L.; Fan, T.; Jia, P.; Wang, S.; Zheng, X. Effects of Carthamus tinctorius L.-Glycyrrhizaradixonadenoine Acids and Energy Charge Levels in Plasma and Brain of Rats with Cold Coagulation and Blood Stasis. Chin. J. Hosp. Pharm. 2019, 39, 2507–2511. [Google Scholar]
- Wang, Q.; Gao, W.; Song, F. Preliminary Study on the Acute Toxicity of Mongolian Medicine Eerdun—Wurile in Rats. China Pharm. 2023, 32, 77–81. [Google Scholar]
- Chandradevan, M.; Simoh, S.; Mediani, A.; Ismail, N.H.; Ismail, I.S.; Abas, F. Uhplc-Esi-Orbitrap-Ms Analysis of Biologically Active Extracts from Gynura procumbens (Lour.) Merr. And Cleome gynandra L. Leaves. Evid.-Based Complement. Altern. Med. 2020, 2020, 3238561. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Zheng, B.; Guan, Y.; Wang, L.; Chen, L.; Cai, W. Rapid Characterization of Chlorogenic Acids in Duhaldea Nervosa Based on Ultra-High-Performance Liquid Chromatography–Linear Trap Quadropole-Orbitrap-Mass Spectrometry and Mass Spectral Trees Similarity Filter Technique. J. Sep. Sci. 2018, 41, 1764–1774. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Wang, Y.; Zhao, J.; Wang, M.; Avula, B.; Peng, Q.; Ouyang, H.; Lingyun, Z.; Zhang, J.; Khan, I.A. Identification and Characterization of Key Chemical Constituents in Processed Gastrodia Elata Using Uhplc-Ms/Ms and Chemometric Methods. J. Anal. Methods Chem. 2019, 2019, 4396201. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Wei, J.; Tian, X.Y.; Wang, B.; Chan, W.; Li, S.; Tang, Z.; Zhang, H.; Cheang, W.S.; Zhao, Q.; et al. Comprehensive Analysis of Acylcarnitine Species in Db/Db Mouse Using a Novel Method of High-Resolution Parallel Reaction Monitoring Reveals Widespread Metabolic Dysfunction Induced by Diabetes. Anal. Chem. 2017, 89, 10368–10375. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.C.; Qin, S.H.; Li, K.L.; Liu, Y.N.; Wu, J.L.; Yan, F.; Cai, W. A Systematic Method for the Identification of Aporphine Alkaloid Constituents in Sabia Schumanniana Diels Using Uhplc-Q-Exactive Orbitrap/Mass Spectrometry. Molecules 2022, 27, 7643. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Cong, Z.; Wang, C.; Wang, S.; Yan, Z.; Wang, B.; Liu, X.; Li, Z.; Gao, P.; Kang, H. Comprehensive Metabolism Study of Tangeretin in Rat Plasma, Urine and Faeces Using Ultra-High Performance Liquid Chromatography-Q Exactive Hybrid Quadrupole-Orbitrap High-Resolution Accurate Mass Spectrometry. Curr. Drug Metab. 2022, 23, 973–990. [Google Scholar] [CrossRef]
- Zhang, J.; Cai, W.; Zhou, Y.; Liu, Y.; Wu, X.; Li, Y.; Lu, J.; Qiao, Y. Profiling and Identification of the Metabolites of Baicalin and Study on Their Tissue Distribution in Rats by Ultra-High-Performance Liquid Chromatography with Linear Ion Trap-Orbitrap Mass Spectrometer. J. Chromatogr. B Analyt. Technol. Biomed. Life 2015, 985, 91–102. [Google Scholar] [CrossRef]
- Li, Y.; Cai, W.; Cai, Q.; Che, Y.; Zhao, B.; Zhang, J. Comprehensive Characterization of the in Vitro and in Vivo Metabolites of Geniposide in Rats Using Ultra-High-Performance Liquid Chromatography Coupled with Linear Ion Trap–Orbitrap Mass Spectrometer. Xenobiotica 2015, 46, 357–368. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Wang, Z.J.; Zhang, Q.; Wang, F.; Ma, Q.; Lin, Z.Z.; Lu, J.Q.; Qiao, Y.J. Rapid Screening and Identification of Target Constituents Using Full Scan-Parent Ions List-Dynamic Exclusion Acquisition Coupled to Diagnostic Product Ions Analysis on a Hybrid Ltq-Orbitrap Mass Spectrometer. Talanta 2014, 124, 111–122. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, Y.; Chen, S.; Fu, Y. Characterization and Identification of the Chemical Constituents in the Root of Lindera reflexa Hemsl. Using Ultra-High Performance Liquid Chromatography Coupled with Linear Trap Quadrupole Orbitrap Mass Spectrometry. J. Pharm. Biomed. Anal. 2016, 126, 34–47. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, D.; Li, H.H.; Wang, H.; Tan, H.S.; Xu, H.X. Diagnostic Filtering to Screen Polycyclic Polyprenylated Acylphloroglucinols from Garcinia Oblongifolia by Ultrahigh Performance Liquid Chromatography Coupled with Ion Mobility Quadrupole Time-of-Flight Mass Spectrometry. Anal. Chim. Acta 2016, 912, 85–96. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Zhang, Q.; Li, N.; Wang, Z.J.; Lu, J.Q.; Qiao, Y.J. Diagnostic Fragment-Ion-Based and Extension Strategy Coupled to Dfis Intensity Analysis for Identification of Chlorogenic Acids Isomers in Flos Lonicerae Japonicae by Hplc-Esi-Msn. Talanta 2013, 104, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.-Q.; Wang, C.-X.; Kuang, X.-J.; Li, Y.; Sun, C. Advance in Flavonoids Biosynthetic Pathway and Synthetic Biology. China J. Chin. Mater. Medica 2016, 41, 4124. [Google Scholar]
- Li, H.; Lyv, Y.; Zhou, S.; Yu, S.; Zhou, J. Microbial Cell Factories for the Production of Flavonoids–Barriers and Opportunities. Bioresour. Technol. 2022, 360, 127538. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Yang, D.; Wu, L.; Wusa, L.; Song, L. Study on the Mechanism of Sugemule-4 Decoction in Treating Insomnia by Lc-Ms and Network Pharmacology. Mod. Tradit. Chin. Med. Mater. Medica World Sci. Technol. 2023, 25, 3866–3889. [Google Scholar]
- Fang, S.; Qu, Q.; Zheng, Y.; Zhong, H.; Shan, C.; Wang, F.; Li, C.; Peng, G. Structural Characterization and Identification of Flavonoid Aglycones in Three Glycyrrhiza Species by Liquid Chromatography with Photodiode Array Detection and Quadrupole Time-of-Flight Mass Spectrometry. J. Sep. Sci. 2016, 39, 2068–2078. [Google Scholar] [CrossRef]
- Qiao, X.; Liu, C.-F.; Ji, S.; Lin, X.-H.; Guo, D.-A.; Ye, M. Simultaneous Determination of Five Minor Coumarins and Flavonoids in Glycyrrhiza Uralensis by Solid-Phase Extraction and High-Performance Liquid Chromatography/Electrospray Ionization Tandem Mass Spectrometry. Planta Medica 2014, 80, 237–242. [Google Scholar] [CrossRef]
- Song, L.; Wang, X.; Zheng, X.; Huang, D. Polyphenolic Antioxidant Profiles of Yellow Camellia. Food Chem. 2011, 129, 351–357. [Google Scholar] [CrossRef]
- Yang, R.; Guan, Y.; Wang, W.; Chen, H.; He, Z.; Jia, A.Q. Antioxidant Capacity of Phenolics in Camellia Nitidissima Chi Flowers and Their Identification by Hplc Triple Tof Ms/Ms. PLoS ONE 2018, 13, e0195508. [Google Scholar] [CrossRef]
- Yang, R.; Guan, Y.; Zhou, J.; Sun, B.; Wang, Z.; Chen, H.; He, Z.; Jia, A. Phytochemicals from Camellia Nitidissima Chi Flowers Reduce the Pyocyanin Production and Motility of Pseudomonas Aeruginosa Pao1. Front. Microbiol. 2018, 8, 2640. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, J.; Su, S.; Huang, L. Hepatoprotective Effects of Camellia Nitidissima Aqueous Ethanol Extract against Ccl4-Induced Acute Liver Injury in Sd Rats Related to Nrf2 and Nf-Κb Signalling. Pharm. Biol. 2020, 58, 239–246. [Google Scholar] [CrossRef]
- Yang, F.Y.; Xu, R.L.; Niu, W.; Huo, J.G.; Ju, J.M. Uplc-Q-Tof-Ms Analysis of Chemical Constituents of Classical Prescription Yiguanjian Standard Decoction. China J. Chin. Mater. Medica 2022, 47, 2134–2147. [Google Scholar]
- Rakkhitawatthana, V.; Sillapachaiyaporn, C.; Nilkhet, S.; Brimson, J.; Tencomnao, T. Effect of Thai Medicinal Plants Acanthus ebracteatus Vahl Carthamus tinctorius L. and Streblus asper lour. on Neurite Outgrowth Activity in Neuro-2a Cells. J. Assoc. Med Sci. 2023, 56, 71–84. [Google Scholar]
- Umehara, K.; Nemoto, K.; Matsushita, A.; Terada, E.; Monthakantirat, O.; De-Eknamkul, W.; Miyase, T.; Warashina, T.; Degawa, M.; Noguchi, H. Flavonoids from the Heartwood of the Thai Medicinal Plant Dalbergia Parviflora and Their Effects on Estrogenic-Responsive Human Breast Cancer Cells. J. Nat. Prod. 2009, 72, 2163–2168. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, J.; Avula, B.; Lee, J.; Upton, R.; Khan, I.A. Chemical Characterization and Quantitative Determination of Flavonoids and Phenolic Acids in Yerba Santa (Eriodictyon Spp.) Using Uhplc/Dad/Q-Tof. J. Pharm. Biomed. Anal. 2023, 234, 115570. [Google Scholar] [CrossRef] [PubMed]
- Sharifi-Rad, J.; Song, S.; Ali, A.; Subbiah, V.; Taheri, Y.; Suleria, H.A. LC-ESI-QTOF-MS/MS Characterization of Phenolic Compounds from Pyracantha coccinea M.Roem. and Their Antioxidant Capacity. Cell. Mol. Biol. 2021, 67, 201–211. [Google Scholar] [CrossRef]
- Zhang, L.; Qin, S.; Tang, S.; E, S.; Li, K.; Li, J.; Cai, W.; Sun, L.; Li, H. Qualitative Analysis of Multiple Phytochemical Compounds in Tojapride Based on Uhplc Q-Exactive Orbitrap Mass Spectrometry. Molecules 2022, 27, 6639. [Google Scholar] [CrossRef]
- Ginz, M.; Engelhardt, U.H. Identification of Proline-Based Diketopiperazines in Roasted Coffee. J. Agric. Food Chem. 2000, 48, 3528–3532. [Google Scholar] [CrossRef]
- Nakamura, T.; Nagaki, H.; Ohki, Y.; Kinoshita, T. Differentiation of Leucine and Isoleucine Residues in Peptides by Consecutive Reaction Mass Spectrometry. Anal. Chem. 1990, 62, 311–313. [Google Scholar] [CrossRef]
- Simpson, J.P.; Olson, J.; Dilkes, B.; Chapple, C. Identification of the Tyrosine-and Phenylalanine-Derived Soluble Metabolomes of Sorghum. Front. Plant Sci. 2021, 12, 714164. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, X.; Yin, X.; Wang, H.; Fu, C.; Wang, H.; Li, K.; Li, Y.; Zhang, X.; Liang, H.; et al. Metabolomic Profiling Reveals Serum L-Pyroglutamic Acid as a Potential Diagnostic Biomarker for Systemic Lupus Erythematosus. Rheumatology 2021, 60, 598–606. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).