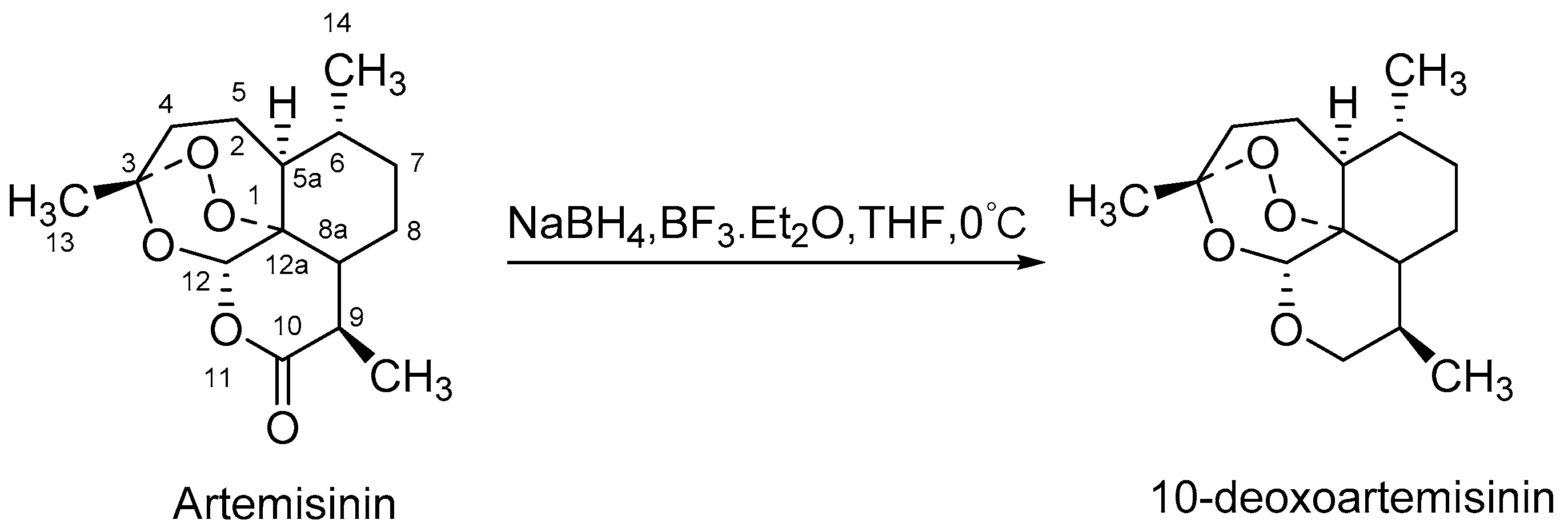Abstract
10-deoxoartemisinin is a semisynthetic derivative of artemisinin that lacks a lactone carbonyl group at the 10-position, and has stronger antimalarial properties than artemisinin. However, 10-deoxoartemisinin has limited utility as a therapeutic agent because of its low solubility and bioavailability. Hydroxylated 10-deoxoartemisinins are a series of properties-improved derivatives. Via microbial transformation, which can hydroxylate 10-deoxoartemisinin at multiple sites, the biotransformation products of 10-deoxoartemisinin have been investigated in this paper. Using ultra-performance liquid chromatography-electrospray ionization-quadrupole time-of-flight mass spectrometry (UPLC-ESI-Q-TOF-MSE) combined with UNIFI software, products of microbial transformation of 10-deoxoartemisinin were rapidly and directly analyzed. The hydroxylation abilities of nine microorganisms were compared using this method. All of the microorganisms evaluated were able to hydroxylate 10-deoxoartemisinin, and a total of 35 hydroxylated products were identified. These can be grouped into dihydroxylated 10-deoxoartemisinins, monohydroxylated 10-deoxoartemisinins, hydroxylated dehydrogenated 10-deoxoartemisinins, and hydroxylated hydrogenated 10-deoxoartemisinins. Cunninghamella echinulata and Cunninghamella blakesleeana are able to hydroxylate 10-deoxoartemisinin, and their biotransformation products are investigated here for the first time. Cunninghamella elegans CICC 40250 was shown to most efficiently hydroxylate 10-deoxoartemisinin, and could serve as a model organism for microbial transformation. This method could be used to generate additional hydroxylated 10-deoxoartemisinins for further research.
1. Introduction
Artemisinin is a first-line antimalarial agent, which is first derived from Chinese herb by Professor Tu [1]. 10-deoxoartemisinin is a bioactive derivative of artemisinin, first prepared by the simple transformation in 1990 [2]. It lacks the C-10 lactone carbonyl group of artemisinin, but retains its antimalarial endoperoxide activity [3]. Compared with artemisinin, the derivate was eight times more active against multidrug-resistant malaria in vitro [2]. However, poor solubility and bioavailability are the barriers for further application in the clinic. Moreover, 10-deoxoartemisinin also has impressive antitumor and antiangiogenesis activities [4,5,6]. As a part of our ongoing projects [7,8,9,10], novel 10-deoxoartemisinin derivatives with improved the properties are attractive to develop for further research. Hydroxylation, as the main modification route, could not only improve chemical properties, but provide candidates of novel drug development.
In recent years, microbial transformation has become an efficient tool to generate novel active products. Microorganisms can achieve predetermined selectivity to add functional groups in an environment-friendly manner because of their abundant cytochrome P450 enzymes [11,12,13]. Several fungi have been reported to exhibit particularly strong hydroxylation capabilities, including Cunninghamella, Aspergillus, Mucor, etc. [14,15,16]. Up to now, five microbial transformation products of 10-deoxoartemisinin from four strains have been documented. These include 5β-hydroxy-10-deoxoartemisinin from Mucor ramannianus [17], 4α-hydroxy-1, 10-deoxoartemisinin and 7β-hydroxy-10-deoxoartemisinin from both Mucor ramannianus and Cunninghamella elegans [18], 15-hydroxy-10-deoxoartemisinin from Aspergillus Niger [19], and a 13-carbon rearranged product from Aspergillus ochraceus [20], none of which were reported to be bioactive. Even though, 10-deoxoartemisinin provides an excellent biotransformation substrate for the production of new potentially antimalarial active, and there remains a need for more exploited. The scheme synthesis of 10-deoxoartemisinin is shown in Figure 1.
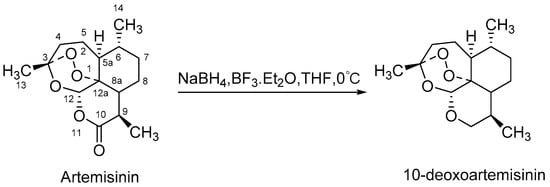
Figure 1.
Scheme synthesis of 10-deoxoartemisinin.
Different microbial strains may use distinct pathways to accomplish biotransformation, making a rapid and direct method to evaluate hydroxylation products is imperative. Here, ultra-performance liquid chromatography-electrospray ionization-quadrupole time-of-flight mass spectrometry (UPLC-ESI-Q-TOF-MSE) is established as the method to predict the hydroxylation of 10-deoxoartemisinin by nine different microorganisms. These microorganisms studied in this research were Cunninghamella elegans CICC 40250 (MT1), Mucor circinelloides CGMCC 3.3421 (MT2), Cunninghamella elegans CGMCC 3.4832 (MT3), Cunninghamella echinulata CGMCC 3.4879 (MT4), Mucor circinelloides CGMCC 3.49 (MT5), Cunninghamella echinulata CGMCC 3.5771 (MT6), Cunninghamella blakesleeana CGMCC 3.5802 (MT7), Cunninghamella blakesleeana CGMCC 3.910 (MT8), and Cunninghamella elegans ATCC 9245 (MT9), which have all been reported to perform hydroxylation reactions [21,22,23,24,25]. The biotransformation products of 10-deoxoartemisinin were analyzed and identified using UNIFI software. Among these organisms, Cunninghamella elegans CICC 40250 (MT1) exhibited the best hydroxylation profile, and holds promise for generating novel bioactive hydroxylated products for further research.
2. Results
2.1. Identification of 10-deoxoartemisinin
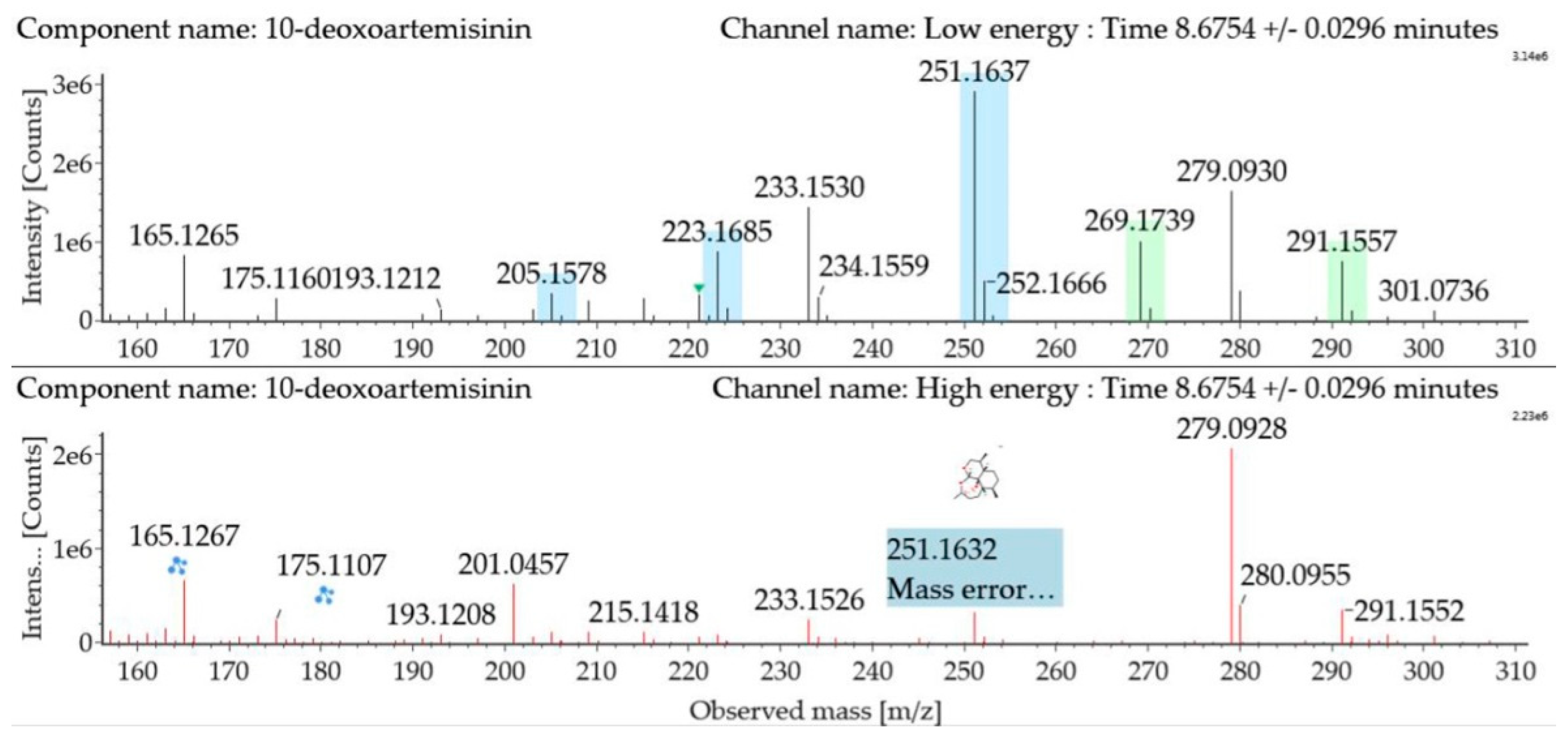
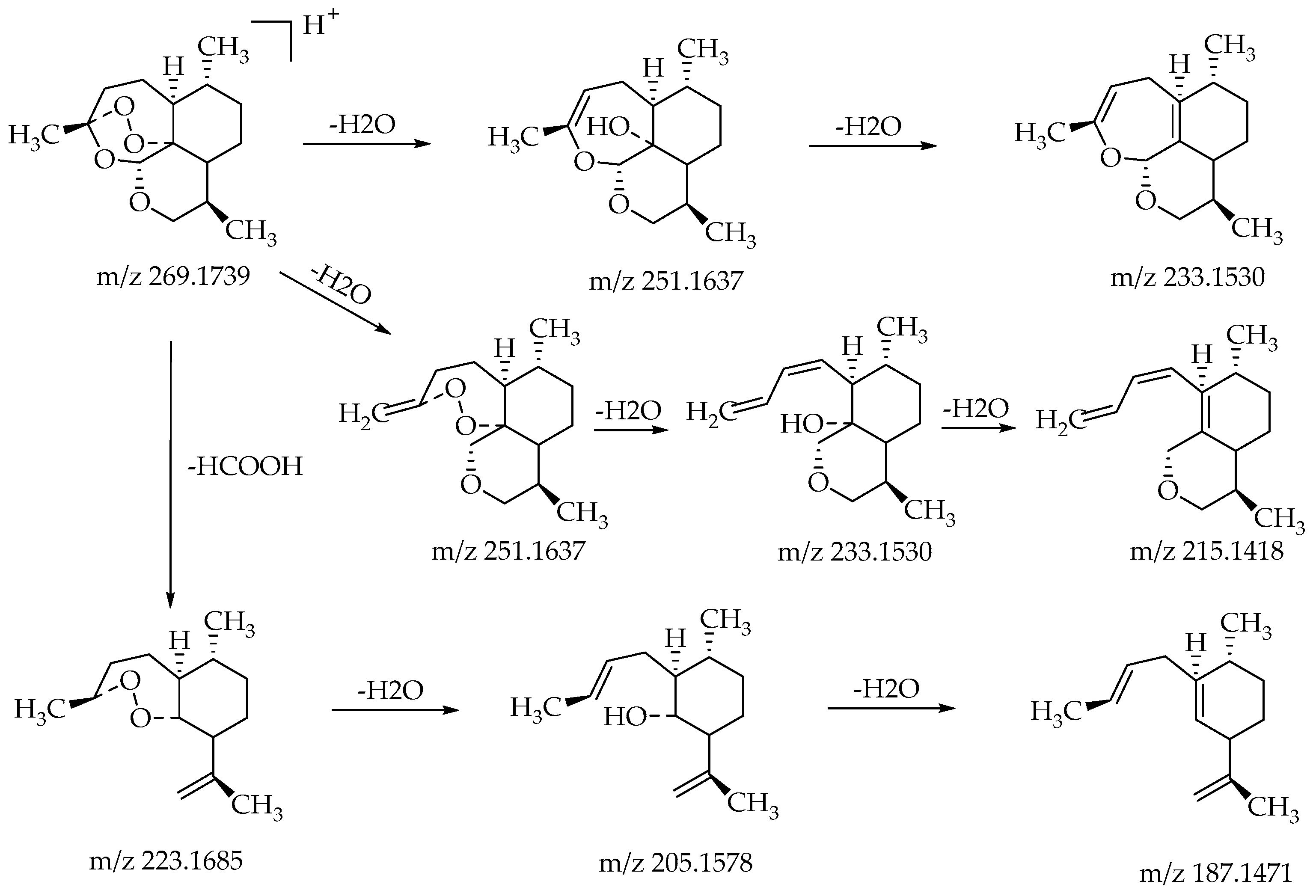
The substrate 10-deoxoartemisinin was used to establish a reference fragmentation pattern. Its molecular ions [M + H]+ and [M + Na]+ were observed at m/z 269.1739 and m/z 291.1557. The fragment ions at m/z 251.1637, 233.1530, and 215.1418 were generated by the successive loss of water from m/z 269.1739. The fragment ion at m/z 223.1685 represented the loss of CO from m/z 251.1637 or the loss of HCOOH from m/z 269.1739. The fragment ion at m/z 205.1578 resulted from the loss of water from m/z 223.1685. Mass spectra and the fragmentation diagram of 10-deoxoartemisinin are shown in Figure 2, and the proposed fragmentation pathways are shown in Figure 3.
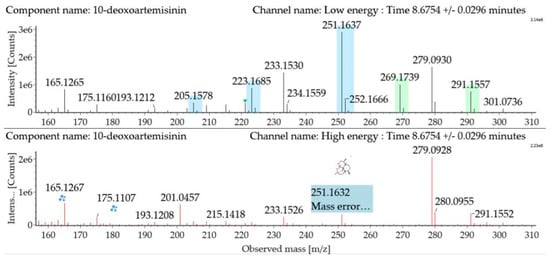
Figure 2.
Ultra-performance liquid chromatography-electrospray ionization-quadrupole time-of-flight mass spectrometry (UPLC-ESI-Q-TOF-MSE) spectra and fragmentation diagram of 10-deoxoartemisinin.

Figure 3.
Proposed fragmentation pathways of 10-deoxoartemisinin.
2.2. Identification of Microbial Transformation Products
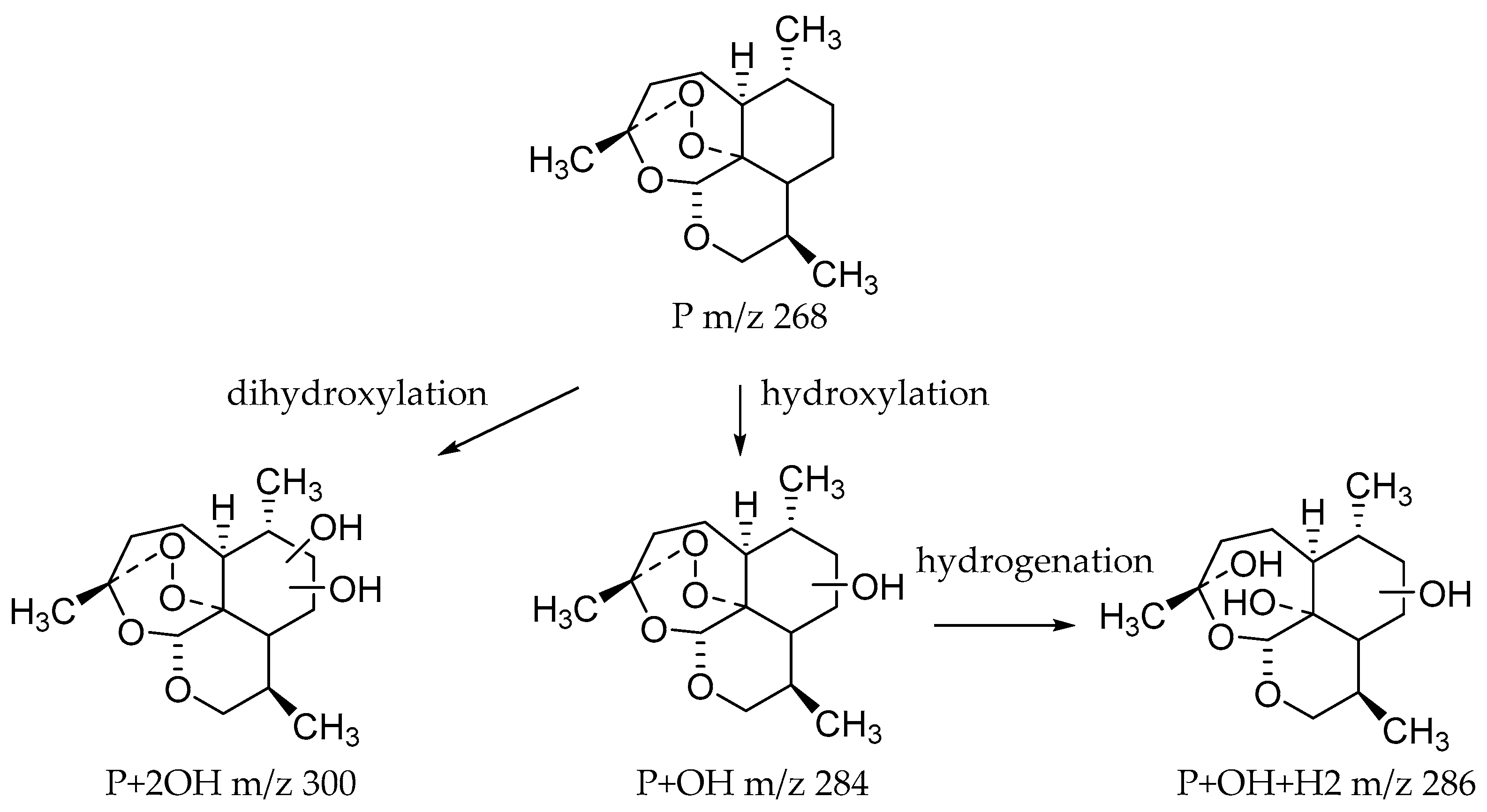
Microbial transformation products were identified using the characteristic mass spectrometric behavior of 10-deoxoartemisinin, including parent ions, internal cleavage of the ion, and retention times. Altogether, 35 transformation products were identified; primarily dihydroxylated 10-deoxoartemisinin, monohydroxylated 10-deoxoartemisinin, hydroxylated dehydrogenated 10-deoxoartemisinin, and hydroxylated hydrogenated 10-deoxoartemisinin. While all strains tested were able to produce hydroxylated 10-deoxoartemisinins, MT1 produced the most derivatives, with 18 distinct products. The 18 hydroxylated products from MT1 all had a peroxide bridge. These results demonstrate that MT1 is an ideal microorganism model to obtain hydroxylated 10-deoxoartemisinin for further research. Meanwhile, the biotransformation products from Cunninghamella echinulata and Cunninghamella blakesleeana are first to be investigated. The identified microbial transformation products are shown in Table 1, and the structures of predicted transformation products are shown in Figure 4.

Table 1.
Summary of microbial transformation products of 10-deoxoartemisinin.
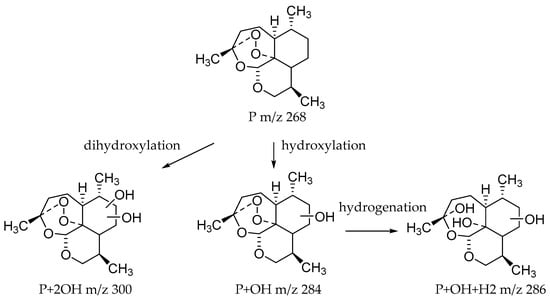
Figure 4.
Structures of predicted transformation products.
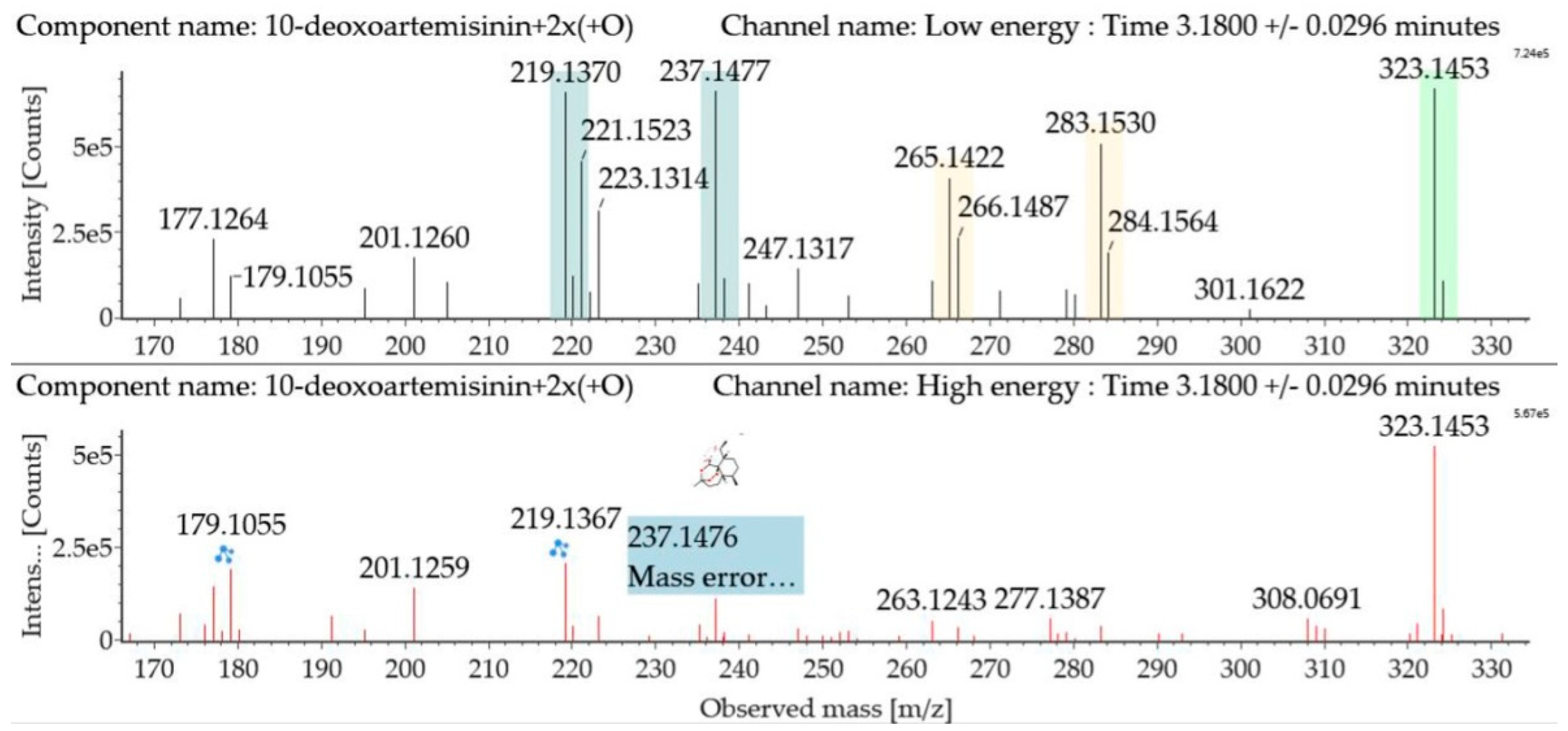
Dihydroxylated 10-deoxoartemisinin transformation products were detected between 2.83 and 4.41 min, and the parent ions were detected at m/z 323 ([M + Na]+) and m/z 301 ([M + H]+). The products were identified by comparing their parent ions to 10-deoxoartemisinin, which showed a series of fragment ions resulting from H2O and CO loss. The dihydroxylated products were more likely to lose two water molecules compared with 10-deoxoartemisinin, and the fragment ions at m/z 265 and 247 were generated by successive water loss from m/z 283. The fragment ion at m/z 237 was generated by CO loss from m/z 265 or HCOOH loss from m/z 283. The fragment ion at m/z 219 was generated by water loss from m/z 237. Mass spectra and the fragmentation diagram for dihydroxylated 10-deoxoartemisinin products are shown in Figure 5.
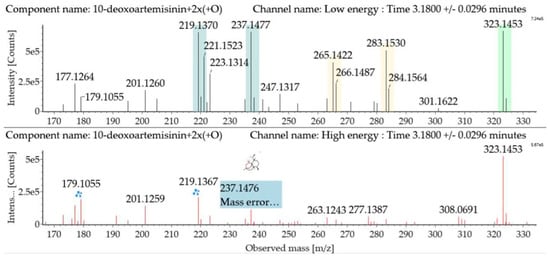
Figure 5.
UPLC-ESI-Q-TOF-MSE spectra and the fragmentation diagram of dihydroxylated 10-deoxoartemisinin.
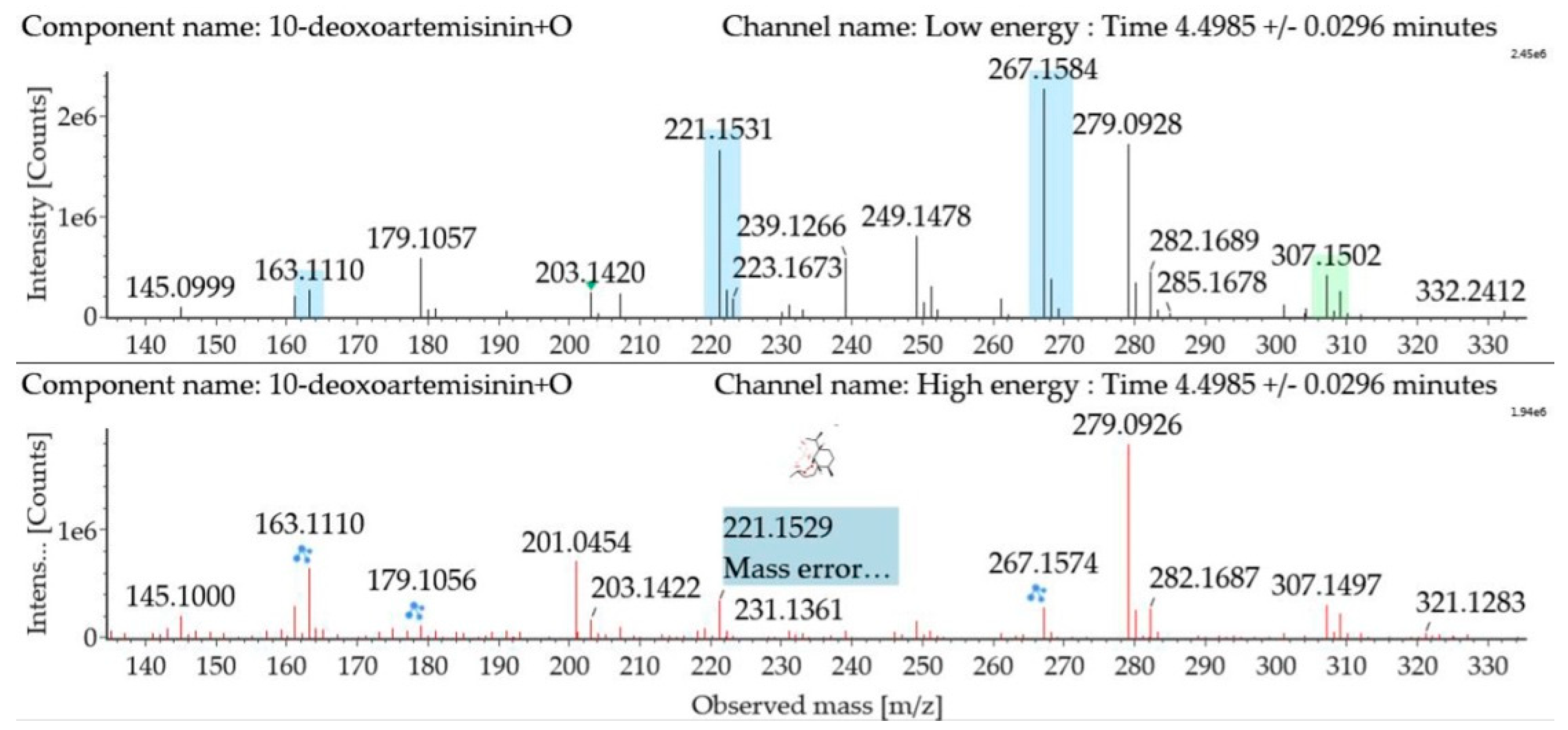
Monohydroxylated 10-deoxoartemisinin transformation products were detected between 3.14 and 6.91 min. The products were identified by comparing their parent ions to 10-deoxoartemisinin, with a 16 Da mass shift attributed to an oxygen atom. The molecular ion at m/z 307 ([M + Na]+) was observed. The fragment ion at m/z 267 resulted from water loss from m/z 285. The fragment ions at m/z 249 and m/z 231 were generated by water loss from m/z 267 and m/z 249, respectively. The fragment ion at m/z 221 resulted from CO loss from m/z 249. Mass spectra and the fragmentation diagram for monohydroxylated 10-deoxoartemisinin products are shown in Figure 6.
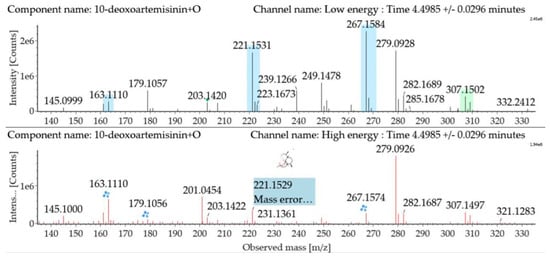
Figure 6.
UPLC-ESI-Q-TOF-MSE spectra and the fragmentation diagram of monohydroxylated 10-deoxoartemisinin.
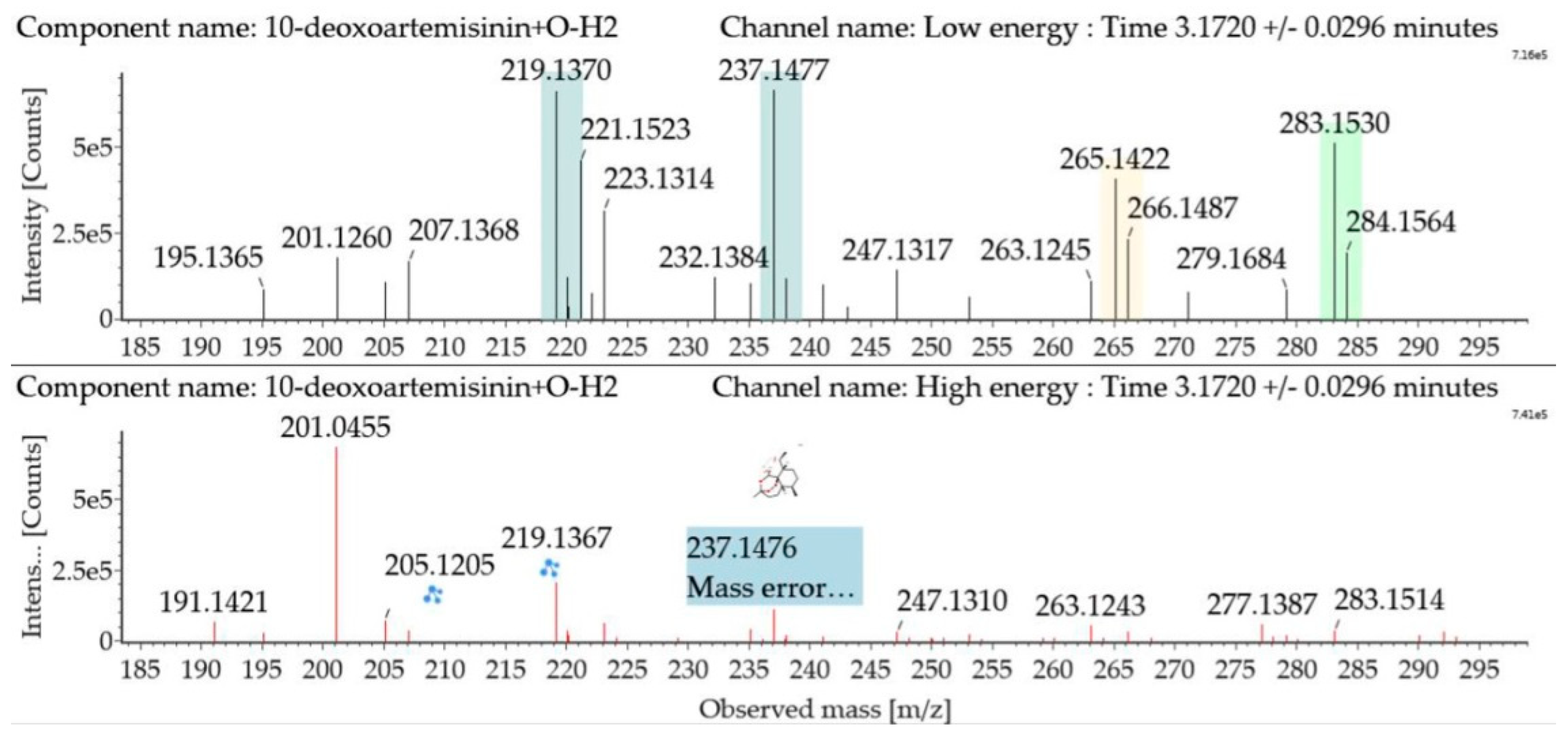
Hydroxylated dehydrogenated 10-deoxoartemisinin products were detected between 2.83 and 5.57 min. The molecular ion at m/z 283 ([M + H]+) was observed. Consistent with 10-deoxoartemisinin, these products showed a series of fragment ions resulting from the loss of H2O, CO, or HCOOH. Mass spectra and the fragmentation diagram for hydroxylated dehydrogenated 10-deoxoartemisinin products are shown in Figure 7.
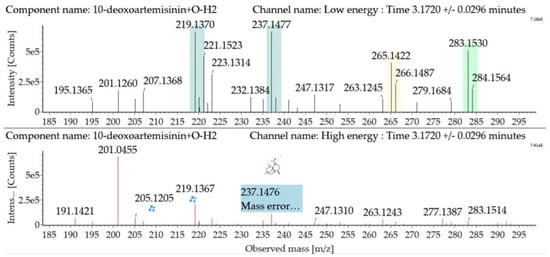
Figure 7.
UPLC-ESI-Q-TOF-MSE spectra and the fragmentation diagram of hydroxylated dehydrogenated 10-deoxoartemisinin.
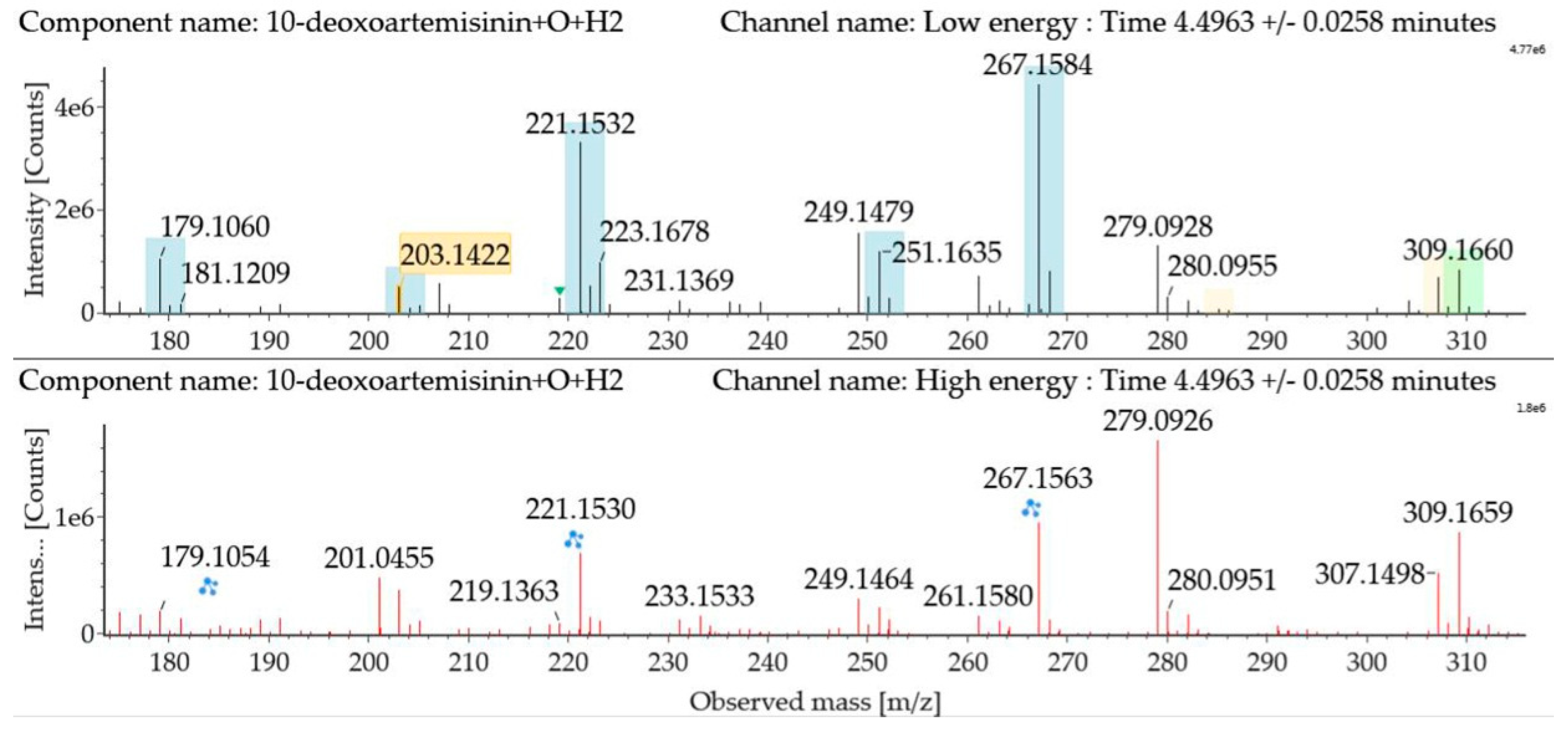
Hydroxylated hydrogenated 10-deoxoartemisinin products were detected at about 4.50 min. The fragment ions were similar to monohydroxylation products of 10-deoxoartemisinin. The parent ion was detected at m/z 309 ([M + Na]+). Fragment ions at m/z 249 and 231 were generated by successive water loss from m/z 267. The fragment ion at m/z 221 was generated by CO loss from m/z 249 or HCOOH loss from m/z 267. The fragment ion at m/z 203 resulted from water loss from m/z 221. The mass spectra and the fragmentation diagram for hydroxylated hydrogenated 10-deoxoartemisinin products are shown in Figure 8.
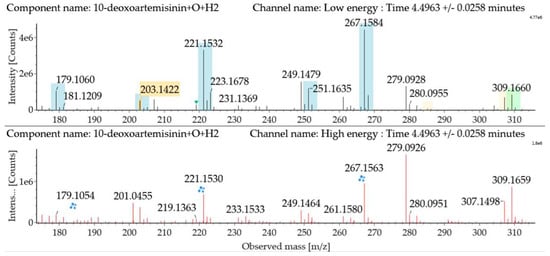
Figure 8.
UPLC-ESI-Q-TOF-MSE spectra and the fragmentation diagram of hydroxylated hydrogenated 10-deoxoartemisinin.
3. Discussion
Hydroxylation is the main route for molecular modification and also a primary metabolic reaction in vivo involving artemisinin and its derivatives [20,26]. Hydroxylated 10-deoxoartemisinins are a series of products that have a peroxide bridge and may retain antimalarial activity, with better physiological solubility compared with unmodified 10-deoxoartemisinin. Meanwhile, these hydroxylated compounds provide the new reactive site to link another functional group for developing a novel active drug.
Microbial transformation is an increasingly popular method used to modify small molecules, particularly adept at generating hydroxylated derivatives, because many enzymes catalyze hydroxylation reactions, and a variety of microorganisms can be used in this method. The microbial transformation was used in this study to find out novel hydroxylated 10-deoxoartemisinins. The hydroxylation abilities of nine microorganisms from the genera Cunninghamella and Mucor was evaluated, because previous studies have shown that species in these two genera are rich in hydroxylases [27]. Additionally, C. elegans, C. echinulata, C.blakesleeana, and M. circinelloides have been successfully employed to hydroxylate other substrates [28,29,30]. The hydroxylation ability of these microorganisms was estimated by identifying and comparing their hydroxylation products. A total of 35 hydroxylated products were identified from the nine microorganisms.
Strains MT1-MT9 produced 18, 1, 4, 15, 1, 9, 14, 7, and 3 distinct hydroxylated 10-deoxoartemisinins, respectively. These hydroxylated 10-deoxoartemisinins were categorized as ehydroxylated 10-deoxoartemisinins, monohydroxylated 10-deoxoartemisinins, hydroxylated dehydrogenated 10-deoxoartemisinins, and hydroxylated hydrogenated 10-deoxoartemisinins. Strains MT1, MT4, MT6, and MT7 had the best hydroxylation abilities, generating more than ten hydroxylated products each, with products from all four of the above categories. Strains MT2 and MT5 showed only weak hydroxylation, producing only one monohydroxylated 10-deoxoartemisinin each. In addition, strains MT3 and MT9 only produced monohydroxylated 10-deoxoartemisinins. MT1 (C. elegans CICC 40250) exhibited the best hydroxylation ability, with 18 hydroxylated products identified. MT1 could serve as a model organism for this process and could also produce novel hydroxylated 10-deoxoartemisinins. Overall, Cunninghamella hydroxylates 10-deoxoartemisinin better than Mucor.
The rapid and direct analysis method developed in this paper is significant for efficiently screening numerous of Microorganisms transformation. The results could be the guidance for further isolation research to obtain more derivates. This work is foundational for producing novel, potentially bioactive, hydroxylated 10-deoxoartemisinins.
4. Materials and Methods
4.1. Materials and Reagents
10-deoxoartemisinin was synthesized from artemisinin, as previously described [2]. M. circinelloides CGMCC 3.3421, M. circinelloides CGMCC 3.49, C. blakesleeana CGMCC 3.910 and 3.5802, C. echinulata CGMCC 3.5771, C. echinulata 3.4879, and C. elegans CGMCC 3.4832 were obtained from the China General Microbiological Culture Collection Center (Beijing, China). C. elegans CICC 40250 was obtained from the China Center of Industrial Culture Collection (Beijing, China). C. elegans ATCC 9245 was obtained from the American Type Culture Collection (Virginia, United States). Acetonitrile and methanol were purchased from Fisher (Geel, Belgium), and formic acid, EtOAc (ethyl acetate), and other chemicals used in this research were purchased from Beijing Chemical Works (Beijing, China).
4.2. Culture and Biotransformation Procedure
Culture and biotransformation experiments were conducted in a medium composed of 20 g Sabouraud Dextrose Broth (Oxoid, Basingstoke, UK), 10 g peptone (Solarbio, Beijing, China), 15 g sucrose (Solarbio, Beijing, China), and 1000 mL deionized water [31,32].
Two-stage fermentation was employed in this study [33,34]. The substrate was dissolved in MeOH at a concentration of 25 mg/mL, and 2 mL of the solution was added into each flask after the second fermentation stage, producing a 10-deoxoartemisinin concentration of 0.5 mg/mL. Cultures were incubated at 28 °C and shaken at 180 rpm for 14 days. Mycelia and broth were then separated by filtration. The filtrate was extracted with EtOAc (1:1, v/v) three times, and the extract was evaporated under vacuum to produce a brown residue, which was dissolved in MeOH for LC-MS analysis.
4.3. LC–MSE Conditions and Data Processing
Mass spectrometry conditions were established in previous studies [32]. The UPLC-ESI-Q-TOF-MSE system consisted of a Waters ACQUITY I-class UPLC and Xevo G2-XS Q-TOF Mass Spectrometer (Waters, Manchester, UK). Chromatography was conducted using an Acquity UPLC BEH C18 (2.1 mm × 100 mm, 1.7 µm (Waters, Manchester, UK)). The mobile phase consisted of solvent A (H2O containing 0.1% formic acid, v/v) and solvent B (acetonitrile containing 0.1% formic acid, v/v). The gradient program included three segments: 5%–100% B from 0 to 15 min; 100%–5% B from 15 to 17 min; followed by a 3 min post-run for column equilibration. The flow rate was 0.4 mL/min, and the temperature was 25 °C throughout the analysis.
The MS was operated in positive ionization mode across a scan range of 50 to 1200 m/z. The low collision energy was set at 6 eV, and the high collision energy was ramped from 12 to 25 eV. MSE analysis was conducted using multiple reactions monitoring positive-ion electrospray ionization.
All data processing was performed using UNIFI 1.9 (Waters, Manchester, UK). Components were identified using the following 3D peak detection features: Low-energy limits of 150 and high-energy limits of 20, isotope clustering, and high-to-low energy association within a 0.5 fraction of the chromatographic and drift peak width, with a mass accuracy of ± 2 mDa. The maximum number of allowed fragment ions per match was set to 10.
5. Conclusions
This research establishes microbial transformation as a method for obtaining various hydroxylated products. Transformation products from nine microorganisms were identified using UPLC-ESI-Q-TOF-MSE, and the hydroxylation ability of the nine microorganisms was evaluated. Transformation products of 10-deoxoartemisinin were categorized as hydroxylated 10-deoxoartemisinins, hydroxylated dehydrogenated 10-deoxoartemisinins, and hydroxylated hydrogenated 10-deoxoartemisinins. MT1 (C. elegans CICC 40250) exhibited the best hydroxylation ability among the selected microorganisms, and could be used as a transformation model to prepare hydroxylated 10-deoxoartemisinins for further research. Additionally, this study was the first to investigate the transformation products of 10-deoxoartemisinin from C. echinulata and C. blakesleeana.
Author Contributions
L.Y. and Y.M. conceived the idea for this project, designed, and conducted experiments. Y.B. collected and analyzed the data and wrote the manuscript with contributions from other authors. D.Z. participated in research design and assisted in the data analysis. Y.Z. and X.C. participated in performing the microbial transformation experiments. P.S. contributed to the writing.
Funding
This work was supported by the emergency management program of the National Natural Science Foundation of China (Grant No. 81841001), major science and technology project for ‘Significant New Drugs Creation’ (Grant No. 2017ZX09101002-002), the Fundamental Research Funds for the Central public welfare research institutes (Grant No. ZXKT19023).
Acknowledgments
Thanks to Huimin Gao from the Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences for critical review of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Collaboration Research Group for Qinghaosu. A new sesquiterpene lactone—qinghaosu. KexueTongbao 1977, 3, 142. (In Chinese) [Google Scholar]
- Jung, M.; Li, X.; Bustos, D.A.; ElSohly, H.N.; McChesney, J.D.; Milhous, W.K. Synthesis and Antimalarial Activity of (+)-Deoxoartemisinin. J. Med. Chem. 1990, 33, 1516–1518. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Li, X.; Bustos, D.A.; ElSohly, H.N.; McChesney, J.D. A short and stereospecific synthesis of (+)-deoxoartemisinin and (−)-deoxodesoxyartemisinin. Tetrahedron Lett. 1989, 30, 5973–5976. [Google Scholar] [CrossRef]
- Lee, C.H.; Hong, H.; Shin, J.; Jung, M.; Shin, I.; Yoon, J.; Lee, W. NMR Studies on Novel Antitumor Drug Candidates Deoxoartemisinin and Carboxypropyldeoxoartemisinin. Biochem. Biophys. Res. Commun. 2000, 274, 359–369. [Google Scholar] [CrossRef]
- Jung, M.; Tak, J.; Chuang, W.Y.; Park, K.-K. Antiangiogenic activity of deoxoartemisinin derivatives on chorioallantoic membrane. Bioorg. Med. Chem. Lett. 2006, 16, 1227–1230. [Google Scholar] [CrossRef]
- Jung, M.; Lee, S.; Ham, J.; Lee, K.; Kim, H.; Kim, S.K. Antitumor Activity of Novel Deoxoartemisinin Monomers, Dimers, and Trimer. J. Med. Chem. 2003, 46, 987–994. [Google Scholar] [CrossRef]
- Tu, Y.Y. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat. Med. 2011, 17, 1217–1220. [Google Scholar] [CrossRef]
- Tu, Y.Y. Artemisinin and Artemisinin Drugs; Chemical Industry Press: Beijing, China, 2009; pp. 34–56. [Google Scholar]
- Wang, J.; Xu, C.; Liao, F.L.; Jiang, T.; Krishna, S.; Tu, Y. A Temporizing Solution to “Artemisinin Resistance”. N. Engl. J. Med. 2019, 380, 2087–2089. [Google Scholar] [CrossRef]
- Ma, Y.; Zhu, Y.P.; Zhang, D.; Meng, Y.; Tang, T.; Wang, K.; Ma, J.; Wang, J.; Sun, P. Eco-friendly decarboxylative cyclization in water: practical access to the anti-malarial 4-quinolones. Green Chem. 2019, 21, 478–482. [Google Scholar] [CrossRef]
- Guan, S.-L.; Wu, Y.-X.; Sun, H.; Zhang, Y.C.; Zhao, R.Q.; Wang, H.; Sun, L.J.; Su, L. Application of microbial transformation technology in the development of Chinese medicine. Microbiol. China. 2018, 45, 900–906. [Google Scholar]
- Chen, D.-J.; Zhu, B.-Q. Application of microbial transformation in modern pharmaceutical industry. Chin. J. Antibiot. 2012, 31, 112–118. [Google Scholar]
- Niu, H.-J.; Wang, P.; Yang, G.-E. Application of Microbial Transformation in Research of Chinese Medicine. Chin. J. Exp. Trad. Med. Formulae. 2013, 19, 346–349. [Google Scholar]
- Zhan, J.X.; Guo, H.Z.; Dai, J.G.; Zhang, Y.; Guo, D. Microbial transformations of artemisinin by Cunninghamella echinulata and Aspergillus niger. Tetrahedron Lett. 2002, 43, 4519–4521. [Google Scholar] [CrossRef]
- Zhan, Y.L.; Liu, H.; Wu, Y.S.; Wei, P.; Chen, Z.; Williamson, J.S. Biotransformation of artemisinin by Aspergillus niger. Appl. Microbiol. Biotechnol. 2015, 99, 3443–3446. [Google Scholar] [CrossRef]
- Parshikov, I.A.; Miriyala, B.; Muraleedharan, K.M.; Illendula, A.; Avery, M.A.; Williamson, J.S. Biocatalysis of the Antimalarial Artemisinin by Mucor ramannianus Strains. Pharm. Biol. 2005, 43, 579–582. [Google Scholar] [CrossRef]
- Parshikov, I.A.; Muraleedharan, K.M.; Miriyala, B.; Avery, M.A.; Williamson, J.S. Hydroxylation of 10-Deoxoartemisinin by Cunninghamella elegans. Nat. Prod. 2004, 67, 1595–1597. [Google Scholar] [CrossRef]
- De Medeiros, S.F.; Avery, M.A.; Avery, B.; Leite, S.G.; Freitas, A.C.C.; Williamson, J.S. Biotransformation of 10-deoxoartemisinin to its 7β-hydroxy derivative by Mucor ramannianus. Biotechnol. Lett. 2002, 24, 937–941. [Google Scholar] [CrossRef]
- Parshikov, I.A.; Miriyala, B.; Avery, M.A.; Williamson, J.S. Hydroxylation of 10-deoxoartemisinin to 15-hydroxy-10-deoxoartemisinin by Aspergillus niger. Biotechnol. Lett. 2004, 26, 607–610. [Google Scholar] [CrossRef]
- Khalifa, S.I.; Baker, J.K.; Jung, M.; McChesney, J.D.; Hufford, C.D. Microbial and Mammalian Metabolism Studies on the Semisynthetic Antimalarial, Deoxoartemisinin. Pharm. Res. 1995, 12, 1493–1498. [Google Scholar] [CrossRef]
- Adachi, T.; Saito, M.; Sasaki, J.; Karasawa, Y.; Araki, H.; Hanada, K.; Omura, S. Microbial Hydroxylation of (-)-Eburnamonine by Mucor circinelloides and Streptomyces violens. Chem. Pharm. Bull. 1993, 41, 611–613. [Google Scholar] [CrossRef][Green Version]
- Ma, Y.; Xie, D.; Wang, Z.H.; Dai, J.-G.; An, X.-Q.; Gu, Z.-Y. Microbial transformation of glycyrrhetinic acid by Cunninghamella blakesleeana. Chin. J. Trad. Chin. Med. 2015, 40, 4212–4217. [Google Scholar]
- Dong, T.; Wu, G.W.; Wang, X.N.; Gao, J.-M.; Chen, J.-G.; Lee, S.-S. Microbiological transformation of diosgenin by resting cells of filamentous fungus, Cunninghamella echinulata CGMCC 3.2716. J. Mol. Catal. B Enzym. 2010, 67, 251–256. [Google Scholar] [CrossRef]
- Qin, S.; Zhou, C.-L. Application of Microbial Transformationin Medicine Metabolization Model in vitro by Cunninghammella Matruchot. Strait Pharm. 2004, 1, 4–8. [Google Scholar]
- Weidner, S.; Goeke, K.; Trinks, U.; Traxler, P.; Ucci-Stoll, K.; Ghisalba, O. Preparation of 4-(4′-Hydroxyanilino)-5-anilinophthalimide and 4, 5-Bis-(4′-hydroxyanilino)-phthalimide by Microbial Hydroxylation. Biosci. Biotechnol. Biochem. 1999, 63, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.S.; ElSohly, H.N.; Croom, E.M.; Hufford, C.D. Microbial metabolism studies of the antimalarial sesquiterpene artemisinin. J. Nat. Prod. 1989, 52, 337–341. [Google Scholar] [CrossRef]
- Ye, M.; Han, J.; Tu, G.; An, D.; Guo, D. Microbial hydroxylation of bufalin by Cunninghamella blakesleana and Mucor spinosus. J. Nat. Prod. 2005, 68, 626–628. [Google Scholar] [CrossRef]
- Parshikov, I.A.; Muraleedharan, K.M.; Avery, M.A.; Williamson, J.S. Transformation of artemisinin by Cunninghamella elegans. Appl. Microbiol. Biotechnol. 2004, 64, 782–786. [Google Scholar] [CrossRef]
- Baydoun, E.; Ahmad, M.S.; Mehmood, H.; Ahmad, M.S.; Malik, R.; Smith, C.; Choudhary, M.I. Microbial transformation of danazol with Cunninghamella blakesleeana and anti-cancer activity of danazol and its transformed products. Steroids 2016, 105, 121–127. [Google Scholar] [CrossRef]
- Sasaki, J.; Mizoue, K.; Morimoto, S.; Adachi, T.; Omura, S. Microbial transformation of 6-O-methylerythromycin derivatives. J. Antibiot. Tokyo. 1988, 41, 908–915. [Google Scholar] [CrossRef]
- Zhan, Y.L.; Wu, Y.S.; Xu, F.F.; Bai, Y.; Guan, Y.; Williamson, J.S.; Liu, B. A novel dihydroxylated derivative of artemisinin from microbial transformation. Fitoterapia 2017, 120, 93–97. [Google Scholar] [CrossRef]
- Ma, Y.; Sun, P.; Zhao, Y.F.; Wang, K.; Chang, X.; Bai, Y.; Zhang, D.; Yang, L. A Microbial Transformation Model for Simulating Mammal Metabolism of Artemisinin. Molecules 2019, 24, 315. [Google Scholar] [CrossRef] [PubMed]
- Betts, R.E.; Walters, D.E.; Rosazza, J.P. Microbial Transformations of Antitumor Compounds. 1. Conversion of Acronycine to 9-Hydroxyacronycine by Cunninghamella echinulata. J. Med. Chem. 1974, 17, 599–602. [Google Scholar] [CrossRef] [PubMed]
- Elmarakby, S.A.; Clark, A.M.; Baker, J.K.; Hufford, C.D. Microbial Metabolism of Bornaprine, 3-(Diethylamino)propyl 2-Phenylbicyclo [2.2.1] heptane-2-carboxylate. Pharm. Sci. 1986, 75, 614–618. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).