Feature Papers in ‘Organelle Function’
A topical collection in Cells (ISSN 2073-4409).
Submission Status:
Closed
|
Viewed by 55912
Share This Topical Collection
Editor
 Prof. Dr. Paolo Bernardi
Prof. Dr. Paolo Bernardi
 Prof. Dr. Paolo Bernardi
Prof. Dr. Paolo Bernardi
E-Mail
Collection Editor
Department of Biomedical Sciences, University of Padova, Via Ugo Bassi 58/B, I-35131 Padova, Italy
Interests: mitochondria; calcium; channels; permeability transition; ATP synthase; cell death
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
This Topical Collection entitled “Feature Papers in Organelle Function” aims to collect high-quality research articles, communications, and review articles in the key field of organelle function. Since the aim of this Topical Collection is to illustrate frontier research in organelle function through selected works, we encourage Editorial Board Members of Cells to contribute papers reflecting the latest progress in their research field, or relevant experts and colleagues to do so. Please kindly note that all invited papers will be published online with discounts or free of charge once accepted.
We welcome the submission of manuscripts that deal with specific aspects of organelle physiology or that help deciphering how subcellular structures are maintained in the face of membrane flow and how individual organelles send and receive specific information to maintain intracellular homeostasis.
Topics of interest include, but are not limited to:
- cell membrane
- cell wall
- centrioles
- chloroplasts
- cytoplasm
- cytoskeleton
- endoplasmic reticulum
- Golgi apparatus
- intracellular vesicles
- lysosomes
- microfilaments
- microtubules
- mitochondria
- nuclei
- nucleoli
- peroxisomes
- ribosomes
- vacuoles
- Lipid
Prof. Paolo Bernardi
Guest Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cells is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Published Papers (9 papers)
Open AccessReview
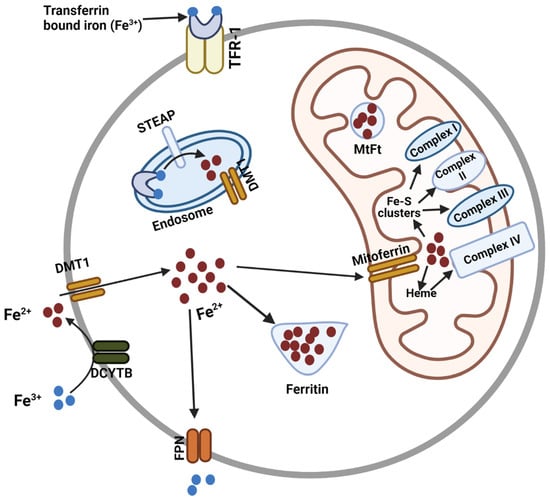
Mitoferrin, Cellular and Mitochondrial Iron Homeostasis
by
Md Yousuf Ali, Claudia R. Oliva, Susanne Flor and Corinne E. Griguer
Cited by 45 | Viewed by 6571
Abstract
Iron is essential for many cellular processes, but cellular iron homeostasis must be maintained to ensure the balance of cellular signaling processes and prevent disease. Iron transport in and out of the cell and cellular organelles is crucial in this regard. The transport
[...] Read more.
Iron is essential for many cellular processes, but cellular iron homeostasis must be maintained to ensure the balance of cellular signaling processes and prevent disease. Iron transport in and out of the cell and cellular organelles is crucial in this regard. The transport of iron into the mitochondria is particularly important, as heme and the majority of iron-sulfur clusters are synthesized in this organelle. Iron is also required for the production of mitochondrial complexes that contain these iron-sulfur clusters and heme. As the principal iron importers in the mitochondria of human cells, the mitoferrins have emerged as critical regulators of cytosolic and mitochondrial iron homeostasis. Here, we review the discovery and structure of the mitoferrins, as well as the significance of these proteins in maintaining cytosolic and mitochondrial iron homeostasis for the prevention of cancer and many other diseases.
Full article
►▼
Show Figures
Open AccessArticle
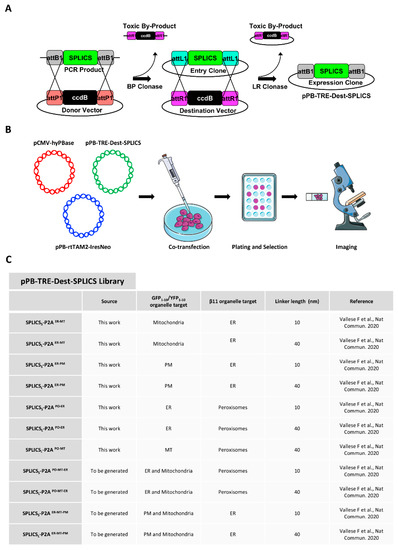
Stable Integration of Inducible SPLICS Reporters Enables Spatio-Temporal Analysis of Multiple Organelle Contact Sites upon Modulation of Cholesterol Traffic
by
Flavia Giamogante, Lucia Barazzuol, Elena Poggio, Marta Tromboni, Marisa Brini and Tito Calì
Cited by 8 | Viewed by 4512
Abstract
The study of organelle contact sites has received a great impulse due to increased interest in the understanding of their involvement in many disease conditions. Split-GFP-based contact sites (SPLICS) reporters emerged as essential tools to easily detect changes in a wide range of
[...] Read more.
The study of organelle contact sites has received a great impulse due to increased interest in the understanding of their involvement in many disease conditions. Split-GFP-based contact sites (SPLICS) reporters emerged as essential tools to easily detect changes in a wide range of organelle contact sites in cultured cells and in vivo, e.g., in zebrafish larvae. We report here on the generation of a new vector library of SPLICS cloned into a piggyBac system for stable and inducible expression of the reporters in a cell line of interest to overcome any potential weakness due to variable protein expression in transient transfection studies. Stable HeLa cell lines expressing SPLICS between the endoplasmic reticulum (ER) and mitochondria (MT), the ER and plasma membrane (PM), peroxisomes (PO) and ER, and PO and MT, were generated and tested for their ability to express the reporters upon treatment with doxycycline. Moreover, to take advantage of these cellular models, we decided to follow the behavior of different membrane contact sites upon modulating cholesterol traffic. Interestingly, we found that the acute pharmacological inhibition of the intracellular cholesterol transporter 1 (NPC1) differently affects membrane contact sites, highlighting the importance of different interfaces for cholesterol sensing and distribution within the cell.
Full article
►▼
Show Figures
Open AccessArticle
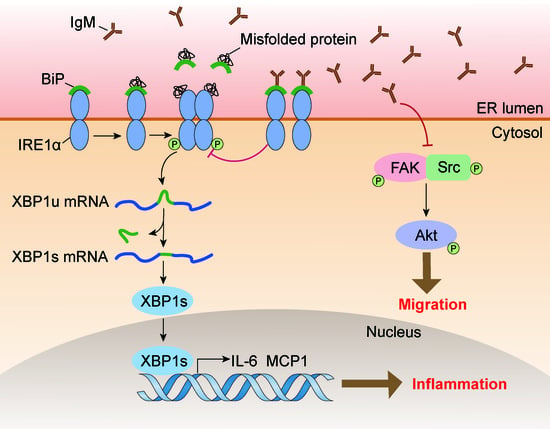
Macrophage-Derived Immunoglobulin M Inhibits Inflammatory Responses via Modulating Endoplasmic Reticulum Stress
by
Xiaoting Gong, Huige Yan, Junfan Ma, Zhu Zhu, Shenghua Zhang, Weiyan Xu, Jing Huang and Xiaoyan Qiu
Cited by 10 | Viewed by 4644
Abstract
Immunoglobulin (Ig), a characteristic marker of B cells, is a multifunctional evolutionary conserved antibody critical for maintaining tissue homeostasis and developing fully protective humoral responses to pathogens. Increasing evidence revealed that Ig is widely expressed in non-immune cells; moreover, Ig produced by different
[...] Read more.
Immunoglobulin (Ig), a characteristic marker of B cells, is a multifunctional evolutionary conserved antibody critical for maintaining tissue homeostasis and developing fully protective humoral responses to pathogens. Increasing evidence revealed that Ig is widely expressed in non-immune cells; moreover, Ig produced by different lineages cells plays different biological roles. Recently, it has been reported that monocytes or macrophages also express Ig. However, its function remains unclear. In this study, we further identified that Ig, especially Ig mu heavy chain (IgM), was mainly expressed in mice macrophages. We also analyzed the IgM repertoire characteristic in macrophages and found that the V
HDJ
H rearrangements of macrophage-derived IgM showed a restricted and conservative V
HDJ
H pattern, which differed from the diverse V
HDJ
H rearrangement pattern of the B cell-expressed IgM in an individual. Functional investigation showed that IgM knockdown significantly promoted macrophage migration and FAK/Src-Akt axis activation. Furthermore, some inflammatory cytokines such as MCP1 and IL-6 increased after IgM knockdown under LPS stimulation. A mechanism study revealed that the IgM interacted with binding immunoglobulin protein (Bip) and inhibited inflammatory response and unfolded protein response (UPR) activation in macrophages. Our data elucidate a previously unknown function of IgM in macrophages that explains its ability to act as a novel regulator of Bip to participate in endoplasmic reticulum stress and further regulate the inflammatory response.
Full article
►▼
Show Figures
Open AccessArticle
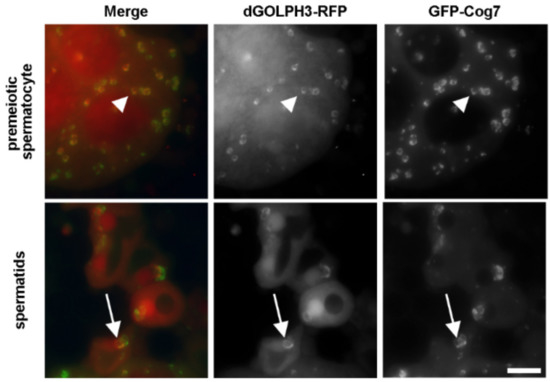
Identification of GOLPH3 Partners in Drosophila Unveils Potential Novel Roles in Tumorigenesis and Neural Disorders
by
Stefano Sechi, Angela Karimpour-Ghahnavieh, Anna Frappaolo, Laura Di Francesco, Roberto Piergentili, Eugenia Schininà, Pier Paolo D’Avino and Maria Grazia Giansanti
Cited by 10 | Viewed by 5009
Abstract
Golgi phosphoprotein 3 (GOLPH3) is a highly conserved peripheral membrane protein localized to the Golgi apparatus and the cytosol. GOLPH3 binding to Golgi membranes depends on phosphatidylinositol 4-phosphate [PI(4)P] and regulates Golgi architecture and vesicle trafficking. GOLPH3 overexpression has been correlated with poor
[...] Read more.
Golgi phosphoprotein 3 (GOLPH3) is a highly conserved peripheral membrane protein localized to the Golgi apparatus and the cytosol. GOLPH3 binding to Golgi membranes depends on phosphatidylinositol 4-phosphate [PI(4)P] and regulates Golgi architecture and vesicle trafficking. GOLPH3 overexpression has been correlated with poor prognosis in several cancers, but the molecular mechanisms that link GOLPH3 to malignant transformation are poorly understood. We recently showed that PI(4)P-GOLPH3 couples membrane trafficking with contractile ring assembly during cytokinesis in dividing
Drosophila spermatocytes. Here, we use affinity purification coupled with mass spectrometry (AP-MS) to identify the protein-protein interaction network (interactome) of
Drosophila GOLPH3 in testes. Analysis of the GOLPH3 interactome revealed enrichment for proteins involved in vesicle-mediated trafficking, cell proliferation and cytoskeleton dynamics. In particular, we found that dGOLPH3 interacts with the
Drosophila orthologs of Fragile X mental retardation protein and Ataxin-2, suggesting a potential role in the pathophysiology of disorders of the nervous system. Our findings suggest novel molecular targets associated with GOLPH3 that might be relevant for therapeutic intervention in cancers and other human diseases.
Full article
►▼
Show Figures
Open AccessBrief Report
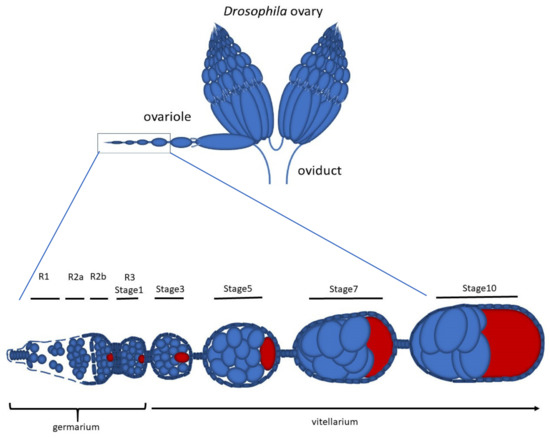
Early Drosophila Oogenesis: A Tale of Centriolar Asymmetry
by
Maria Giovanna Riparbelli, Veronica Persico and Giuliano Callaini
Cited by 4 | Viewed by 5430
Abstract
Among the morphological processes that characterize the early stages of Drosophila oogenesis, the dynamic of the centrioles deserves particular attention. We re-examined the architecture and the distribution of the centrioles within the germarium and early stages of the vitellarium. We found that most
[...] Read more.
Among the morphological processes that characterize the early stages of Drosophila oogenesis, the dynamic of the centrioles deserves particular attention. We re-examined the architecture and the distribution of the centrioles within the germarium and early stages of the vitellarium. We found that most of the germ cell centrioles diverge from the canonical model and display notable variations in size. Moreover, duplication events were frequently observed within the germarium in the absence of DNA replication. Finally, we report the presence of an unusually long centriole that is first detected in the cystoblast and is always associated with the developing oocyte. This centriole is directly inherited after the asymmetric division of the germline stem cells and persists during the process of oocyte selection, thus already representing a marker for oocyte identification at the beginning of its formation and during the ensuing developmental stages.
Full article
►▼
Show Figures
Open AccessReview
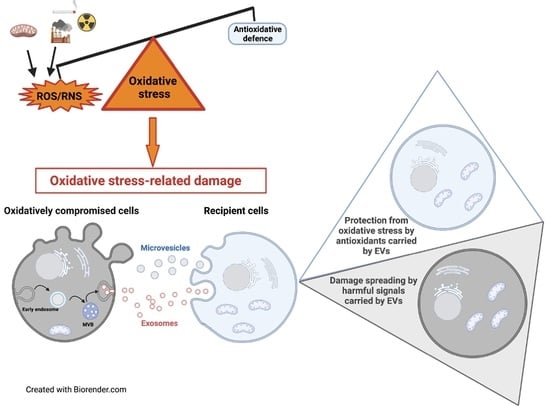
Extracellular Vesicles under Oxidative Stress Conditions: Biological Properties and Physiological Roles
by
Elisabetta Chiaradia, Brunella Tancini, Carla Emiliani, Federica Delo, Roberto Maria Pellegrino, Alessia Tognoloni, Lorena Urbanelli and Sandra Buratta
Cited by 172 | Viewed by 9743
Abstract
Under physio-pathological conditions, cells release membrane-surrounded structures named Extracellular Vesicles (EVs), which convey their molecular cargo to neighboring or distant cells influencing their metabolism. Besides their involvement in the intercellular communication, EVs might represent a tool used by cells to eliminate unnecessary/toxic material.
[...] Read more.
Under physio-pathological conditions, cells release membrane-surrounded structures named Extracellular Vesicles (EVs), which convey their molecular cargo to neighboring or distant cells influencing their metabolism. Besides their involvement in the intercellular communication, EVs might represent a tool used by cells to eliminate unnecessary/toxic material. Here, we revised the literature exploring the link between EVs and redox biology. The first proof of this link derives from evidence demonstrating that EVs from healthy cells protect target cells from oxidative insults through the transfer of antioxidants. Oxidative stress conditions influence the release and the molecular cargo of EVs that, in turn, modulate the redox status of target cells. Oxidative stress-related EVs exert both beneficial or harmful effects, as they can carry antioxidants or ROS-generating enzymes and oxidized molecules. As mediators of cell-to-cell communication, EVs are also implicated in the pathophysiology of oxidative stress-related diseases. The review found evidence that numerous studies speculated on the role of EVs in redox signaling and oxidative stress-related pathologies, but few of them unraveled molecular mechanisms behind this complex link. Thus, the purpose of this review is to report and discuss this evidence, highlighting that the analysis of the molecular content of oxidative stress-released EVs (reminiscent of the redox status of originating cells), is a starting point for the use of EVs as diagnostic and therapeutic tools in oxidative stress-related diseases.
Full article
►▼
Show Figures
Open AccessArticle
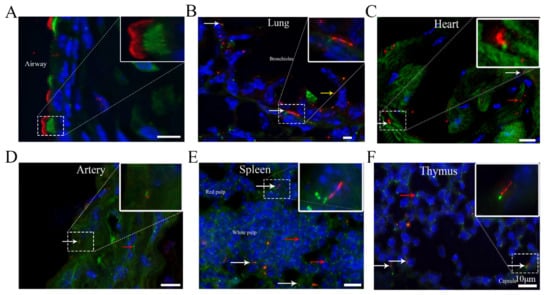
Identification of Cilia in Different Mouse Tissues
by
Xinhua Li, Shuting Yang, Vishwa Deepak, Zahra Chinipardaz and Shuying Yang
Cited by 27 | Viewed by 6350
Abstract
Cilia are microtubule-based hair-like organelles that extend from the cell surface. However, the existence and distribution of cilia in each organ and tissue at the postnatal stage in vivo remain largely unknown. In this study, we defined cilia distribution and arrangement and measured
[...] Read more.
Cilia are microtubule-based hair-like organelles that extend from the cell surface. However, the existence and distribution of cilia in each organ and tissue at the postnatal stage in vivo remain largely unknown. In this study, we defined cilia distribution and arrangement and measured the ciliary lengths and the percentage of ciliated cells in different organs and tissues in vivo by using cilium dual reporter-expressing transgenic mice. Cilia were identified by the presence of ARL13B with an mCherry+ signal, and the cilium basal body was identified by the presence of Centrin2 with a GFP+ signal. Here, we provide in vivo evidence that chondrocytes and cells throughout bones have cilia. Most importantly, we reveal that: 1. primary cilia are present in hepatocytes; 2. no cilia but many centrioles are distributed on the apical cell surface in the gallbladder, intestine, and thyroid epithelia; 3. cilia on the cerebral cortex are well oriented, pointing to the center of the brain; 4. ARL13B+ inclusion is evident in the thyroid and islets of Langerhans; and 5. approximately 2% of cilia show irregular movement in nucleus pulposus extracellular fluid. This study reveals the existence and distribution of cilia and centrioles in different tissues and organs, and provides new insights for further comprehensive study of ciliary function in these organs and tissues.
Full article
►▼
Show Figures
Open AccessFeature PaperArticle
A Novel FRET Approach Quantifies the Interaction Strength of Peroxisomal Targeting Signals and Their Receptor in Living Cells
by
Bernhard Hochreiter, Cheng-Shoong Chong, Andreas Hartig, Sebastian Maurer-Stroh, Johannes Berger, Johannes A. Schmid and Markus Kunze
Cited by 10 | Viewed by 4785
Abstract
Measuring Förster–resonance–energy–transfer (FRET) efficiency allows the investigation of protein–protein interactions (PPI), but extracting quantitative measures of affinity necessitates highly advanced technical equipment or isolated proteins. We demonstrate the validity of a recently suggested novel approach to quantitatively analyze FRET-based experiments in living mammalian
[...] Read more.
Measuring Förster–resonance–energy–transfer (FRET) efficiency allows the investigation of protein–protein interactions (PPI), but extracting quantitative measures of affinity necessitates highly advanced technical equipment or isolated proteins. We demonstrate the validity of a recently suggested novel approach to quantitatively analyze FRET-based experiments in living mammalian cells using standard equipment using the interaction between different type-1 peroxisomal targeting signals (PTS1) and their soluble receptor peroxin 5 (PEX5) as a model system. Large data sets were obtained by flow cytometry coupled FRET measurements of cells expressing PTS1-tagged EGFP together with mCherry fused to the PTS1-binding domain of PEX5, and were subjected to a fitting algorithm extracting a quantitative measure of the interaction strength. This measure correlates with results obtained by in vitro techniques and a two-hybrid assay, but is unaffected by the distance between the fluorophores. Moreover, we introduce a live cell competition assay based on this approach, capable of depicting dose- and affinity-dependent modulation of the PPI. Using this system, we demonstrate the relevance of a sequence element next to the core tripeptide in PTS1 motifs for the interaction strength between PTS1 and PEX5, which is supported by a structure-based computational prediction of the binding energy indicating a direct involvement of this sequence in the interaction.
Full article
►▼
Show Figures
Open AccessArticle
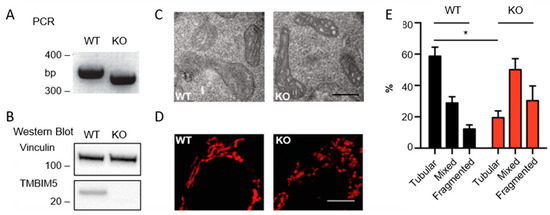
Transmembrane BAX Inhibitor-1 Motif Containing Protein 5 (TMBIM5) Sustains Mitochondrial Structure, Shape, and Function by Impacting the Mitochondrial Protein Synthesis Machinery
by
Bruno Seitaj, Felicia Maull, Li Zhang, Verena Wüllner, Christina Wolf, Philipp Schippers, Rita La Rovere, Ute Distler, Stefan Tenzer, Jan B. Parys, Geert Bultynck and Axel Methner
Cited by 17 | Viewed by 6439
Abstract
The Transmembrane Bax Inhibitor-1 motif (TMBIM)-containing protein family is evolutionarily conserved and has been implicated in cell death susceptibility. The only member with a mitochondrial localization is TMBIM5 (also known as GHITM or MICS1), which affects cristae organization and associates with the Parkinson’s
[...] Read more.
The Transmembrane Bax Inhibitor-1 motif (TMBIM)-containing protein family is evolutionarily conserved and has been implicated in cell death susceptibility. The only member with a mitochondrial localization is TMBIM5 (also known as GHITM or MICS1), which affects cristae organization and associates with the Parkinson’s disease-associated protein CHCHD2 in the inner mitochondrial membrane. We here used CRISPR-Cas9-mediated knockout HAP1 cells to shed further light on the function of TMBIM5 in physiology and cell death susceptibility. We found that compared to wild type,
TMBIM5-knockout cells were smaller and had a slower proliferation rate. In these cells, mitochondria were more fragmented with a vacuolar cristae structure. In addition, the mitochondrial membrane potential was reduced and respiration was attenuated, leading to a reduced mitochondrial ATP generation. TMBIM5 did not associate with Mic10 and Mic60, which are proteins of the mitochondrial contact site and cristae organizing system (MICOS), nor did
TMBIM5 knockout affect their expression levels.
TMBIM5-knockout cells were more sensitive to apoptosis elicited by staurosporine and BH3 mimetic inhibitors of Bcl-2 and Bcl-XL. An unbiased proteomic comparison identified a dramatic downregulation of proteins involved in the mitochondrial protein synthesis machinery in TMBIM5-knockout cells. We conclude that TMBIM5 is important to maintain the mitochondrial structure and function possibly through the control of mitochondrial biogenesis.
Full article
►▼
Show Figures