New Advances in Proteomics in Cancer
A special issue of Cells (ISSN 2073-4409). This special issue belongs to the section "Cell Methods".
Deadline for manuscript submissions: closed (30 November 2023) | Viewed by 12945
Special Issue Editors
2. Department of Bioengineering and Therapeutic Sciences, University of California, San Francisco, CA, USA
Interests: surface proteomics; drug target discovery; cellular therapy development; mechanisms of therapy resistance in cancer
Interests: clinical proteomics; biomarker discovery in infectious diseases and cancer
Special Issues, Collections and Topics in MDPI journals
Special Issue Information
Dear Colleagues,
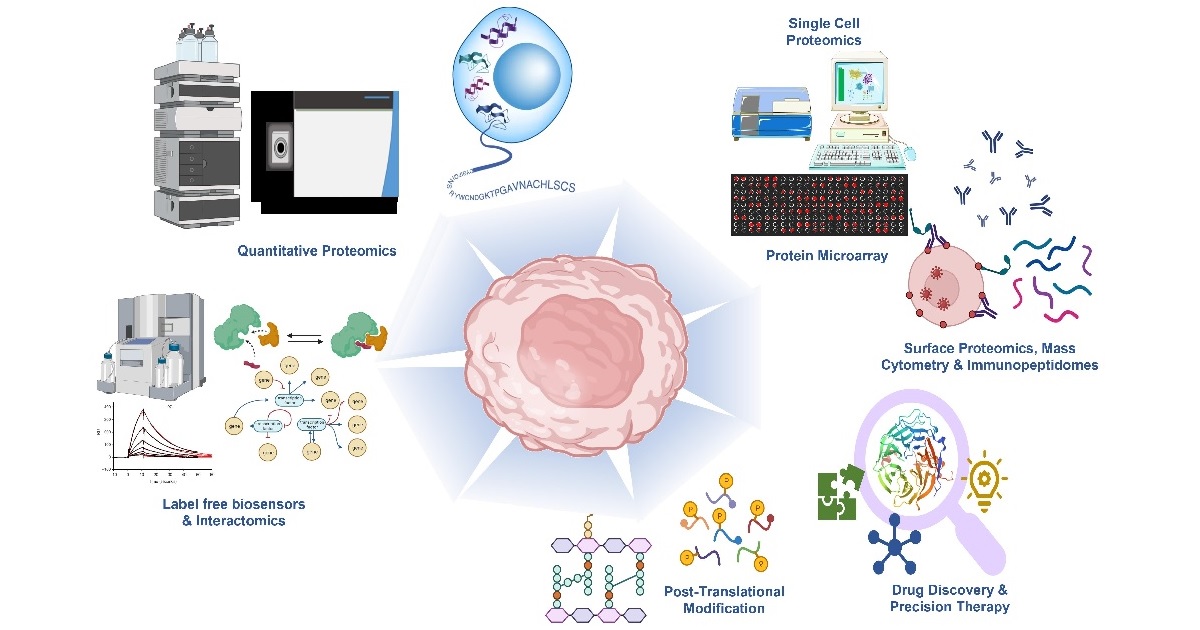
Proteomic technologies have rapidly expanded over the past two decades to address the crucial observation: Biology Happens at the Protein Level. In cancer biology, we have seen proteomic approaches play a foundational role in uncovering potential mechanisms of cancer initiation, metastasis, and treatment, as well as biomarker discovery and precision medicine strategies. These achievements have come through the systematic and extensive high-throughput analysis of protein expression profiles, protein sequences, structures, variants, PTMs, and functional effects. The emergence of fields featuring multi-omics integration, such as proteogenomics, as well as the incorporation of artificial intelligence into data analysis, have unlocked new capabilities of acquired datasets to provide biological insight into cancer. The goal of this Special Issue is to showcase key advances on applying proteomic approaches to the study of cancer, both related to advances in underlying technologies and key biological discoveries. The topics to be included, but not limited to, are proteomic research with a cancer focus involving the development of high-throughput proteomic methods, proteomics-based biological and clinical findings, small molecule and immunotherapeutic target discovery, therapeutic mechanism of action, cancer immunology, advancements in sample preparation techniques, PTM analysis, single cell analysis, comprehensive metadata analysis, AI/statistics technology, etc. While tandem mass spectrometry-based strategies will be emphasized, other important technologies such as protein microarrays, mass cytometry, and direct protein sequencing will also be included. Interdisciplinary research is highly valued and encouraged. We welcome the submission of both original research articles and reviews.
Dr. Arun Wiita
Prof. Dr. Sanjeeva Srivastava
Guest Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the special issue website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cells is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- proteomics
- cancer
- biomarkers
- surface proteomics
- immunotherapeutic
- cancer immunology
- proteogenomics
- protein microarrays
- mass cytometry
- single cell proteomics
- immunopeptidomics
- label-free detection platforms
- sample prep for HT cancer proteomics
Benefits of Publishing in a Special Issue
- Ease of navigation: Grouping papers by topic helps scholars navigate broad scope journals more efficiently.
- Greater discoverability: Special Issues support the reach and impact of scientific research. Articles in Special Issues are more discoverable and cited more frequently.
- Expansion of research network: Special Issues facilitate connections among authors, fostering scientific collaborations.
- External promotion: Articles in Special Issues are often promoted through the journal's social media, increasing their visibility.
- Reprint: MDPI Books provides the opportunity to republish successful Special Issues in book format, both online and in print.
Further information on MDPI's Special Issue policies can be found here.







