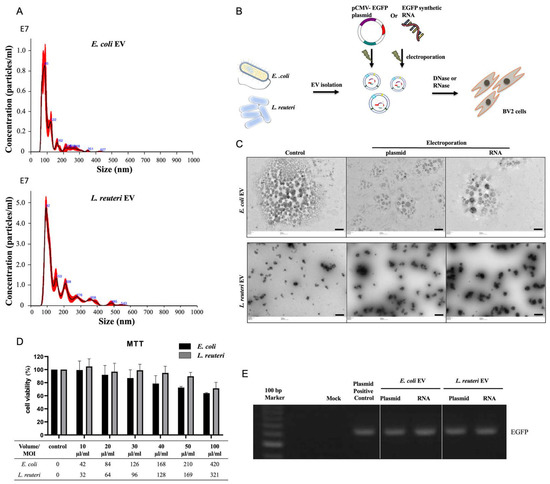
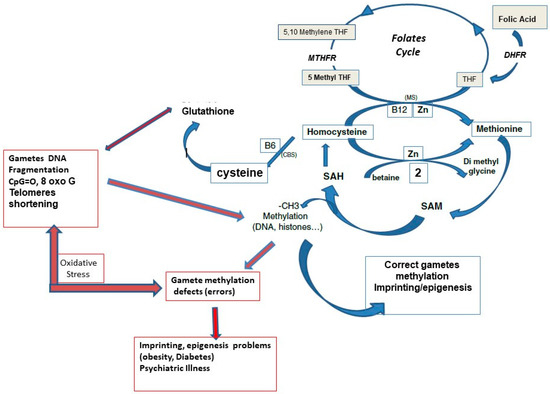
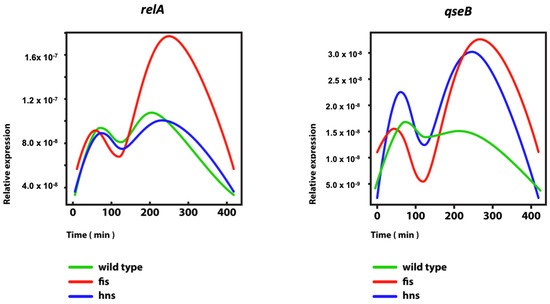
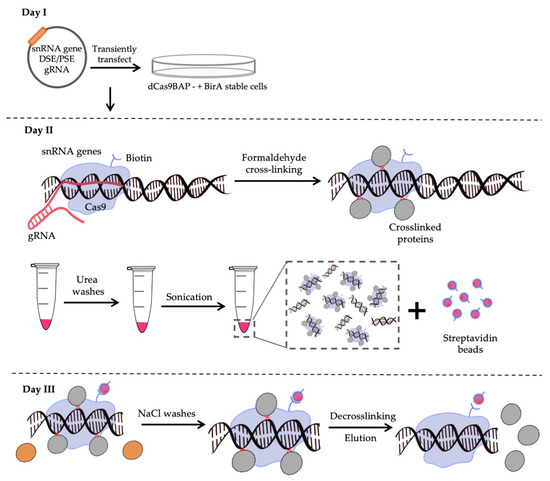
Feature Papers in Molecular Genetics
A topical collection in Biomolecules (ISSN 2218-273X). This collection belongs to the section "Molecular Genetics".
Viewed by 121608Editor
Interests: gene regulation; genomics; transcriptomics; non-coding RNAs; genome function and evolution; fission yeast; cellular quiescence and ageing
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
This Topical Collection, “Feature Papers in Molecular Genetics”, will bring together high-quality research articles, review articles, and communications on all aspects of molecular genetics. It is dedicated to diverse recent advances in genetics research, as highlighted in the topics below, and comprises a selection of exclusive papers from the Editorial Board Members (EBMs) of the Molecular Genetics Section as well as invited papers from relevant experts. We also welcome established experts in the field to make contributions to this Topical Collection. Please note that all invited papers will be published online once accepted. We aim to represent our Section as an attractive open access publishing platform for molecular genetics research.
Topics include, without being limited to:
Chromosome biology;
Epigenetics;
Gene editing and genetic variants;
Gene regulation and transcriptomics;
Genetic assays and methods;
Genetic screens and gene-function analyses;
Genome function and evolution;
Genotype-phenotype relationships;
Medical genetics and disease biology;
Molecular evolution;
Non-coding RNAs;
Population genetics and complex traits;
Synthetic biology.
Prof. Dr. Jürg Bähler
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Biomolecules is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.