Divergent Pharmacology and Biased Signaling of the Four Melanocortin-4 Receptor Isoforms in Rainbow Trout (Oncorhynchus mykiss)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ligands and Plasmids
2.2. Homology, Phylogenetic, and Chromosome Synteny Analyses
2.3. Cell Culture and Transfection
2.4. Flow Cytometry Assay
2.5. Ligand-Binding Assay
2.6. cAMP Assay
2.7. ERK1/2 Phosphorylation Assay
2.8. Statistical Analysis
3. Results
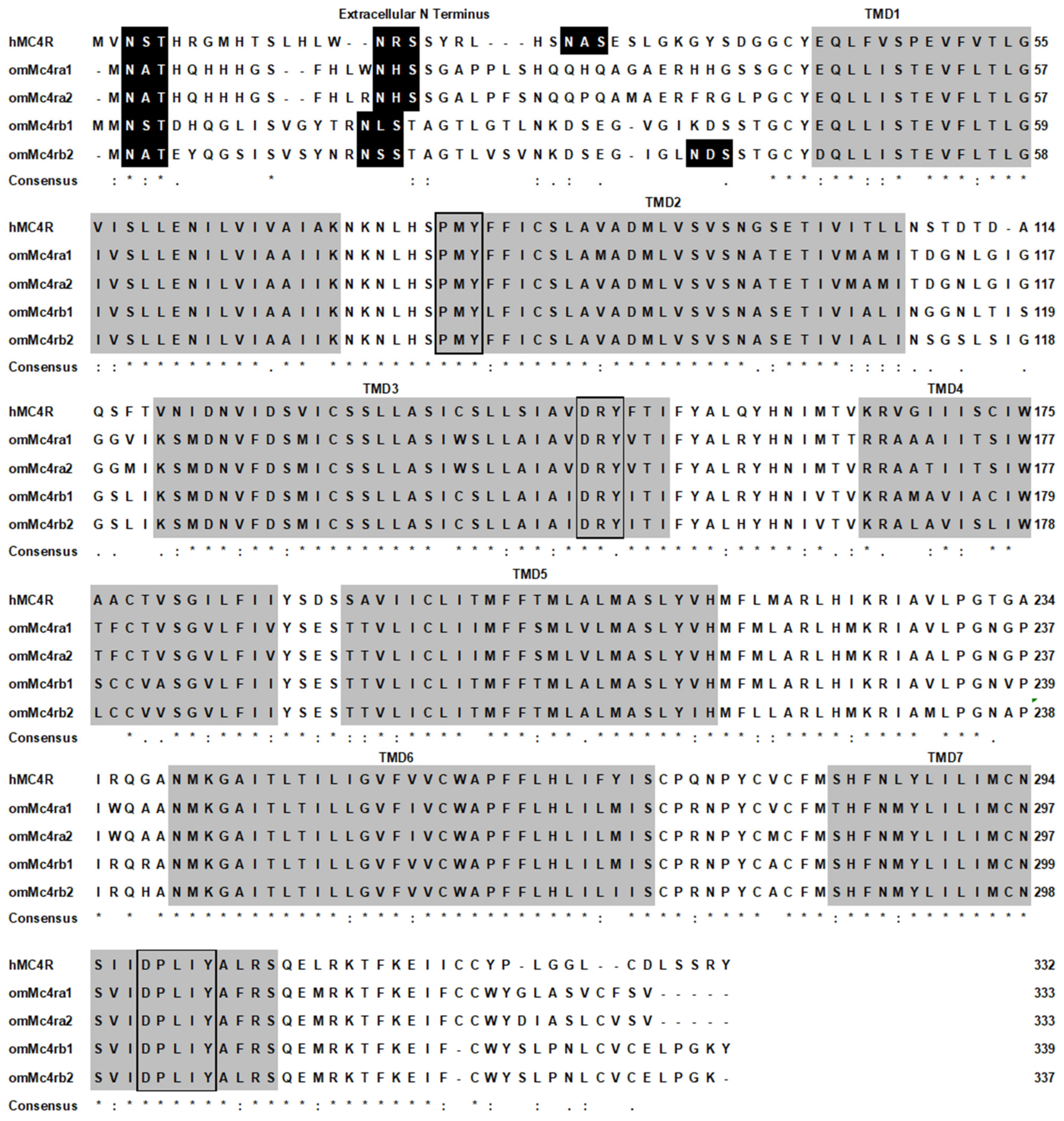
3.1. Nucleotide and Deduced Amino Acid Sequences of Trout Mc4rs
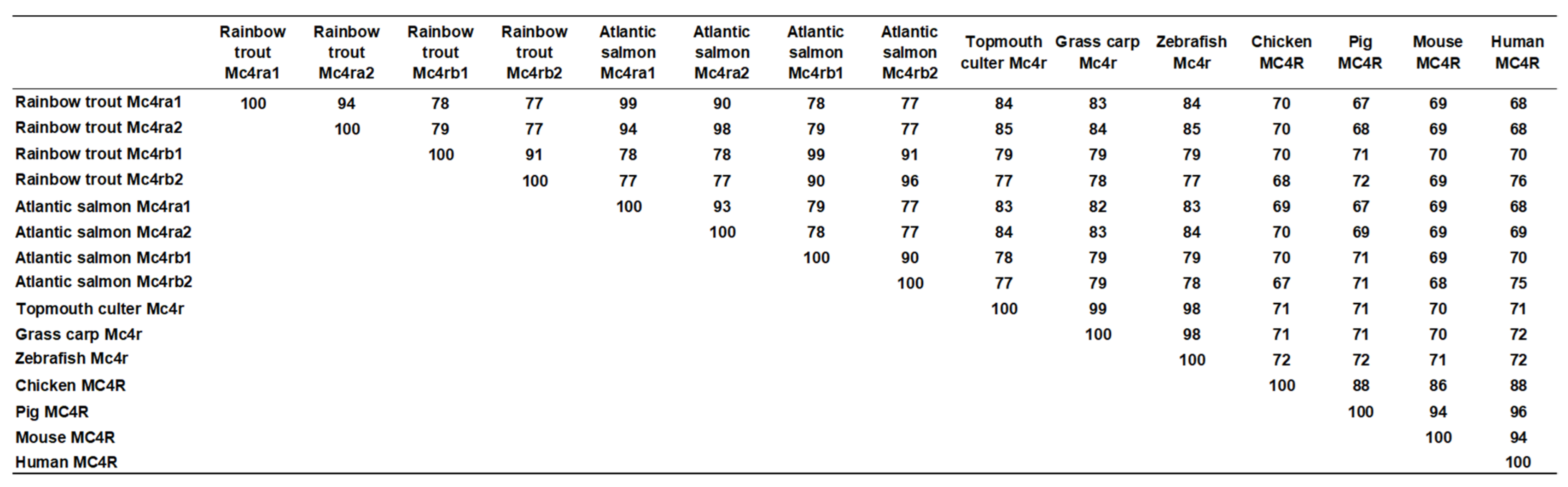
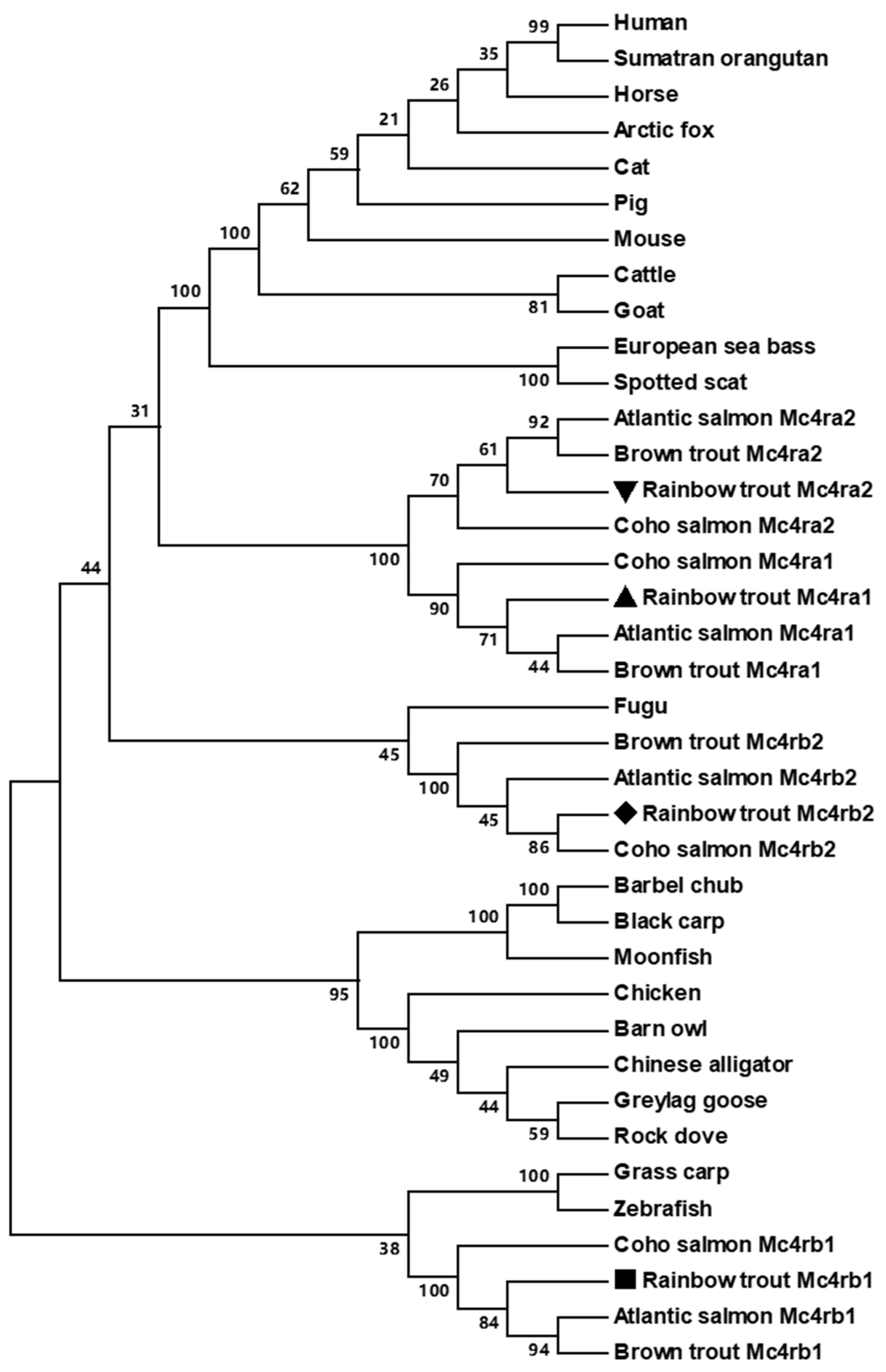
3.2. Chromosome Synteny and Phylogenetic Analyses of Trout Mc4rs
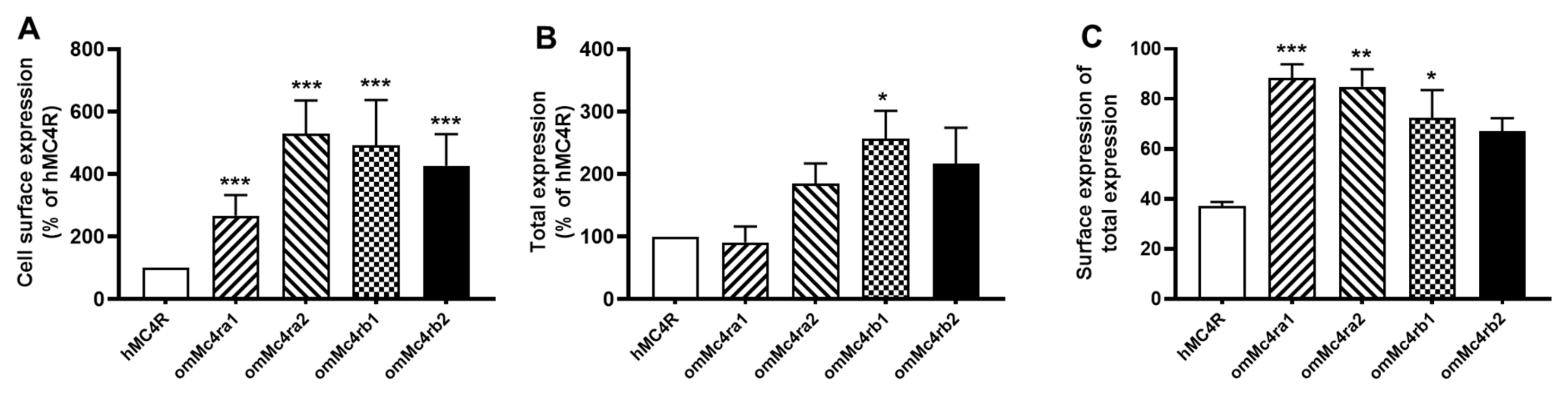
3.3. Cell-Surface and Total Expression of omMc4rs
3.4. Ligand-Binding Properties of omMc4rs
3.5. Signaling Properties of omMc4rs
3.6. Constitutive Activities of omMc4rs in Response to Four Antagonists
3.7. ERK1/2 Signaling Properties of omMc4rs in Response to Five Ligands
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tao, Y.X. The melanocortin-4 receptor: Physiology, pharmacology, and pathophysiology. Endocr. Rev. 2010, 31, 506–543. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.X. Molecular mechanisms of the neural melanocortin receptor dysfunction in severe early onset obesity. Mol. Cell. Endocrinol. 2005, 239, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Ji, R.L.; Tao, Y.X. Naturally occurring mutations in G protein-coupled receptors associated with obesity and type 2 diabetes mellitus. Pharmacol. Ther. 2022, 234, 108044. [Google Scholar] [CrossRef]
- Huszar, D.; Lynch, C.A.; Fairchild-Huntress, V.; Dunmore, J.H.; Fang, Q.; Berkemeier, L.R.; Gu, W.; Kesterson, R.A.; Boston, B.A.; Cone, R.D.; et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 1997, 88, 131–141. [Google Scholar] [CrossRef]
- Balthasar, N.; Dalgaard, L.T.; Lee, C.E.; Yu, J.; Funahashi, H.; Williams, T.; Ferreira, M.; Tang, V.; McGovern, R.A.; Kenny, C.D.; et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell 2005, 123, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, I.S.; Keogh, J.M.; Yeo, G.S.; Lank, E.J.; Cheetham, T.; O’Rahilly, S. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N. Engl. J. Med. 2003, 348, 1085–1095. [Google Scholar] [CrossRef]
- Tao, Y.X. Mutations in melanocortin-4 receptor and human obesity. Prog. Mol. Biol. Transl. Sci. 2009, 88, 173–204. [Google Scholar] [PubMed]
- Hinney, A.; Volckmar, A.L.; Knoll, N. Melanocortin-4 receptor in energy homeostasis and obesity pathogenesis. Prog. Mol. Biol. Transl. Sci. 2013, 114, 147–191. [Google Scholar] [CrossRef]
- Tao, Y.X. Mutations in melanocortin-4 receptor: From fish to men. Prog. Mol. Biol. Transl. Sci. 2022, 189, 215–257. [Google Scholar] [CrossRef]
- Ringholm, A.; Fredriksson, R.; Poliakova, N.; Yan, Y.L.; Postlethwait, J.H.; Larhammar, D.; Schioth, H.B. One melanocortin 4 and two melanocortin 5 receptors from zebrafish show remarkable conservation in structure and pharmacology. J. Neurochem. 2002, 82, 6–18. [Google Scholar] [CrossRef]
- Cerdá-Reverter, J.M.; Ringholm, A.; Schioth, H.B.; Peter, R.E. Molecular cloning, pharmacological characterization, and brain mapping of the melanocortin 4 receptor in the goldfish: Involvement in the control of food intake. Endocrinology 2003, 144, 2336–2349. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Tsuchiya, K.; Yamanome, T.; Schioth, H.B.; Kawauchi, H.; Takahashi, A. Food deprivation increases the expression of melanocortin-4 receptor in the liver of barfin flounder, Verasper moseri. Gen. Comp. Endocrinol. 2008, 155, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Li, J.T.; Yang, Z.; Chen, H.P.; Zhu, C.H.; Deng, S.P.; Li, G.L.; Tao, Y.X. Molecular cloning, tissue distribution, and pharmacological characterization of melanocortin-4 receptor in spotted scat, Scatophagus argus. Gen. Comp. Endocrinol. 2016, 230–231, 143–152. [Google Scholar] [CrossRef]
- Li, L.; Yang, Z.; Zhang, Y.P.; He, S.; Liang, X.F.; Tao, Y.X. Molecular cloning, tissue distribution, and pharmacological characterization of melanocortin-4 receptor in grass carp (Ctenopharyngodon idella). Domest. Anim. Endocrinol. 2017, 59, 140–151. [Google Scholar] [CrossRef]
- Yi, T.L.; Yang, L.K.; Ruan, G.L.; Yang, D.Q.; Tao, Y.X. Melanocortin-4 receptor in swamp eel (Monopterus albus): Cloning, tissue distribution, and pharmacology. Gene 2018, 678, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.Z.; Chen, R.; Zhang, Y.; Tao, Y.X. Orange-spotted grouper melanocortin-4 receptor: Modulation of signaling by MRAP2. Gen. Comp. Endocrinol. 2019, 284, 113234. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, Y.J.; Zhu, M.; Xu, B.X.; Guo, W.X.; Lyu, Y.S.; Zhang, C. Pharmacological modulation of melanocortin-4 receptor by melanocortin receptor accessory protein 2 in Nile tilapia. Gen. Comp. Endocrinol. 2019, 282, 113219. [Google Scholar] [CrossRef]
- Tao, M.; Ji, R.L.; Huang, L.; Fan, S.Y.; Liu, T.; Liu, S.J.; Tao, Y.X. Regulation of melanocortin-4 receptor pharmacology by two isoforms of melanocortin receptor accessory protein 2 in topmouth culter (Culter alburnus). Front. Endocrinol. 2020, 11, 538. [Google Scholar] [CrossRef]
- Wen, Z.Y.; Liu, T.; Qin, C.J.; Zou, Y.C.; Wang, J.; Li, R.; Tao, Y.X. MRAP2 interaction with melanocortin-4 receptor in snakehead (Channa argus). Biomolecules 2021, 11, 481. [Google Scholar] [CrossRef]
- Cerdá-Reverter, J.M.; Peter, R.E. Endogenous melanocortin antagonist in fish: Structure, brain mapping, and regulation by fasting of the goldfish agouti-related protein gene. Endocrinology 2003, 144, 4552–4561. [Google Scholar] [CrossRef]
- Cerdá-Reverter, J.M.; Schioth, H.B.; Peter, R.E. The central melanocortin system regulates food intake in goldfish. Regul. Pept. 2003, 115, 101–113. [Google Scholar] [CrossRef]
- Schjolden, J.; Schioth, H.B.; Larhammar, D.; Winberg, S.; Larson, E.T. Melanocortin peptides affect the motivation to feed in rainbow trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 2009, 160, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Aspiras, A.C.; Rohner, N.; Martineau, B.; Borowsky, R.L.; Tabin, C.J. Melanocortin 4 receptor mutations contribute to the adaptation of cavefish to nutrient-poor conditions. Proc. Natl. Acad. Sci. USA 2015, 112, 9668–9673. [Google Scholar] [CrossRef] [PubMed]
- Lampert, K.P.; Schmidt, C.; Fischer, P.; Volff, J.N.; Hoffmann, C.; Muck, J.; Lohse, M.J.; Ryan, M.J.; Schartl, M. Determination of onset of sexual maturation and mating behavior by melanocortin receptor 4 polymorphisms. Curr. Biol. 2010, 20, 1729–1734. [Google Scholar] [CrossRef]
- Smith, C.C.; Harris, R.M.; Lampert, K.P.; Schartl, M.; Hofmann, H.A.; Ryan, M.J. Copy number variation in the melanocortin 4 receptor gene and alternative reproductive tactics the swordtail Xiphophorus multilineatus. Environ. Biol. Fishes 2015, 98, 23–33. [Google Scholar] [CrossRef]
- Jiang, D.N.; Li, J.T.; Tao, Y.X.; Chen, H.P.; Deng, S.P.; Zhu, C.H.; Li, G.L. Effects of melanocortin-4 receptor agonists and antagonists on expression of genes related to reproduction in spotted scat, Scatophagus argus. J. Comp. Physiol. B 2017, 187, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wen, H.S.; Li, Y.; Lyu, L.K.; Zhang, Z.X.; Wang, X.J.; Li, J.S.; Tao, Y.X.; Qi, X. Melanocortin-4 receptor regulation of reproductive function in black rockfish (Sebastes schlegelii). Gene 2020, 741, 144541. [Google Scholar] [CrossRef]
- Nijenhuis, W.A.; Oosterom, J.; Adan, R.A. AgRP(83–132) acts as an inverse agonist on the human melanocortin-4 receptor. Mol. Endocrinol. 2001, 15, 164–171. [Google Scholar]
- Tao, Y.X. Constitutive activity in melanocortin-4 receptor: Biased signaling of inverse agonists. Adv. Pharmacol. 2014, 70, 135–154. [Google Scholar] [CrossRef]
- Srinivasan, S.; Lubrano-Berthelier, C.; Govaerts, C.; Picard, F.; Santiago, P.; Conklin, B.R.; Vaisse, C. Constitutive activity of the melanocortin-4 receptor is maintained by its N-terminal domain and plays a role in energy homeostasis in humans. J. Clin. Investig. 2004, 114, 1158–1164. [Google Scholar] [CrossRef]
- Tao, Y.X.; Segaloff, D.L. Functional analyses of melanocortin-4 receptor mutations identified from patients with binge eating disorder and nonobese or obese subjects. J. Clin. Endocrinol. Metab. 2005, 90, 5632–5638. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.X. Constitutive activation of G protein-coupled receptors and diseases: Insights into mechanism of activation and therapeutics. Pharmacol. Ther. 2008, 120, 129–148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Song, Y.; Thompson, D.A.; Madonna, M.A.; Millhauser, G.L.; Toro, S.; Varga, Z.; Westerfield, M.; Gamse, J.; Chen, W.; et al. Pineal-specific agouti protein regulates teleost background adaptation. Proc. Natl. Acad. Sci. USA 2010, 107, 20164–20171. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.Q.; Hou, Z.S.; Wen, H.S.; Li, Y.; Qi, X.; Li, W.J.; Tao, Y.X. Melanocortin-4 receptor in spotted sea bass, Lateolabrax maculatus: Cloning, tissue distribution, physiology, and pharmacology. Front. Endocrinol. 2019, 10, 705. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Forlano, P.M.; Cone, R.D. AgRP and POMC neurons are hypophysiotropic and coordinately regulate multiple endocrine axes in a larval teleost. Cell Metab. 2012, 15, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Sebag, J.A.; Zhang, C.; Hinkle, P.M.; Bradshaw, A.M.; Cone, R.D. Developmental control of the melanocortin-4 receptor by MRAP2 proteins in zebrafish. Science 2013, 341, 278–281. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.L.; Huang, L.; Wang, Y.; Liu, T.; Fan, S.Y.; Tao, M.; Tao, Y.X. Topmouth culter melanocortin-3 receptor: Regulation by two isoforms of melanocortin-2 receptor accessory protein 2. Endocr. Connect. 2021, 10, 1489–1501. [Google Scholar] [CrossRef]
- Rajagopal, S.; Rajagopal, K.; Lefkowitz, R.J. Teaching old receptors new tricks: Biasing seven-transmembrane receptors. Nat. Rev. Drug Discov. 2010, 9, 373–386. [Google Scholar] [CrossRef]
- Reiter, E.; Ahn, S.; Shukla, A.K.; Lefkowitz, R.J. Molecular mechanism of β-arrestin-biased agonism at seven-transmembrane receptors. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 179–197. [Google Scholar] [CrossRef]
- Kenakin, T.; Christopoulos, A. Signalling bias in new drug discovery: Detection, quantification and therapeutic impact. Nat. Rev. Drug Discov. 2013, 12, 205–216. [Google Scholar] [CrossRef]
- Smith, J.S.; Lefkowitz, R.J.; Rajagopal, S. Biased signalling: From simple switches to allosteric microprocessors. Nat. Rev. Drug Discov. 2018, 17, 243–260. [Google Scholar] [CrossRef] [PubMed]
- Kenakin, T. Biased receptor signaling in drug discovery. Pharmacol. Rev. 2019, 71, 267–315. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.K.; Hou, Z.S.; Tao, Y.X. Biased signaling in naturally occurring mutations of G protein-coupled receptors associated with diverse human diseases. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 165973. [Google Scholar] [CrossRef] [PubMed]
- Daniels, D.; Patten, C.S.; Roth, J.D.; Yee, D.K.; Fluharty, S.J. Melanocortin receptor signaling through mitogen-activated protein kinase in vitro and in rat hypothalamus. Brain Res. 2003, 986, 1–11. [Google Scholar] [CrossRef]
- Vongs, A.; Lynn, N.M.; Rosenblum, C.I. Activation of MAP kinase by MC4-R through PI3 kinase. Regul. Pept. 2004, 120, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Sutton, G.M.; Duos, B.; Patterson, L.M.; Berthoud, H.R. Melanocortinergic modulation of cholecystokinin-induced suppression of feeding through extracellular signal-regulated kinase signaling in rat solitary nucleus. Endocrinology 2005, 146, 3739–3747. [Google Scholar] [CrossRef] [PubMed]
- Chai, B.; Li, J.Y.; Zhang, W.; Newman, E.; Ammori, J.; Mulholland, M.W. Melanocortin-4 receptor-mediated inhibition of apoptosis in immortalized hypothalamic neurons via mitogen-activated protein kinase. Peptides 2006, 27, 2846–2857. [Google Scholar] [CrossRef]
- Mo, X.L.; Tao, Y.X. Activation of MAPK by inverse agonists in six naturally occurring constitutively active mutant human melanocortin-4 receptors. Biochim. Biophys. Acta 2013, 1832, 1939–1948. [Google Scholar] [CrossRef]
- He, S.; Tao, Y.X. Defect in MAPK signaling as a cause for monogenic obesity caused by inactivating mutations in the melanocortin-4 receptor gene. Int. J. Biol. Sci. 2014, 10, 1128–1137. [Google Scholar] [CrossRef]
- Yang, L.K.; Tao, Y.X. Biased signaling at neural melanocortin receptors in regulation of energy homeostasis. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2486–2495. [Google Scholar] [CrossRef]
- Yang, Z.; Liang, X.F.; Li, G.L.; Tao, Y.X. Biased signaling in fish melanocortin-4 receptors (MC4Rs): Divergent pharmacology of four ligands on spotted scat (Scatophagus argus) and grass carp (Ctenopharyngodon idella) MC4Rs. Mol. Cell. Endocrinol. 2020, 515, 110929. [Google Scholar] [CrossRef] [PubMed]
- Thorgaard, G.H.; Bailey, G.S.; Williams, D.; Buhler, D.R.; Kaattari, S.L.; Ristow, S.S.; Hansen, J.D.; Winton, J.R.; Bartholomew, J.L.; Nagler, J.J.; et al. Status and opportunities for genomics research with rainbow trout. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2002, 133, 609–646. [Google Scholar] [CrossRef] [PubMed]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noel, B.; Bento, P.; Da Silva, C.; Labadie, K.; Alberti, A.; et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 2014, 5, 3657. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Kawauchi, H. Evolution of melanocortin systems in fish. Gen. Comp. Endocrinol. 2006, 148, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Lien, S.; Koop, B.F.; Sandve, S.R.; Miller, J.R.; Kent, M.P.; Nome, T.; Hvidsten, T.R.; Leong, J.S.; Minkley, D.R.; Zimin, A.; et al. The Atlantic salmon genome provides insights into rediploidization. Nature 2016, 533, 200–205. [Google Scholar] [CrossRef]
- Chaki, S.; Hirota, S.; Funakoshi, T.; Suzuki, Y.; Suetake, S.; Okubo, T.; Ishii, T.; Nakazato, A.; Okuyama, S. Anxiolytic-like and antidepressant-like activities of MCL0129 (1-[(S)-2-(4-fluorophenyl)-2-(4-isopropylpiperadin-1-yl)ethyl]-4-[4-(2-met hoxynaphthalen-1-yl)butyl]piperazine), a novel and potent nonpeptide antagonist of the melanocortin-4 receptor. J. Pharmacol. Exp. Ther. 2003, 304, 818–826. [Google Scholar] [CrossRef]
- Vos, T.J.; Caracoti, A.; Che, J.L.; Dai, M.; Farrer, C.A.; Forsyth, N.E.; Drabic, S.V.; Horlick, R.A.; Lamppu, D.; Yowe, D.L.; et al. Identification of 2-[2-[2-(5-bromo-2- methoxyphenyl)-ethyl]-3-fluorophenyl]-4,5-dihydro-1H-imidazole (ML00253764), a small molecule melanocortin 4 receptor antagonist that effectively reduces tumor-induced weight loss in a mouse model. J. Med. Chem. 2004, 47, 1602–1604. [Google Scholar] [CrossRef]
- Poitout, L.; Brault, V.; Sackur, C.; Bernetiere, S.; Camara, J.; Plas, P.; Roubert, P. Identification of a novel series of benzimidazoles as potent and selective antagonists of the human melanocortin-4 receptor. Bioorg. Med. Chem. Lett. 2007, 17, 4464–4470. [Google Scholar] [CrossRef]
- Steiner, A.L.; Kipnis, D.M.; Utiger, R.; Parker, C. Radioimmunoassay for the measurement of adenosine 3’,5’-cyclic phosphate. Proc. Natl. Acad. Sci. USA 1969, 64, 367–373. [Google Scholar] [CrossRef]
- Mo, X.L.; Yang, R.; Tao, Y.X. Functions of transmembrane domain 3 of human melanocortin-4 receptor. J. Mol. Endocrinol. 2012, 49, 221–235. [Google Scholar] [CrossRef]
- Tao, Y.X.; Segaloff, D.L. Functional characterization of melanocortin-4 receptor mutations associated with childhood obesity. Endocrinology 2003, 144, 4544–4551. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.X. Functional characterization of novel melanocortin-3 receptor mutations identified from obese subjects. Biochim. Biophys. Acta 2007, 1772, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Okayama, H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987, 7, 2745–2752. [Google Scholar]
- Ji, R.L.; Tao, Y.X. Regulation of melanocortin-3 and -4 receptors by isoforms of melanocortin-2 receptor accessory protein 1 and 2. Biomolecules 2022, 12, 244. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.X.; Fan, Z.C.; Tao, Y.X. Functions of acidic transmembrane residues in human melanocortin-3 receptor binding and activation. Biochem. Pharmacol. 2008, 76, 520–530. [Google Scholar] [CrossRef]
- Yang, L.K.; Zhang, Z.R.; Wen, H.S.; Tao, Y.X. Characterization of channel catfish (Ictalurus punctatus) melanocortin-3 receptor reveals a potential network in regulation of energy homeostasis. Gen. Comp. Endocrinol. 2019, 277, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Tao, Y.X. Pleiotropic functions of the transmembrane domain 6 of human melanocortin-4 receptor. J. Mol. Endocrinol. 2012, 49, 237–248. [Google Scholar] [CrossRef]
- Kalananthan, T.; Lai, F.; Gomes, A.S.; Murashita, K.; Handeland, S.; Ronnestad, I. The melanocortin system in Atlantic salmon (Salmo salar L.) and its role in appetite control. Front. Neuroanat. 2020, 14, 48. [Google Scholar] [CrossRef]
- Klovins, J.; Haitina, T.; Ringholm, A.; Lowgren, M.; Fridmanis, D.; Slaidina, M.; Stier, S.; Schioth, H.B. Cloning of two melanocortin (MC) receptors in spiny dogfish: MC3 receptor in cartilaginous fish shows high affinity to ACTH-derived peptides while it has lower preference to γ-MSH. Eur. J. Biochem. 2004, 271, 4320–4331. [Google Scholar] [CrossRef]
- Klovins, J.; Haitina, T.; Fridmanis, D.; Kilianova, Z.; Kapa, I.; Fredriksson, R.; Gallo-Payet, N.; Schioth, H.B. The melanocortin system in Fugu: Determination of POMC/AGRP/MCR gene repertoire and synteny, as well as pharmacology and anatomical distribution of the MCRs. Mol. Biol. Evol. 2004, 21, 563–579. [Google Scholar] [CrossRef]
- Metz, J.R.; Peters, J.J.; Flik, G. Molecular biology and physiology of the melanocortin system in fish: A review. Gen. Comp. Endocrinol. 2006, 148, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Smit, M.J.; Vischer, H.F.; Bakker, R.A.; Jongejan, A.; Timmerman, H.; Pardo, L.; Leurs, R. Pharmacogenomic and structural analysis of constitutive g protein-coupled receptor activity. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 53–87. [Google Scholar] [CrossRef] [PubMed]
- Ersoy, B.A.; Pardo, L.; Zhang, S.; Thompson, D.A.; Millhauser, G.; Govaerts, C.; Vaisse, C. Mechanism of N-terminal modulation of activity at the melanocortin-4 receptor GPCR. Nat. Chem. Biol. 2012, 8, 725–730. [Google Scholar] [CrossRef]
- Nishi, S.; Nakabayashi, K.; Kobilka, B.; Hsueh, A.J.W. The ectodomain of the luteinizing hormone receptor interacts with exoloop 2 to constrain the transmembrane region. Studies using chimeric human and fly receptors. J. Biol. Chem. 2002, 277, 3958–3964. [Google Scholar] [CrossRef]
- Parma, J.; Van Sande, J.; Swillens, S.; Tonacchera, M.; Dumont, J.; Vassart, G. Somatic mutations causing constitutive activity of the thyrotropin receptor are the major cause of hyperfunctioning thyroid adenomas: Identification of additional mutations activating both the cyclic adenosine 3’,5’-monophosphate and inositol phosphate-Ca2+ cascades. Mol. Endocrinol. 1995, 9, 725–733. [Google Scholar] [PubMed]
- Zhang, M.; Tong, K.P.; Fremont, V.; Chen, J.; Narayan, P.; Puett, D.; Weintraub, B.D.; Szkudlinski, M.W. The extracellular domain suppresses constitutive activity of the transmembrane domain of the human TSH receptor: Implications for hormone-receptor interaction and antagonist design. Endocrinology 2000, 141, 3514–3517. [Google Scholar] [CrossRef]
- Seifert, R.; Wenzel-Seifert, K. Constitutive activity of G-protein-coupled receptors: Cause of disease and common property of wild-type receptors. Naunyn Schmiedebergs Arch. Pharmacol. 2002, 366, 381–416. [Google Scholar] [CrossRef]
- Cortes, R.; Navarro, S.; Agulleiro, M.J.; Guillot, R.; Garcia-Herranz, V.; Sanchez, E.; Cerdá-Reverter, J.M. Evolution of the melanocortin system. Gen. Comp. Endocrinol. 2014, 209, 3–10. [Google Scholar] [CrossRef]
- Chaki, S.; Okuyama, S. Involvement of melanocortin-4 receptor in anxiety and depression. Peptides 2005, 26, 1952–1964. [Google Scholar] [CrossRef]
- Tao, Y.X.; Huang, H.; Wang, Z.Q.; Yang, F.; Williams, J.N.; Nikiforovich, G.V. Constitutive activity of neural melanocortin receptors. Methods Enzymol. 2010, 484, 267–279. [Google Scholar]
- Yang, Z.; Tao, Y.X. Biased signaling initiated by agouti-related peptide through human melanocortin-3 and -4 receptors. Biochim. Biophys. Acta 2016, 1862, 1485–1494. [Google Scholar] [CrossRef]
- Yu, H.X.; Li, Y.; Zhong, D.B.; Ren, X.; Mo, H.L.; Jiang, Z.B.; Yu, J.J.; Xiong, D.M.; Liu, H.X.; Wang, L.X. The interaction of MC3R and MC4R with MRAP2a in rainbow trout (Oncorhynchus mykiss). Fish Physiol. Biochem. 2023, 49, 61–74. [Google Scholar] [CrossRef]

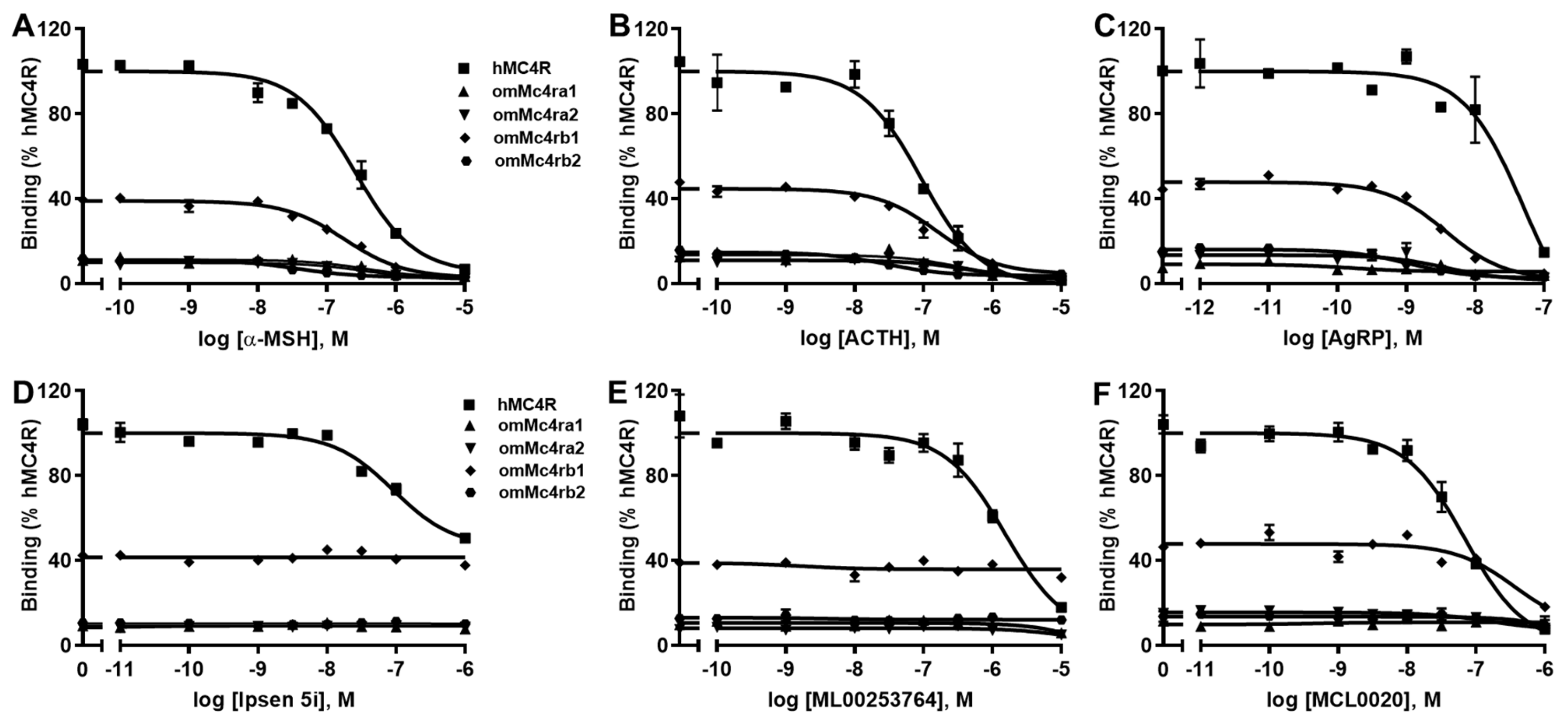

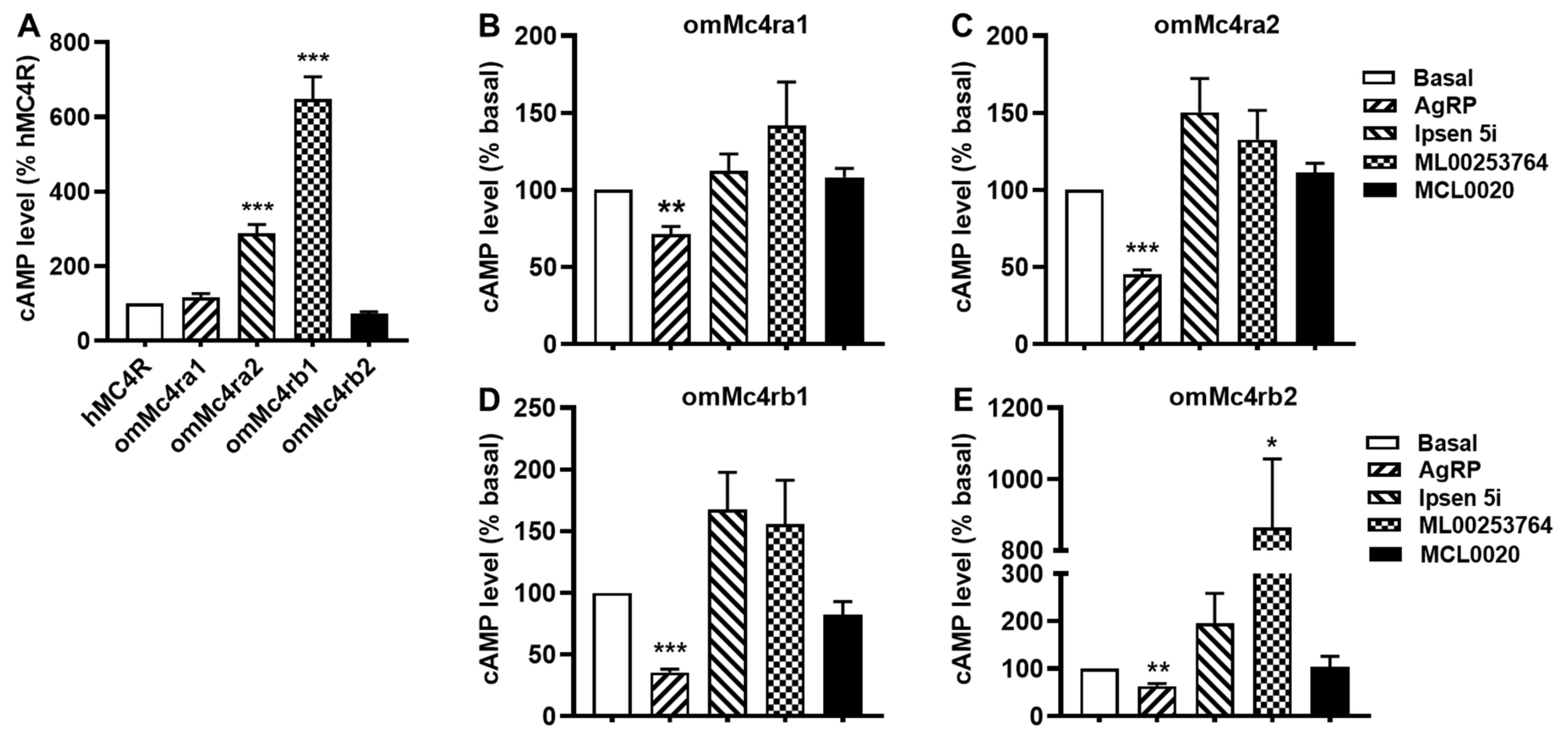
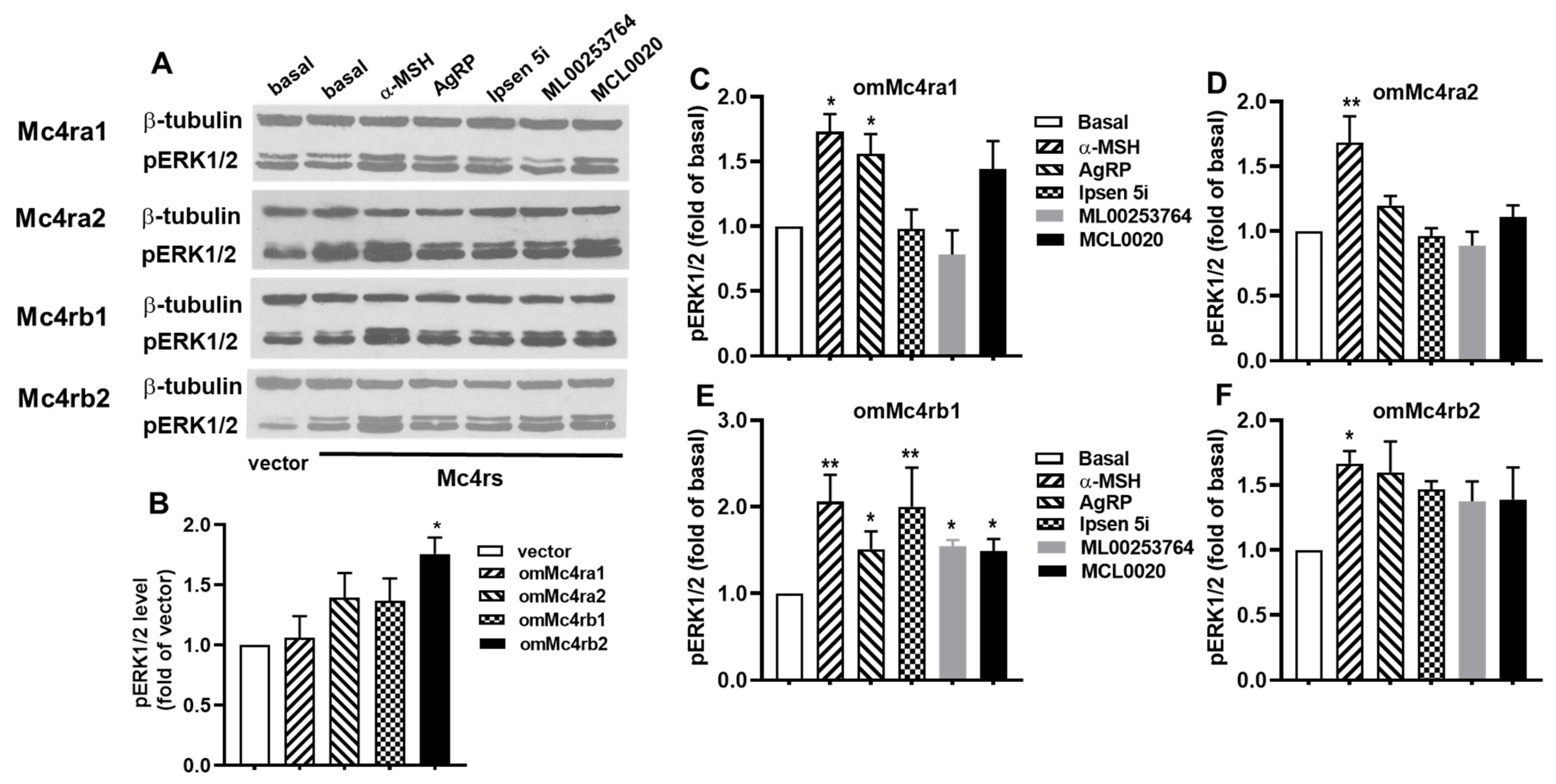
| hMC4R | omMc4ra1 | omMc4ra2 | omMc4rb1 | omMc4rb2 | ||
|---|---|---|---|---|---|---|
| Bmax (%) | 100 | 15.51 ± 3.62 c | 10.80 ± 0.49 c | 39.66 ± 2.06 c | 14.32 ± 1.25 c | |
| α-MSH | IC50 (nM) | 306.69 ± 45.40 | 413.39 ± 64.70 | 384.86 ± 100.62 | 222.50 ± 42.74 | 64.20 ± 22.25 b |
| ACTH (1–24) | IC50 (nM) | 114.70 ± 27.56 | 260.83 ± 61.91 | 375.67 ± 88.27 a | 100.46 ± 20.98 | 22.95 ± 5.62 a |
| AgRP | IC50 (nM) | 32.48 ± 7.76 | 0.42 ± 0.15 a | 2.87 ± 1.07 a | 3.28 ± 0.63 a | 1.54 ± 0.65 a |
| Ipsen 5i | IC50 (nM) | 399.86 ± 122.11 | N/A | N/A | N/A | N/A |
| ML00253764 | IC50 (nM) | 1644.57 ± 215.24 | N/A | N/A | N/A | N/A |
| MCL0020 | IC50 (nM) | 62.33 ± 4.82 | N/A | N/A | 305.05 ± 66.19 a | N/A |
| α-MSH | ACTH (1–24) | |||
|---|---|---|---|---|
| EC50 (nM) | Rmax (%) | EC50 (nM) | Rmax (%) | |
| hMC4R | 4.34 ± 1.04 | 100 | 4.19 ± 0.60 | 100 |
| omMc4ra1 | 11.71 ± 2.59 a | 43.78 ± 4.16 c | 13.54 ± 2.60 b | 48.64 ± 4.32 c |
| omMc4ra2 | 24.51 ± 2.26 c | 48.30 ± 3.82 c | 25.66 ± 2.36 c | 48.47 ± 4.56 c |
| omMc4rb1 | 0.04 ± 0.02 a | 54.63 ± 4.94 c | 0.07 ± 0.02 c | 48.52 ± 2.40 c |
| omMc4rb2 | 0.32 ± 0.07 a | 54.15 ± 3.76 c | 0.13 ± 0.03 c | 53.86 ± 1.42 c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, R.-L.; Liu, T.; Hou, Z.-S.; Wen, H.-S.; Tao, Y.-X. Divergent Pharmacology and Biased Signaling of the Four Melanocortin-4 Receptor Isoforms in Rainbow Trout (Oncorhynchus mykiss). Biomolecules 2023, 13, 1248. https://doi.org/10.3390/biom13081248
Ji R-L, Liu T, Hou Z-S, Wen H-S, Tao Y-X. Divergent Pharmacology and Biased Signaling of the Four Melanocortin-4 Receptor Isoforms in Rainbow Trout (Oncorhynchus mykiss). Biomolecules. 2023; 13(8):1248. https://doi.org/10.3390/biom13081248
Chicago/Turabian StyleJi, Ren-Lei, Ting Liu, Zhi-Shuai Hou, Hai-Shen Wen, and Ya-Xiong Tao. 2023. "Divergent Pharmacology and Biased Signaling of the Four Melanocortin-4 Receptor Isoforms in Rainbow Trout (Oncorhynchus mykiss)" Biomolecules 13, no. 8: 1248. https://doi.org/10.3390/biom13081248
APA StyleJi, R.-L., Liu, T., Hou, Z.-S., Wen, H.-S., & Tao, Y.-X. (2023). Divergent Pharmacology and Biased Signaling of the Four Melanocortin-4 Receptor Isoforms in Rainbow Trout (Oncorhynchus mykiss). Biomolecules, 13(8), 1248. https://doi.org/10.3390/biom13081248








