Dynamic Equilibrium of Protein Phosphorylation by Kinases and Phosphatases Visualized by Phos-Tag SDS-PAGE
Abstract
1. Introduction
2. Results
2.1. Selection of Target Proteins
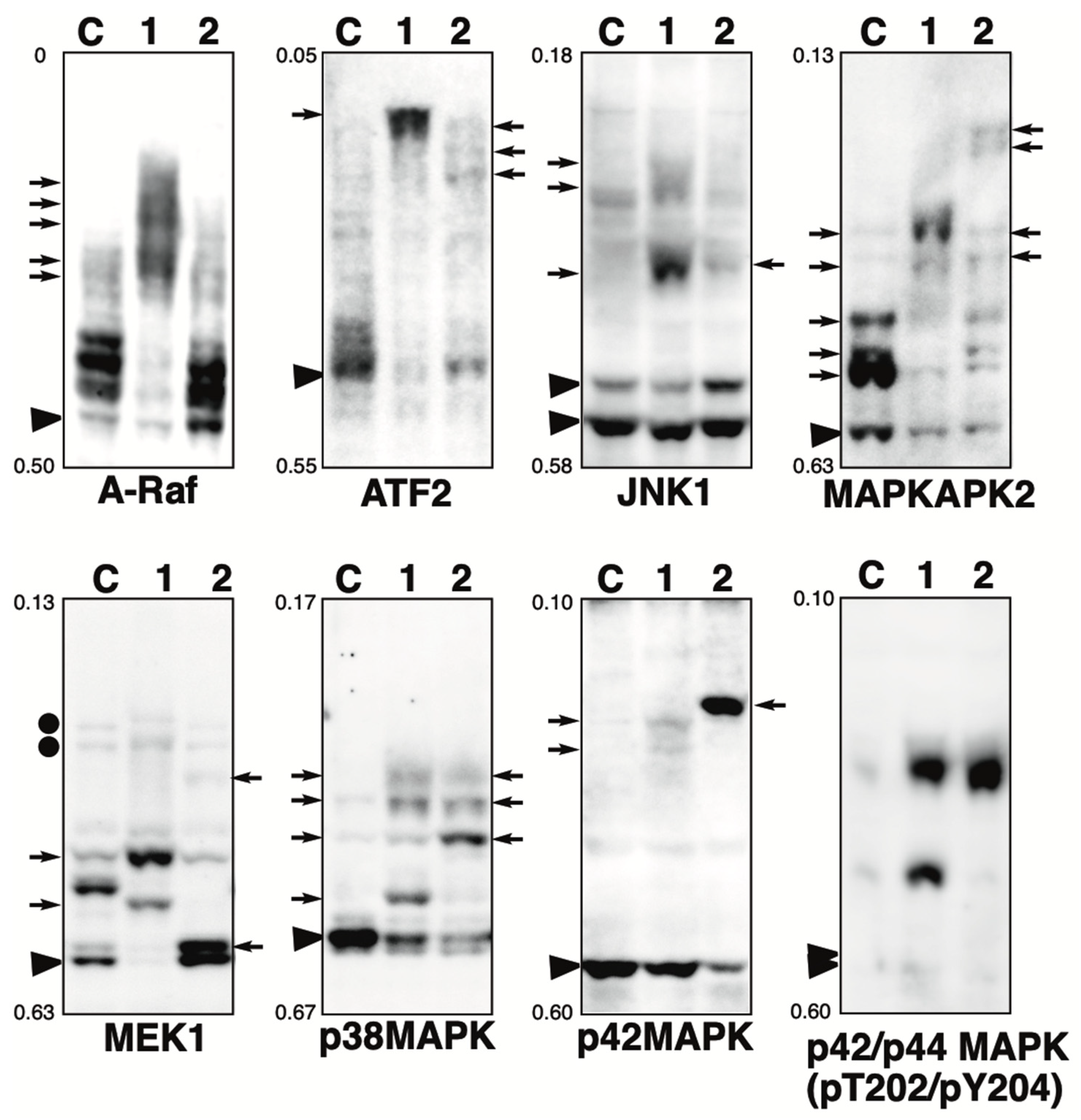
2.2. Phosphorylation State of MAPK Pathway-Related Proteins in the Presence of the Phosphatase Inhibitor
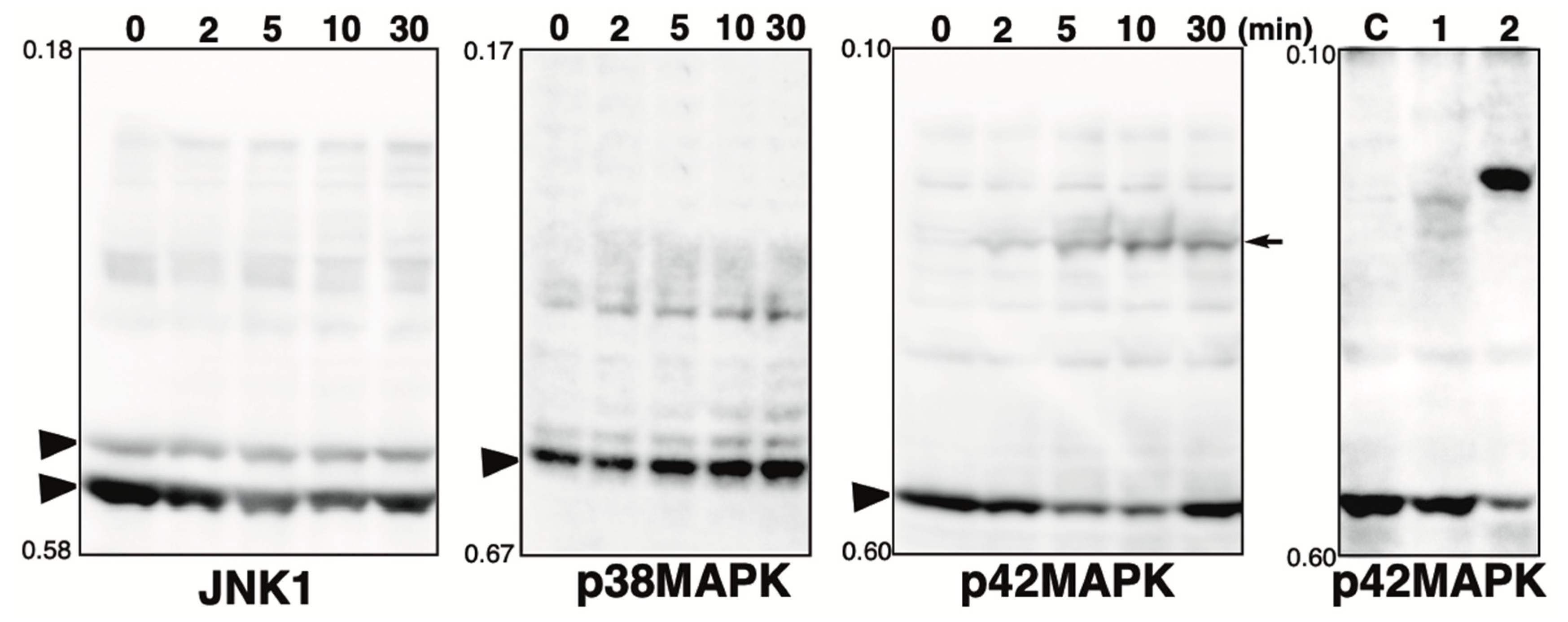
2.3. Effect of Hydrogen Peroxide Treatment on MAPKs
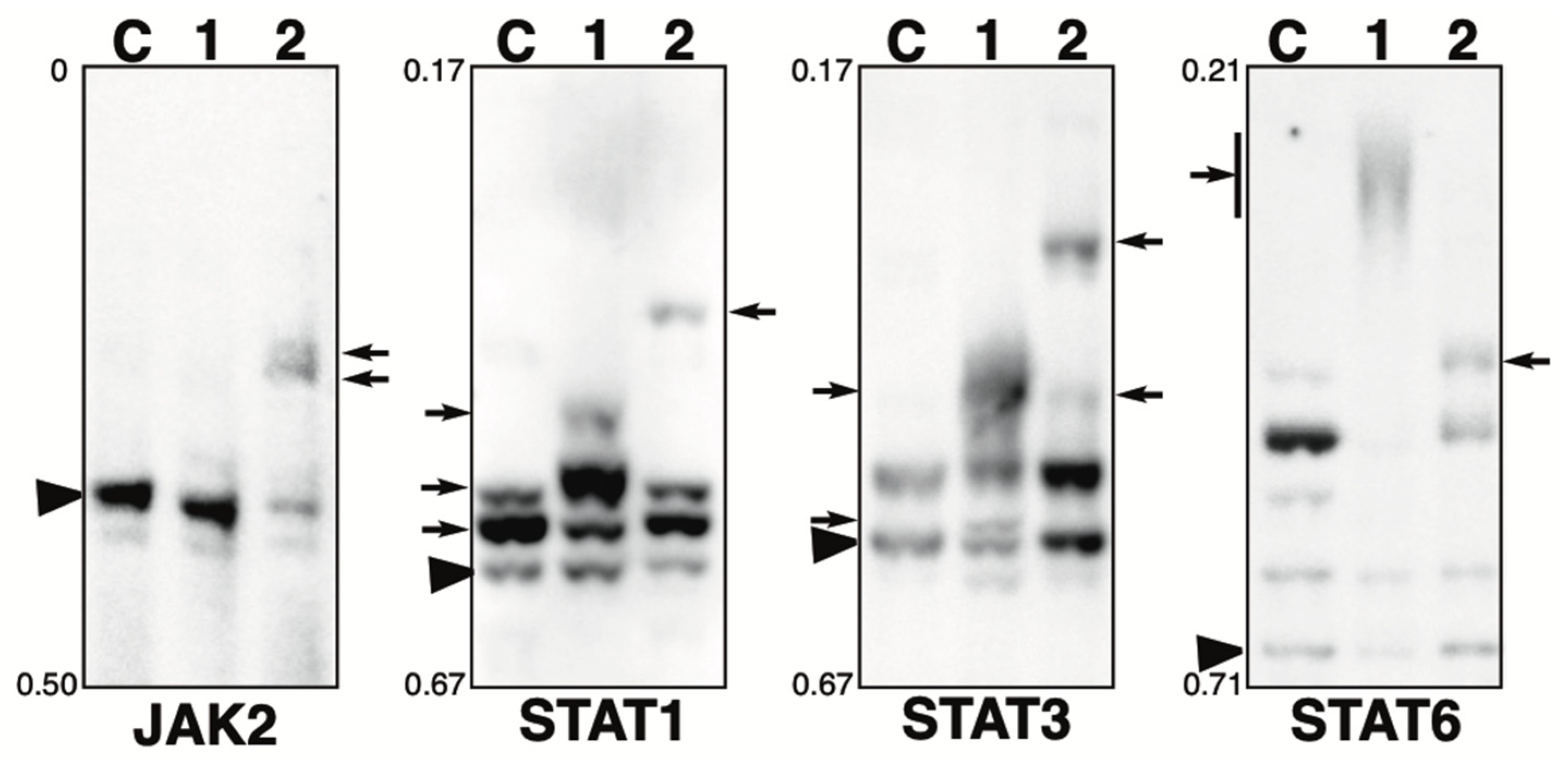
2.4. Phosphorylation State of JAK-STAT Pathway-Related Proteins in the Presence of the Phosphatase Inhibitors
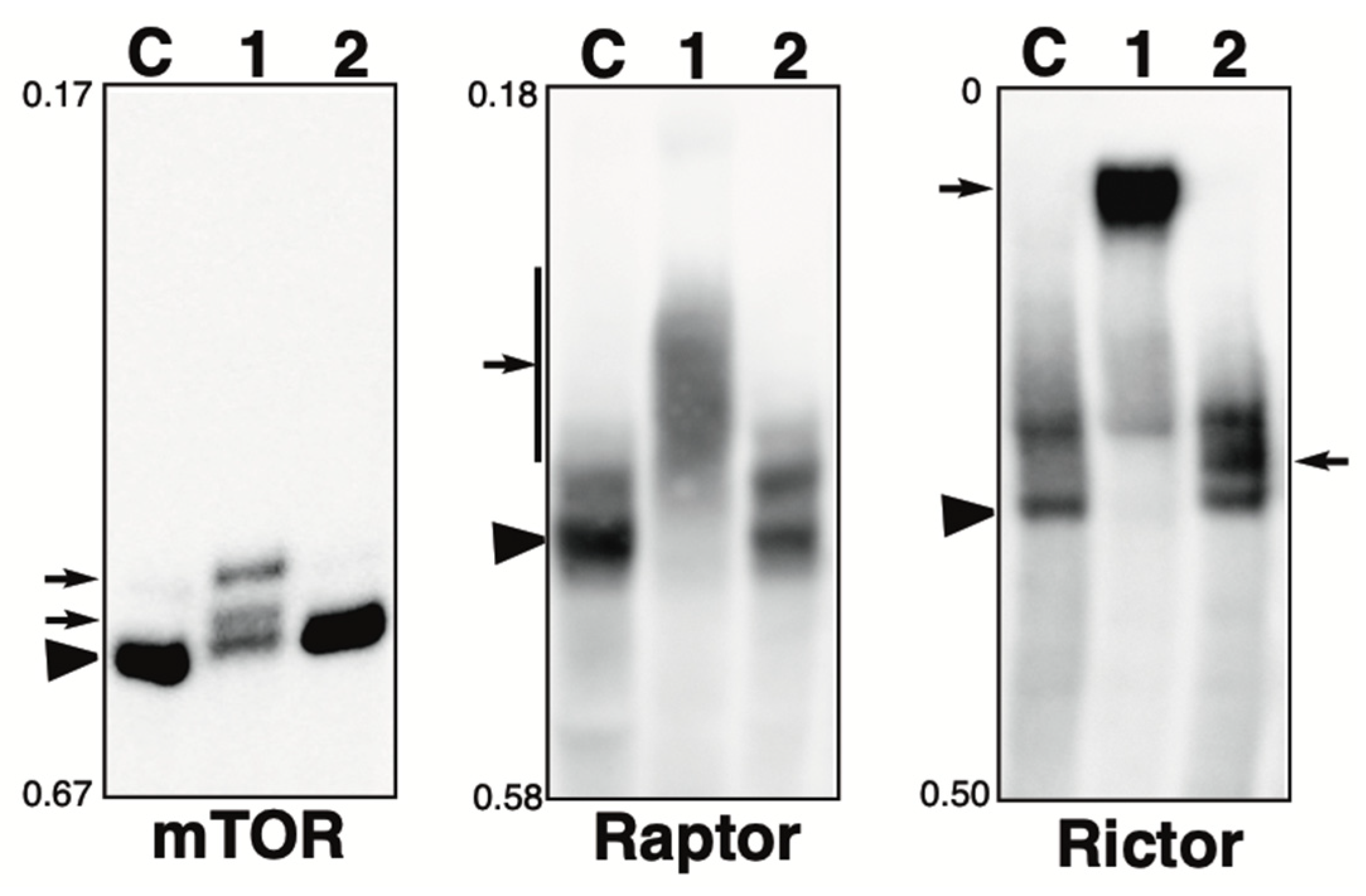
2.5. Phosphorylation State of mTOR Pathway-Related Proteins in the Presence of the Phosphatase Inhibitors
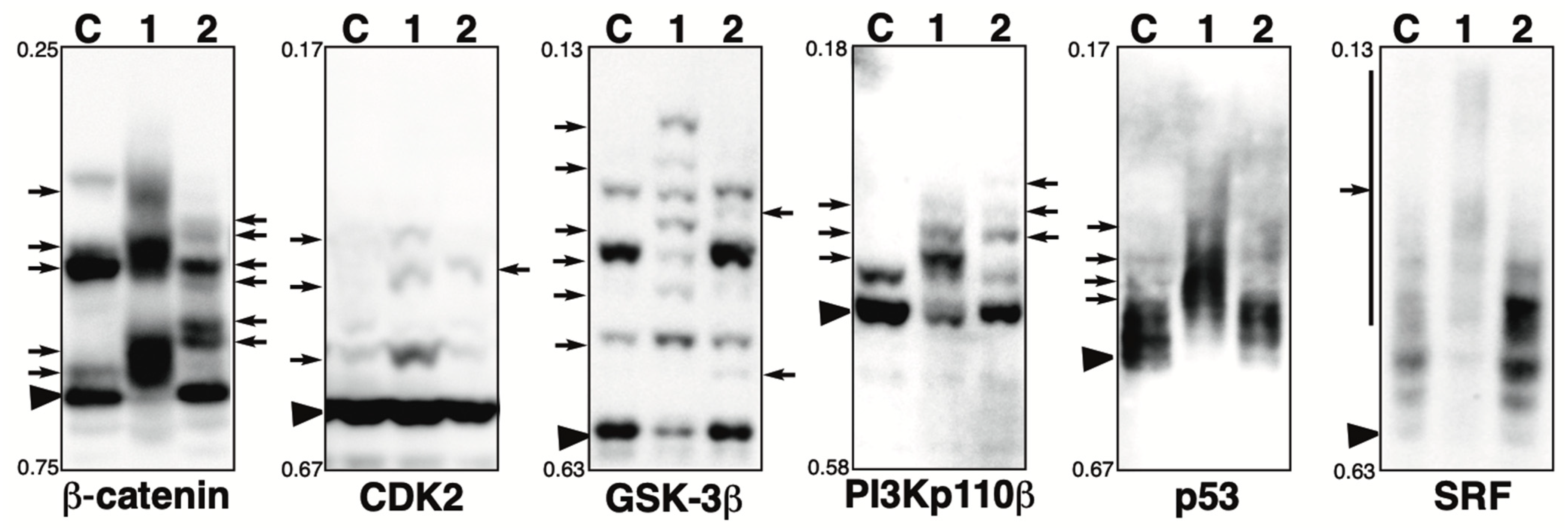
2.6. Phosphorylation State of Other Proteins in the Presence of the Phosphatase Inhibitor
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture and Phosphatase Inhibitor Treatment
4.3. Phos-Tag SDS-PAGE
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ATF | cyclic AMP-dependent transcription factor |
| CDK | cyclin-dependent kinase |
| DMEM | Dulbecco’s modified Eagle’s medium |
| EGF | epidermal growth factor |
| FBS | fetal bovine serum |
| GSK | glycogen synthase kinase |
| JAK | Janus kinase |
| JNK | Jun-amino-terminal kinase |
| MAPK | mitogen-activated protein kinases |
| MAPKAPK | mitogen-activated protein kinase-activated protein kinase |
| MEK | mitogen-activated ERK-regulated kinase |
| MKK | mitogen-activated protein kinase |
| mTOR | mammalian target of rapamycin |
| mTORC | mTOR complex |
| PI3 kinase | phosphoinositide 3-kinase |
| PP1 | protein phosphatase 1 |
| PP2A | protein phosphatase 2A |
| Raf | rapidly accelerated fibrosarcoma |
| SAPK | stress-activated protein kinase |
| SDS-PAGE | sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| STAT | signal transduction and activator of transcription |
| SRF | serum response factor |
| SUMO | small ubiquitin-related modifier |
| 2D DIGE | two-dimensional fluorescence difference gel electrophoresis |
References
- Arshad, O.A.; Danna, V.; Petyuk, V.A.; Piehowski, P.D.; Liu, T.; Rodland, K.D.; McDermott, J.E. An integrative analysis of tumor proteomic and phosphoproteomic profiles to examine the relationships between kinase activity and phosphorylation. Mol. Cell. Proteom. 2019, 18, S26–S36. [Google Scholar] [CrossRef] [PubMed]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Turdo, A.; D’Accardo, C.; Glaviano, A.; Porcelli, G.; Colarossi, C.; Colarossi, L.; Mare, M.; Faldetta, N.; Modica, C.; Pistonek, G.; et al. Targeting Phosphatases and Kinases: How to Checkmate Cancer. Front. Cell Dev. Biol. 2021, 9, 690306. [Google Scholar] [CrossRef] [PubMed]
- Licheva, M.; Raman, B.; Kraft, C.; Reggiori, F. Phosphoregulation of the autophagy machinery by kinases and phosphatases. Autophagy 2022, 18, 104–123. [Google Scholar] [CrossRef]
- Kinoshita, E.; Kinoshita-Kikuta, E.; Koike, T. Phos-tag SDS-PAGE systems for phosphorylation profiling of proteins with a wide range of molecular masses under neutral pH conditions. Proteomics 2012, 12, 192–202. [Google Scholar] [CrossRef]
- Kita, A.; Matsunaga, S.; Takai, A.; Kataiwa, H.; Wakimoto, T.; Fusetani, N.; Isobe, M.; Miki, K. Crystal structure of the complex between calyculin A and the catalytic subunit of protein phosphatase 1. Structure 2002, 10, 715–724. [Google Scholar] [CrossRef]
- Suganuma, M.; Fujiki, H.; Furuya-Suguri, H.; Yoshizawa, S.; Yasumoto, S.; Kato, Y.; Fusetani, N.; Sugimura, T. Calyculin A, an inhibitor of protein phosphatases, a potent tumor promoter on CD-1 mouse skin. Cancer Res. 1990, 50, 3521–3525. [Google Scholar] [PubMed]
- Ceulemans, H.; Bollen, M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 2004, 84, 1–39. [Google Scholar] [CrossRef]
- Tonks, N.K. Protein tyrosine phosphatases: From genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006, 7, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Sasin, J.; Bottini, N.; Friedberg, I.; Friedberg, I.; Osterman, A.; Godzik, A.; Hunter, T.; Dixon, J.; Mustelin, T. Protein tyrosine phosphatases in the human genome. Cell 2004, 117, 699–711. [Google Scholar] [CrossRef]
- Huyer, G.; Liu, S.; Kelly, J.; Moffat, J.; Payette, P.; Kennedy, B.; Tsaprailis, G.; Gresser, M.J.; Ramachandran, C. Mechanism of inhibition of protein-tyrosine phosphatases by vanadate and pervanadate. J. Biol. Chem. 1997, 272, 843–851. [Google Scholar] [CrossRef]
- Avruch, J.; Zhang, X.F.; Kyriakis, J.M. Raf meets Ras: Completing the framework of a signal transduction pathway. Trends Biochem. Sci. 1994, 19, 279–283. [Google Scholar] [CrossRef]
- Chong, H.; Lee, J.; Guan, K.L. Positive and negative regulation of Raf kinase activity and function by phosphorylation. EMBO J. 2001, 20, 3716–3727. [Google Scholar] [CrossRef] [PubMed]
- van Dam, H.; Wilhelm, D.; Herr, I.; Steffen, A.; Herrlich, P.; Angel, P. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 1995, 14, 1798–1811. [Google Scholar] [CrossRef] [PubMed]
- Kyriakis, J.M.; Avruch, J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 2001, 81, 807–869. [Google Scholar] [CrossRef] [PubMed]
- Ichijo, H. From receptors to stress-activated MAP kinases. Oncogene 1999, 18, 6087–6093. [Google Scholar] [CrossRef] [PubMed]
- Rouse, J.; Cohen, P.; Trigon, S.; Morange, M.; Alonso-Llamazares, A.; Zamanillo, D.; Hunt, T.; Nebreda, A.R. A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAP kinase-2 and phosphorylation of the small heat shock proteins. Cell 1994, 78, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Crews, C.M.; Alessandrini, A.; Erikson, R.L. The primary structure of MEK, a protein kinase that phosphorylates the ERK gene product. Science 1992, 258, 478–480. [Google Scholar] [CrossRef]
- Alessi, D.R.; Saito, Y.; Campbell, D.G.; Cohen, P.; Sithanandam, G.; Rapp, U.; Ashworth, A.; Marshall, C.J.; Cowley, S. Identification of the sites in MAP kinase kinase-1 phosphorylated by p74raf-1. EMBO J. 1994, 13, 1610–1619. [Google Scholar] [CrossRef]
- Rosen, L.B.; Ginty, D.D.; Weber, M.J.; Greenberg, M.E. Membrane depolarization and calcium influx stimulate MEK and MAP kinase via activation of Ras. Neuron 1994, 12, 1207–1221. [Google Scholar] [CrossRef]
- Zheng, C.F.; Guan, K.L. Activation of MEK family kinases requires phosphorylation of two conserved Ser/Thr residues. EMBO J. 1994, 13, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, E.; Kinoshita-Kikuta, E.; Kubota, Y.; Takekawa, M.; Koike, T. A Phos-tag SDS-PAGE method that effectively uses phosphoproteomic data for profiling the phosphorylation dynamics of MEK1. Proteomics 2016, 16, 1825–1836. [Google Scholar] [CrossRef]
- Han, J.; Lee, J.D.; Bibbs, L.; Ulevitch, R.J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 1994, 265, 808–811. [Google Scholar] [CrossRef]
- Lee, J.C.; Laydon, J.T.; McDonnell, P.C.; Gallagher, T.F.; Kumar, S.; Green, D.; McNulty, D.; Blumenthal, M.J.; Heys, J.R.; Land Vatter, S.W.; et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature 1994, 372, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Freshney, N.W.; Rawlinson, L.; Guesdon, F.; Jones, E.; Cowley, S.; Hsuan, J.; Saklatvala, J. Interleukin-1 activates a novel protein kinase cascade that results in the phosphorylation of Hsp27. Cell 1994, 78, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Ando, K.; Uemura, K.; Kuzuya, A.; Maesako, M.; Asada-Utsugi, M.; Kubota, M.; Aoyagi, N.; Yoshioka, K.; Okawa, K.; Inoue, H.; et al. N-cadherin regulates p38 MAPK signaling via association with JNK-associated leucine zipper protein: Implications for neurodegeneration in Alzheimer disease. J. Biol. Chem. 2011, 286, 7619–7628. [Google Scholar] [CrossRef]
- Roux, P.P.; Blenis, J. ERK and p38 MAPK-activated protein kinases: A family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 2004, 68, 320–344. [Google Scholar] [CrossRef] [PubMed]
- Seger, R.; Ahn, N.G.; Posada, J.; Munar, E.S.; Munar, E.S.; Jensen, A.M.; Cooper, J.A.; Cobb, M.H.; Krebs, E.G. Purification and characterization of MAP kinase activator(s) from epidermal growth factor stimulated A431 cells. J. Biol. Chem. 1992, 267, 14373–14381. [Google Scholar] [CrossRef] [PubMed]
- Pearson, G.; Xu, B.; Wright, A.; Vanderbilt, C.; Cobb, M.H.; Chen, Z.; Gibson, T.B.; Robinson, F.; Silvestro, L. MAP kinases. Chem. Rev. 2001, 101, 2449–2476. [Google Scholar]
- Leonard, W.J.; O’Shea, J.J. Jaks and STATs: Biological implications. Annu. Rev. Immunol. 1998, 16, 293–322. [Google Scholar] [CrossRef]
- Darnell, J.E. STATs and gene regulation. Science 1997, 277, 1630–1635. [Google Scholar] [CrossRef]
- Heim, M.H. The Jak-STAT pathway: Cytokine signalling from the receptor to the nucleus. J. Recept. Signal Transduct. Res. 1999, 19, 75–120. [Google Scholar] [CrossRef]
- Durbin, J.E.; Hackenmiller, R.; Simon, M.C.; Levy, D.E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 1996, 84, 443–450. [Google Scholar] [CrossRef]
- Meraz, M.A.; White, J.M.; Sheehan, K.C.; Bach, E.A.; Rodig, S.J.; Dighe, A.S.; Kaplan, D.H.; Riley, J.K.; Greenlund, A.C.; Campbell, D.; et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 1996, 84, 431–442. [Google Scholar] [CrossRef]
- Nelms, K.; Keegan, A.D.; Zamorano, J.; Ryan, J.J.; Paul, W.E. The IL-4 receptor: Signaling mechanisms and biologic functions. Annu. Rev. Immunol. 1999, 17, 701–738. [Google Scholar] [CrossRef] [PubMed]
- Malabarba, M.G.; Rui, H.; Deutsch, H.H.; Chung, J.; Kalthoff, F.S.; Farrar, W.L.; Kirken, R.A. Interleukin-13 is a potent activator of JAK3 and STAT6 in cells expressing interleukin-2 receptor-gamma and interleukin-4 receptor-alpha. Biochem. J. 1996, 319, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Dennis, P.B.; Jaeschke, A.; Saitoh, M.; Fowler, B.; Kozma, S.C.; Thomas, G. Mammalian TOR: A homeostatic ATP sensor. Science 2001, 294, 1102–1105. [Google Scholar] [CrossRef]
- Kinoshita, E.; Kinoshita-Kikuta, E.; Ujihara, H.; Koike, T. Mobility shift detection of phosphorylation on large proteins using a Phos-tag SDS-PAGE gel strengthened with agarose. Proteomics 2009, 9, 4098–4101. [Google Scholar] [CrossRef]
- Hara, K.; Maruki, Y.; Long, X.; Yoshino, K.; Oshiro, N.; Hidayat, S.; Tokunaga, C.; Avruch, J.; Yonezawa, K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 2002, 110, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Sarbassov, D.D.; Ali, S.M.; King, J.E.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002, 110, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Oshiro, N.; Yoshino, K.; Hidayat, S.; Tokunaga, C.; Hara, K.; Eguchi, S.; Avruch, J.; Yonezawa, K. Dissociation of raptor from mTOR is a mechanism of rapamycin-induced inhibition of mTOR function. Genes Cells 2004, 9, 359–366. [Google Scholar] [CrossRef]
- Sarbassov, D.D.; Ali, S.M.; Kim, D.H.; Guertin, D.A.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004, 14, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Cadigan, K.M.; Nusse, R. Wnt signaling: A common theme in animal development. Genes Dev. 1997, 11, 3286–3305. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, E.; Kinoshita-Kikuta, E. Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics 2011, 11, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Morgan, D.O. Principles of CDK regulation. Nature 1995, 374, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Welsh, G.I.; Wilson, C.; Proud, C.G. GSK3: A SHAGGY frog story. Trends Cell Biol. 1996, 6, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Cantley, L.C. The phosphoinositide 3-kinase pathway. Science 2002, 296, 1655–1657. [Google Scholar] [CrossRef]
- Levine, A.J. p53, the cellular gatekeeper for growth and division. Cell 1997, 88, 323–331. [Google Scholar] [CrossRef]
- Meek, D.W. Post-translational modification of p53. Semin. Cancer Biol. 1994, 5, 203–210. [Google Scholar] [PubMed]
- Milczarek, G.J.; Martinez, J.; Bowden, G.T. p53 Phosphorylation: Biochemical and functional consequences. Life Sci. 1997, 60, 1–11. [Google Scholar] [CrossRef]
- Treisman, R. Journey to the surface of the cell: Fos regulation and the SRE. EMBO J. 1995, 14, 4905–4913. [Google Scholar] [CrossRef]
- Janknecht, R.; Cahill, M.A.; Nordheim, A. Signal integration at the c-fos promoter. Carcinogenesis 1995, 16, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 107, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, N.; Imamura, H.; Ishihama, Y. Large-scale discovery of substrates of the human kinome. Sci. Rep. 2019, 9, 10503. [Google Scholar] [CrossRef] [PubMed]
- Farooq, A.; Zhou, M.M. Structure and regulation of MAPK phosphatases. Cell. Signal. 2004, 16, 769–779. [Google Scholar] [CrossRef]
- Plowman, G.D.; Sudarsanam, S.; Bingham, J.; Whyte, D.; Hunter, T. The protein kinases of Caenorhabditis elegans: A model for signal transduction in multicellular organisms. Proc. Natl. Acad. Sci. USA 1999, 96, 13603–13610. [Google Scholar] [CrossRef] [PubMed]
- Yigong, S. Serine/Threonine Phosphatases: Mechanism through Structure. Cell 2009, 139, 468–484. [Google Scholar]
- Gong, C.X.; Grundke-Iqbal, I.; Iqbal, K. Dephosphorylation of Alzheimer’s disease abnormally phosphorylated tau by protein phosphatase-2A. Neuroscience 1994, 61, 765–772. [Google Scholar] [CrossRef]
- Kinoshita, E.; Kinoshita-Kikuta, E.; Koike, T. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat. Protoc. 2009, 9, 513–1521. [Google Scholar] [CrossRef]
| Target Protein of Specific Antibody | Number of Phosphorylation Site Putative Upstream Kinases in Human Cells 2 | ||
|---|---|---|---|
| pS/pT | pY | ||
| A-Raf | 47 - | 8 src | |
| ATF2 | 29 ERK, JNK, p38, VRK1, PKC | 0 | |
| JNK1 | 7 MKK4, MKK7, ASK1 | 3 ASK1, MEKK6, MKK4, MKK7 | |
| MAPKAPK2 | 17 Erk2, p38 | 3 - | |
| MEK1 | 13 A-Raf, B-Raf, Cot, Raf1, PDK1, CDK1, CDK5, Erk1/2, PAK1 | 1 - | |
| P38 MAPK | p38α | 14 ASK1, MEKK6, MKK3/4/6, GRK2 | 2 ASK1, MEKK6, MKK3/4/6, ZAP70 |
| p38β | 7 - | 1 - | |
| p38γ | 4 | 1 | |
| p38δ | 8 - | 2 - | |
| P42 MAPK | 9 CK2A1, Erk2, MEK2 | 7 EGFR, FER, JAK2, MEK1 | |
| JAK2 | 6 JAK2 | 28 JAK2 | |
| STAT1 | 16 CAMK2γ, CDK8, CK2A1, Erk1, p38α, PKCδ | 5 JAK1/2/3, NPM-ALK, Src, Syk, Tyk2 | |
| STAT3 | 20 BUB1, CDK5, Erk1/2, GSK3α/β, JNK1/2, NEK6, p38α, PKCδ/ε, MST2 | 10 ALK, AXL, EEF2K, Fer, Fyn, JAK2, Mer, PKM, Ret, Src, TrkA | |
| STAT6 | 8 JNK1, TBK1 | 1 - | |
| mTOR | 50 p70S6K, Akt1, mTOR, IKKA, TBK1 | 7 - | |
| Rictor | 89 GSK-3α/β, Akt1, p70S6K, SGK1 | 13 - | |
| Raptor | 39 AMPKA1/2, ULK1/2, Erk1/2, JNK1, CDK1, DAPK2, p90RSK, mTOR, ULK1/2, TBK1, ICK, LATS1/2, p38β | 4 - | |
| β-catenin | 44 Akt, CK1, IKKA, PKA, PRKD1, GSK3β, JNK1, PAK, p38, TBK1, PKC, PLK1, PMVK | 12 Abl, JAK3, Src, EGFR, FGFR, TrkA, FAK, Brk | |
| CDK2 | 7 Akt1, CDK20, Erk1/2 | 4 - | |
| GSK-3β | 19 Akt1, AurA, EEF2K, GSK-3β, KHS1, p70S6K, JNK1, PKA, p38, PKC, PKC, RSK2, SGK3, Erk1/2 | 5 MET | |
| PI3Kp110β (PIK3CB) | 12 - | 9 - | |
| P53 | 35 CK1A, CK2B, ATM, ATR, VRK1, CDK1/5/9, Chk1/2, DAPK1, JNK1/2, PLK3, BTK, DNAPK, DYRK1A/2, Erk1/2, LKB1, NuaK1, p38α/γ, PRPK, SMG1, HIPK2, PKCδ, MAPKAPK5, AurA/B, PAK4, NEK2, PKR, Lmr1, GRK5, TAF1, LRRK2 | 3 src | |
| SRF | 23 MAPKAPK2, DMPK1, PKACA, PKCα | 0 - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kinoshita-Kikuta, E.; Nishikawa, K.; Hiraishi, K.; Shimoji, K.; Nagase, K.; Kinoshita, E. Dynamic Equilibrium of Protein Phosphorylation by Kinases and Phosphatases Visualized by Phos-Tag SDS-PAGE. Kinases Phosphatases 2024, 2, 224-239. https://doi.org/10.3390/kinasesphosphatases2030014
Kinoshita-Kikuta E, Nishikawa K, Hiraishi K, Shimoji K, Nagase K, Kinoshita E. Dynamic Equilibrium of Protein Phosphorylation by Kinases and Phosphatases Visualized by Phos-Tag SDS-PAGE. Kinases and Phosphatases. 2024; 2(3):224-239. https://doi.org/10.3390/kinasesphosphatases2030014
Chicago/Turabian StyleKinoshita-Kikuta, Emiko, Kento Nishikawa, Kento Hiraishi, Kaku Shimoji, Kenichi Nagase, and Eiji Kinoshita. 2024. "Dynamic Equilibrium of Protein Phosphorylation by Kinases and Phosphatases Visualized by Phos-Tag SDS-PAGE" Kinases and Phosphatases 2, no. 3: 224-239. https://doi.org/10.3390/kinasesphosphatases2030014
APA StyleKinoshita-Kikuta, E., Nishikawa, K., Hiraishi, K., Shimoji, K., Nagase, K., & Kinoshita, E. (2024). Dynamic Equilibrium of Protein Phosphorylation by Kinases and Phosphatases Visualized by Phos-Tag SDS-PAGE. Kinases and Phosphatases, 2(3), 224-239. https://doi.org/10.3390/kinasesphosphatases2030014








