Abstract
In this work, a total of 97 MRS-grown and 135 M17-grown lactic acid bacteria strains were isolated from 20 goat’s raw milk cheeses, and their antimicrobial, acidifying, and proteolytic activities were determined in vitro. Principal component analyses adjusted to a subset of 84 promising isolates evidenced that, for MRS isolates, antagonism against Staphylococcus aureus correlated well with higher acidification potential, whereas for M17 isolates, the antagonisms against S. aureus and Listeria monocytogenes were more correlated. The outcomes highlighted various strains with pathogen inhibition ability and satisfactory technological properties that may be useful for the development of a customised starter culture.
1. Introduction
Lactic acid bacteria (LAB) are responsible for the cheese fermentation process, but they can also increase the safety of cheeses by acting as biopreservatives, and promote desirable organoleptic properties (such as texture, aroma, and flavour compounds). In this sense, the objective of this work was to collect and assess the antimicrobial, acidifying, and proteolytic capacities of LAB strains from the microflora of artisanal Portuguese goat’s raw milk cheeses for potential development of a customised starter culture and application in cheese manufacture.
2. Materials and Methods
2.1. Isolation and Confirmation of Lactic Acid Bacteria
Samples of four batches of artisanal goat’s raw milk cheeses (n = 20) were collected in Mirandela, Portugal, between November 2020 and March 2021. LAB strains were isolated from cheese samples as described by the ISO standard 15214:1998 [1]. Briefly, after dilution, aliquots were plated in MRS agar (selective medium for enumeration and isolation of lactobacilli) and M17 agar (non-selective medium for enumeration and isolation of lactococci), overlayed with 1.2% bacteriological agar, and plates were incubated at 30 °C for 48 h. Then, eight typical colonies on MRS and M17 agar were selected for purification and incubated at 30 °C for another 48 h in the respective media. Lastly, to eliminate non-LAB isolates, catalase (3% hydrogen peroxide) and Gram tests, as well as morphologic observation, were performed. Cultures presumptively identified as LAB were maintained in MRS broth with 25% glycerol at −80 °C.
2.2. Bacterial Strains and Preparation of Cell Suspensions
Salmonella enterica subsp. Enterica serovar Typhimurium ATCC 43971, Listeria monocytogenes WDCM 00019, and Staphylococcus aureus ATCC 6538, obtained from the Polytechnic Institute of Bragança stock collection, were used for all experiments. A loop of culture kept on Nutrient Agar slants was inoculated separately in 10 mL of BHI broth. Broth tubes were incubated at 37 °C for 16 h, following two successive inoculations, to achieve a concentration of approximately 108 CFU/mL. L. monocytogenes required a pre-activation in 5 mL of BHI at 37 °C for 16 h.
2.3. Screening of LAB for Antimicrobial Capacity
Antimicrobial ability of selected LAB strains was determined by the spot-on-lawn assay as described by Campagnollo et al. [2], with some modifications.
Each LAB strain was cultured separately in MRS broth overnight (37 °C, 24 h), spotted onto the surface of MRS or M17 agar plates (3 μL for LAB strains isolated in MRS and 5 μL for M17 agar, respectively), following incubation at 37 °C for 16 h. Inoculated plates were covered with 10 mL of BHI soft agar (BHI broth with 0.75% bacteriological agar) seeded with 1 mL of each bacterial strain (tested separately). After incubating plates at 37 °C for 16 h (pre-incubation at 4 °C for 2 h), the diameter of the inhibition zones of each pathogen were measured with a calliper.
LAB strains that presented antimicrobial capacity at 37 °C according to the following criteria were also tested at 10 °C for 10 days: distance between halo circumference and LAB colony limit greater than 5 mm for S. aureus, or 8 mm for L. monocytogenes and S. enterica ser. Typhimurium—for MRS agar; or greater than 0.5 mm for S. aureus, 6 mm for L. monocytogenes or 3.5 mm for S. enterica ser. Typhimurium—for M17 agar.
2.4. Screening of LAB for Proteolytic and Acidifying Capacity
Proteolytic activity and acidifying capacity were evaluated according to the protocols of Franciosi et al. [3] and Durlu-Ozkaya et al. [4] respectively, for the subset of LAB strains presenting antimicrobial activity at 37 °C. Proteolytic activity was confirmed qualitatively as clear zones around each LAB colony. Acidifying capacity was assessed by measuring pH of the inoculated milk broth at 30 °C during 8 h (t = 0, 2, 4, 6, 8 h), and after 24 h. LAB strains are considered good acidifiers when able reduce the pH below 5.3 after 6 h at 30 °C [5].
2.5. Statistical Analysis
Data were separated into two subsets (one for MRS-isolated LAB; another for M17-isolated LAB) and each subset subjected to principal component analysis (PCA). From the antimicrobial assays, only the data referring to L. monocytogenes and S. aureus inhibition was used, as these are the pathogens of greater concern (among the three tested) in cheese. The function prcomp from the factoextra package was used in R software (version 3.6.2, R Foundation for Statistical Computing, Vienna, Austria), where a varimax-rotated solution for three principal components was obtained. From the three-dimensional PCA, maps of antimicrobial, acidifying and proteolytic characteristics of cheeses were built from the projection of sample scores onto the span of the principal components.
3. Results and Discussion
3.1. Technological Properties of LAB Strains: Antimicrobial, Acidifying and Proteolytic Capacities
In total, 232 LAB strains (97 isolated in MRS agar and 135 isolated in M17 agar) were isolated. Antimicrobial tests at 37 °C revealed that 98%, 100% and 100% of LAB isolated in MRS agar presented antagonism against L. monocytogenes, S. aureus and S. enterica ser. Typhimurium, respectively. In contrast, only 13.3% and 28.1% of LAB isolated in M17 agar revealed antagonism against L. monocytogenes and S. enterica ser. Typhimurium, respectively (no antagonism was observed against S. aureus).
After selecting isolates with considerable antimicrobial activity at 37 °C, 84 strains (58 isolated in MRS agar and 26 isolated in M17 agar) were subjected to the spot-on-lawn assay at 10 °C. The results of this assessment revealed that all 84 strains from this subset maintained their antimicrobial activity even at 10 °C. The microbial inhibition offered by the isolated LAB may be a consequence of competition against pathogens for the available substrate, production of antimicrobial substances (for example, bacteriocins) and/or production of non-proteinaceous compounds such as H2O2 [6].
Regarding the acidifying capacity, LAB isolated from M17 agar presented better outcomes than LAB isolated from MRS agar. More specifically, 12 out of the 26 strains (46%) isolated in M17 agar were able to reduce the pH of milk broth below 5.3 after 6 h at 30 °C. This result indicates the potential of some strains to contribute to a rapid pH decrease, which is essential in cheese-making to achieve adequate coagulation, curd firmness, and control of bacterial pathogens growth, among other contaminants [2,6]. In this sense, these LAB strains demonstrate potential to be used as starter and/or adjunct cultures to avoid faulty fermentations. On the other hand, no strains isolated from MRS agar provided such a pH reduction under these conditions. Nevertheless, LAB strains with poor acidifying capacity can still be part of a starter mixture, if they possess other technological properties that may benefit cheese production [6].
Regarding the proteolytic capacity, only two strains isolated from MRS agar demonstrated irrefutable clear (transparent) zones around the colonies. However, various LAB also revealed a zone around the colony with less density than the milk agar, but not completely transparent. The clear zone surrounding the colonies is an indicator that proteolytic bacteria hydrolyse casein to form soluble nitrogenous compounds; more clear zones are seen on milk agar if the bacteria also produce acid from fermentable carbohydrates present in the medium [7]. This may explain the two types of zones observed in this assay. From the cheese-making perspective, casein hydrolysis is crucial for texture development, and the released peptides can also accelerate aroma development [2]. In this sense, the results obtained may suggest the potential, even if limited, of some strains to contribute to the improvement of cheese texture and aroma.
3.2. Principal Component Analysis
In each subset of MRS- and M17-isolated LAB, the contribution of the antimicrobial, acidifying and proteolytic attributes to the principal components can be evaluated in Table 1 and Table 2, respectively, by their correlations with the three components extracted. Figure 1 and Figure 2 represent the biplots of variables loadings and observation scores.

Table 1.
Coefficients of correlation of the tested technological properties of MRS-isolated LAB, with the three varimax-rotated factors (PC1, PC2, PC3) along with communalities and explained variances.

Table 2.
Coefficients of correlation of the tested technological properties of M17-isolated LAB, with the three varimax-rotated factors (PC1, PC2, PC3) along with communalities and explained variances.
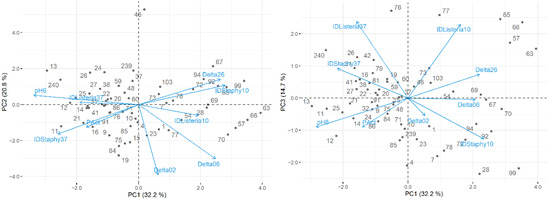
Figure 1.
Maps of the first and second principal components (left) and the first and third principal components (right) of the tested technological properties of MRS-isolated LAB.
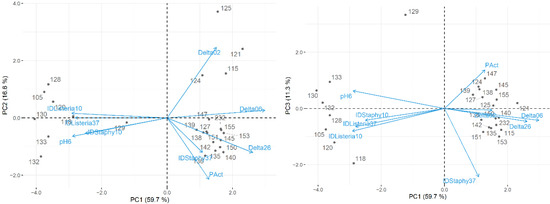
Figure 2.
Maps of the first and second principal components (left) and the first and third principal components (right) of the tested technological properties of M17-isolated LAB.
From Table 1 (MRS subset), a total of 68% of the variability in the nine attributes was jointly explained by the three principal components. The first component (PC1) explained 23% of the total variability and was highly correlated with L. monocytogenes inhibition at 10 °C (R = 0.76), and highly and inversely correlated with milk broth pH value after 6 h (R = −0.73); in contrast, it was weakly correlated with inhibition of L. monocytogenes at 37 °C (R = 0.16). In this sense, PC1 indicates isolates with different inhibitory capacity against L. monocytogenes at 10 °C and 37 °C and distinguishes the ability of isolates to promote a reduced pH value in milk broth after 6 h at 30 °C (Figure 1).
The second component (PC2) explained 22% of the data variability and presented high loadings on two pH-related variables: ΔpH02 (R = 0.95) and ΔpH06 (R = 0.88). For this reason, dissimilarities across the PC2 axis (Figure 1) indicate LAB with distinct acidification profiles, specifically between t = 0 h and t = 2 h (ΔpH02), and t = 0 h and t = 6 h (ΔpH06).
Finally, the third component (PC3) explained another 22% of the total variability and was highly correlated with inhibition of L. monocytogenes and S. aureus at 37 °C (R = 0.77 and R = 0.70, respectively), and highly and inversely correlated with inhibition of S. aureus at 10 °C (R = −0.77). Thus, PC3 reveals LAB strains with distinctive antimicrobial capacities, namely against L. monocytogenes and S. aureus at 37 °C, and S. aureus at 10 °C (Figure 1).
The properties of M17 strains presented stronger relationships between variables than MRS ones, since higher total variability could be explained (88% in Table 2). PC1 explained most (55%) of the total variability and was highly correlated with antimicrobial inhibition (R = 0.91 and R = 0.83 for L. monocytogenes and S. aureus, respectively, at 10 °C; R = 0.89 for L. monocytogenes at 37 °C), and the pH decrease of milk broth (R = 0.91 and R = −0.91 for pH6 and ΔpH06, respectively), respectively. For this reason, PC1 suggests isolates with varying inhibitory power against L. monocytogenes at 10 °C and 37 °C, and against S. aureus at 10 °C; it also provides insight on the isolates capacity to reduce the pH of milk broth after 6 h and their acidification profile between t = 0 h and t = 6 h (Figure 2).
PC2 and PC3 explained 17% and 16% of the total variability, respectively. The first was highly correlated with proteolytic activity (R = 0.88), whereas the second was well correlated with antimicrobial inhibition of S. aureus at 37 °C (R = 0.94) (Figure 2).
Further analysing the figures produced, from MRS isolates (Figure 1), clusters were not easily identified, hence suggesting strains of similar antimicrobial capacity and technological characteristics. However, from Figure 2, two clusters of M17 strains can be distinguished: one with greater acidification and proteolytic capacities and related to higher S. aureus inhibition at 37 °C; and another with better antimicrobial activity against S. aureus (at 10 °C) and L. monocytogenes (at 10 °C and 37 °C).
The joint in-vitro information of the LAB strains will be very helpful in selecting a particular set with desirable characteristics to produce cheeses. More specifically, this PCA may assist in the design of a tailored starter culture that can offer antimicrobial protection against pathogens and assist in the development of pleasing aroma and flavour compounds in the product.
4. Conclusions
This study showed that the grouping of isolates by principal component analysis may be useful for strain selection based on advantageous characteristics for cheese production. In fact, the outcomes obtained indicate that application of indigenous LAB selected in this work, as a customised starter culture, may help prevent pathogen growth (biopreservation potential) and contribute to the development of texture and attractive organoleptic properties in cheeses.
Author Contributions
Conceptualization, V.C., J.A.T. and U.G.-B.; methodology, A.S.F. and U.G.-B.; software, B.N.S. and U.G.-B.; validation, B.N.S. and U.G.-B.; formal analysis, B.N.S. and U.G.-B.; investigation, B.N.S. and A.S.F.; resources, J.A.T., V.C. and U.G.-B.; data curation, B.N.S. and U.G.-B.; writing—original draft, B.N.S.; writing—review and editing, A.S.F. and U.G.-B.; visualization, B.N.S. and U.G.-B.; supervision, V.C., J.A.T. and U.G.-B.; project administration, V.C., J.A.T. and U.G.-B.; funding acquisition, V.C., J.A.T. and U.G.-B. All authors have read and agreed to the published version of the manuscript.
Funding
The authors are grateful to the EU PRIMA program and the Portuguese Foundation for Science and Technology (FCT) for funding the ArtiSaneFood project (PRIMA/0001/2018) and for financial support through national funds FCT/MCTES to CIMO (UIDB/00690/2020). This study was supported by FCT under the scope of the strategic funding of the UIDB/04469/2020 unit and BioTecNorte operation (NORTE-01-0145-FEDER-000004) funded by the European Regional Development Fund under the scope of Norte2020—Programa Operacional Regional do Norte.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Summary data available upon request.
Acknowledgments
Silva acknowledges the financial support provided by FCT through the Ph.D. grant SFRH/BD/137801/2018. Gonzales-Barron acknowledges the national funding by FCT, P.I., through the Institutional Scientific Employment Program contract.
Conflicts of Interest
The authors declare no conflict of interest.
References
- ISO 152147:1998. Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Mesophilic Lactic Acid Bacteria—Colony-Count Technique at 30 Degrees C; International Organization for Standardization: Geneva, Switzerland, 1998. [Google Scholar]
- Campagnollo, F.; Margalho, L.; Kamimura, B.; Feliciano, M.; Freire, L.; Lopes, L.; Alvarenga, V.; Cadavez, V.; Gonzales-Barron, U.; Schaffner, D.; et al. Selection of indigenous lactic acid bacteria presenting anti-listerial activity, and their role in reducing the maturation period and assuring the safety of traditional Brazilian cheeses. Food Microbiol. 2018, 73, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Franciosi, E.; Settanni, L.; Cavazza, A.; Poznanski, E. Biodiversity and technological potential of wild lactic acid bacteria from raw cows’ milk. Int. Dairy J. 2009, 19, 3–11. [Google Scholar] [CrossRef]
- Durlu-Ozkaya, F.; Xanthopoulos, V.; Tunail, N.; Litopoulou-Tzanetaki, E. Technologically important properties of lactic acid bacteria isolates from Beyaz cheese made from raw ewes’ milk. J. Appl. Microbiol. 2001, 91, 861–870. [Google Scholar] [CrossRef]
- Beresford, T.P.; Fitzsimons, N.A.; Brennan, N.L.; Cogan, T.M. Recent advances in cheese microbiology. Int. Dairy J. 2001, 11, 259–274. [Google Scholar] [CrossRef]
- Ribeiro, S.C.; Coelho, M.C.; Todorov, S.D.; Franco, B.D.G.M.; Dapkevicius, M.L.E.; Silva, C.C.G. Technological properties of bacteriocin-producing lactic acid bacteria isolated from Pico cheese an artisanal cow’s milk cheese. J. Appl. Microbiol. 2014, 116, 573–585. [Google Scholar] [CrossRef] [PubMed]
- SM Agar–Technical Data (HiMedia Laboratories). Available online: https://himedialabs.com/td/m763.pdf (accessed on 9 September 2021).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).