Abstract
The possibility of developing deep-sowing tolerant (DST) maize to absorb moisture from subsoil zones is crucial to maize adaptation to water-stressed environments. The function of the mesocotyl in field emergence of seedlings is established in grasses. However, information is scarce on the extent of genetic variability for mesocotyl length (ML) in maize. Sixty-eight maize genotypes were studied using Completely Randomised Design in a laboratory experiment to investigate the extent of genetic variability for ML, and the relationship of seed biochemical components with ML. Ten seeds of each genotype were germinated for 10 days in the dark. Mesocotyl length was determined by placing cut mesocotyl against a flexible measuring tape. Biochemical contents of seeds were determined at a standard diagnostic laboratory. Analysis of variance revealed highly significant (p ≤ 0.01) genotype mean square, indicating sufficient variability for genetic improvement. Broad-sense heritability and genetic advance were high and implied that ML was heritable. Mean ML for genotypes ranged from 0.58 to 9.02 cm; thus, planned crosses can be made for ML improvement. A dendrogram from cluster analysis based on Ward’s minimum variance cluster analysis classified 65 of the genotypes into clusters I, II, and III with ML (mean ± standard deviation) of 0.49 ± 0.18, 4.25 ± 0.96, and 9.16 ± 0.93 cm, respectively. All the measured biochemical parameters, except selenium, showed significant (p ≤ 0.05/0.01) associations with ML. Crosses can be planned involving genotypes from clusters 1 and III, to exploit heterosis for ML in a hybrid program. The results obtained from this study provide a basis for the development of DST maize for drought-prone environments.
1. Introduction
Different crops have developed characteristic adaptive features for their seedlings to push through the soil. This is particularly essential in the arid and semi-arid regions of the world, which are characterised by dry topsoil owing to a low water table. Farmers in these areas find it difficult or impossible to obtain good field establishment when the water table is low. In crops such as cowpea and Arabidopsis, the seedlings break out of the soil through the hypocotyls while the seedlings of monocots (without hypocotyls growth) such as maize, wheat, and rice do so by means of the mesocotyl [1,2]. However, maize lines/varieties that are able to emerge from soil depths below the common 2–5 cm are scarce or unavailable to farmers, particularly in Africa and other drought-prone areas of the world. Therefore, efforts at developing maize varieties with elongated mesocotyl are strategic to the improvement of the crop’s performance in drought-prone areas. Because it is highly regulated by phytohormones, much of the research on maize mesocotyl elongation has focused on hormonal regulation such as auxin [2], gibberellic acid [3,4], and brassinosteroids [5]. Information on the genetics, variability, and/or mode of inheritance of maize mesocotyl length is thus sparse in the literature or altogether unavailable. According to Niu et al. [1], critical genes determining mesocotyl elongation in maize remain unknown. Discovery of candidate genes for maize mesocotyl elongation, and identification of the mode of inheritance of the genes, will enhance the improvement of deep-sowing tolerance in maize, and provide the possibility of developing maize varieties for sowing in areas with low water tables. Investigating the extent of genetic variability for ML in available maize germplasm and the influence of seed biochemical quality attributes on ML will provide information for determining the gene action controlling ML, and secondary traits for selection of DST maize genotypes. The objectives of this study were to determine: (i) the extent of genetic variability for mesocoty length in maize, and (ii) the influence of seed biochemical quality attributes on mesocotyl elongation in maize.
2. Experiments
2.1. Genetic Materials
Sixty-eight maize genotypes were used in this study, comprising of sixty-six genotypes obtained from the International Institute of Tropical Agriculture and two cultivars obtained from a local seed market.
2.2. Experimental Setup
Ten seeds of each genotype were grown in the dark for 10 days using locally available materials. Jute sack was cut into A4 paper-size and soaked in water. Soaked cut jute materials were laid flat on the laboratory workbench and seeds of each genotype were arranged in two rows of five each. The setup was rolled and held in place at the tips using rubber bands. Completely Randomised Design (CRD) was used with two replicates. Both replicates were placed in a cupboard at the Agronomy Laboratory of the Department of Crop Production and Horticulture, Lagos State Polytechnic, Ikorodu, Nigeria. Seedlings were retrieved to determine mesocotyl lengths at 10 days after setup.
2.3. Data Collection
- Mesocotyl length
Elongated mesocotyl was cut from 10 seedlings per replicate, and placed against a measuring tape to record the length in cm. The arithmetic mean of the ML was then recorded for each maize genotype.
- Seed biochemical attributes
Approximately 10-g seed samples of the genotypes were taken to Tetra ‘A’ Analytical and Diagnostic Laboratory, Abeokuta, Ogun State, to determine biochemical composition with respect to iron (Fe), zinc (Zn), selenium (Se), crude protein (CP), free fatty acid (FFA), oil, linoleic acid, and amylase contents. Seeds of genotypes †PVAEH 33 and †PVAEH 40 were not sufficient, and the genotypes were excluded from the biochemical analysis.
2.4. Data Analysis
Means of the ML were subjected to analysis of variance using the ‘glm’ procedure of SAS (SAS Institute, Cary, NC, USA). Estimates of genetic components were obtained using ‘proc varcomp’ of SAS (SAS Institute, Cary, NC, USA). Values of ML were converted to Euclidean distance estimates, which were subjected to Ward’s minimum variance cluster analysis in SAS (SAS Institute, Cary, NC, USA). The relationship among ML and seed biochemical contents was studied via Pearson correlation analysis performed using SAS (SAS Institute, Cary, NC, USA).
3. Results and Discussion
As shown in Table 1, analysis of variance of revealed a significant (p ≤ 0.01) genotype mean square for mesocotyl length. Moreover, genotypic variance (8.79) was close to phenotypic variance (13.19), leading to high broad-sense heritability (0.67), while genotypic (GCV) and phenotypic coefficients of variation (PCV) were high at 0.57 and 0.70, respectively. In addition, genetic advance (as a % of mean) was extremely high (almost unity). There was also a difference between the computed genotypic and phenotypic variances, indicating the role of environmental factors in the elongation of mesocotyl in maize. The significant genotype mean square, as well as the close correspondence between the genotypic and phenotypic variances, indicated the existence of sufficient variability for mesocotyl length among the maize genotypes. Thus, there is the possibility of genetic improvement of the trait for future breeding programs. The estimate of environmental variance suggested the need to test the maize genotypes in more environments to investigate the effect of genotype × environment interaction in the elongation of mesocotyl. Similar observations have been reported for agronomic traits in maize. Sivasubramanian and Menon [6] ranked GCVs and PCVs as low, moderate and high when the values are <0.10, 0.10–0.20, and >0.20, respectively. The high GCV (0.57) observed indicated potential for improvement of ML within the germplasm. The higher value of PCV than GCV is an indication of the role of the environment in the phenotypic expression of ML, and further suggested the need to test in more environments. According to Johnson et al. [7], heritability values < 0.30, between 0.30 and 0.60, and above 0.60 are classified as low, moderate, and high, respectively. Thus, the observed heritability estimate of ML was high and implied that the trait was majorly controlled by the genotype and is therefore heritable. Olayiwola and Soremi [8] have suggested that broad-sense heritability should not be solely used in determining the genetic potentials of a trait since it (broad-sense heritability) is composed of both additive and non-additive genetic variances, and as a result, high heritability is not always associated with high genetic advance [9]. Earlier, Johnson et al. [7] recommended that estimates of heritability and genetic advance should be jointly considered in predicting the value of selection. The high genetic advance (as a % of mean) of mesocotyl length is an indication that the trait will respond favourably to selection.

Table 1.
Mean squares and genetic components of maize genotypes for mesocotyl length after 10 days in the dark.
Grouping genotypes into clusters has been found effective in minimising the genotype pool and easing the process of selection [10]. Sixty-five of the maize genotypes were clearly delineated into three clusters based on mesocotyl length, while three genotypes, PVAQEH-4, LY1919-14, and A1804-66, were ungrouped (Table 2, Figure 1). Cluster I was composed of 10 genotypes with no or short mesocotyl growth. Cluster II was composed of 20 genotypes with moderate mesocotyl growth ranging from 2.45 cm for †PVAEH 36 to 5.56 cm for PVAEH 29, while cluster 3 had 36 genotypes characterised by long observed mesocotyls ranging from 5.95 cm for †PVAEH 32 to 9.02 cm for LY1901-18. The observed clustering pattern implied the existence of considerable genetic diversity for ML among the maize genotypes. Several past works have classified maize genotypes into clusters based on a single trait. For instance, Oyetunde et al. [11] and Badu-Apraku et al. [12] classified maize genotypes into heterotic groups based on specific combining ability for grain yield. Crosses can be planned involving genotypes from clusters 1 and III (the most divergent groups) to exploit heterosis for ML.

Table 2.
Mean ± standard deviation (Std) of mesocotyl length (ML) and classification into clusters based on ML of maize genotypes germinated in the dark for 10 days.
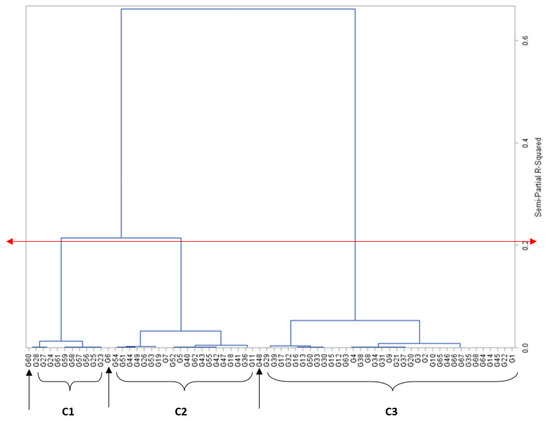
Figure 1.
Dendrogram of relatedness among maize genotypes (X-axis), based on genetic similarity (Y-axis) from Ward’s minimum variance cluster analysis. The red double-arrowed line delineates the genotypes into clusters at approximately 65% level of similarity; C1, C2, C3 are clusters 1, 2, and 3, respectively; single-arrowed line signifies ungrouped genotypes.
Sixty-two of the maize genotypes tested developed measurable mesocotyl. The mean mesocotyl length ranged from 0.58 cm for A1312-12 to 9.02 cm for LY1901-18 (Table 2). The differences in ML lengths of the maize genotypes further indicated the existence of variability of the trait, and genotypes with high ML values, such as LY191-01, †PVAEH 34, and †PVAEH 31 have potential to draw moisture from the subsoil and could be adaptable to drought-prone areas. The standard deviation of a set of numbers is an indication of the spread of the numbers from the mean. A low standard deviation value implies that the majority of the numbers are close to the average, while a high value implies that most numbers are far from the mean. Genotypes with low standard deviation estimates should, therefore, be more reliable for prediction. Hence, genotypes that combined high ML with low standard deviation values, such as LY1901-18 and †PVAEH 34, would be more reliable in predicting the ML of maize. In addition, planned crosses can be made involving the genotypes A1312-12, LY1501-7, and LY1901-14 to develop maize hybrids with potential for adaptation to drought conditions. Moreover, genotypes with high standard deviation values could be useful for development of inbred lines useful in future hybrid programs with focus on developing deep-sowing tolerant maize.
Information on seed micronutrients and biochemical composition which affects the seedling vigor and organ development may provide some insight to the understanding of the mechanism underlying mesocotyl elongation. In this study, significant (p ≤ 0.05/0.01) associations were observed in the relationship of ML with all the measured biochemical parameters except selenium (Table 3). Furthermore, all the significant associations were positive except for the negative association between ML and amylase content. The significant levels of association observed implied the possibility of simultaneous improvement of each of iron, zinc, crude protein, free fatty acid, oil content, linoleic acid, and amylase content with mesocotyl length of maize. The quality attributes could serve as selection criteria for ML. Muhammad [13] employed nutrient priming to reveal the impact of seed reserves on seedling development and root biomass in maize. According to Martinez-Ballesta et al. [14], seed biofortification enhances seed vigour, and can play a role in abiotic stress tolerance.

Table 3.
Pearson correlation coefficients of seed biochemical parameters with mesocotyl length (ML).
4. Conclusions
There was substantial genetic variability among the 68 maize genotypes, allowing the grouping into different clusters. Genotypes †PVAEH 31, †PVAEH 34, and LY1901-18 have potential for improvement as DST maize. Biofortification has the potential to enhance the elongation of mesocotyl in maize during germination. The results obtained from this study provide a basis for the development of DST maize for drought-prone environments.
Supplementary Materials
The poster presentation is available online at https://www.mdpi.com/2673-9976/4/1/26/s1.
Author Contributions
O.A.O. conceived the experiment; O.A.O. and K.A.A. designed the experiment; O.A.O. and K.G.G. performed the experiment; O.A.O. analysed the data and wrote the original draft. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors are grateful to IITA Maize Improvement Program in Ibadan, Nigeria for providing the seeds for the research. We are also grateful to the staff and students of the Department of Crop Production and Horticulture, School of Agriculture, Lagos State Polytechnic, Ikorodu, Nigeria, for assistance with data collection.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Niu, L.; Hao, R.; Wu, X.; Wang, W. Maize mesocotyl: Role in response to stress and deep-sowing tolerance. Plant Breed. 2020, 139, 1–8. [Google Scholar] [CrossRef]
- Zhao, G.W.; Wang, J.H. Effect of auxin on mesocotyl elongation of dark grown maize under different seeding depths. Russ. J. Plant Physiol. 2010, 57, 79–86. [Google Scholar] [CrossRef]
- Pan, B.R.; Zhong, T.L.; Zhao, G.W. Promoting deep-sowing germinability of corn (Zea mays) by seed soaking with gibberellic acid. Arch. Agron. Soil Sci. 2017, 63, 1314–1323. [Google Scholar] [CrossRef]
- Zhao, G.W.; Wang, J.H. Effect of gibberellin and uniconazole on mesocotyl elongation of dark-grown maize under different seeding depths. Plant Prod. Sci. 2008, 11, 423–429. [Google Scholar] [CrossRef]
- Kutschera, U.; Wang, Z.-Y. Growth-limiting proteins in maize coleoptiles and the auxin-brassinosteroid hypothesis of mesocotyl elongation. Protoplasma 2016, 253, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Sivasubramanian, S.; Menon, M. Heterosis and inbreeding depression in rice. Madras Agric. J. 1973, 60, 1139. [Google Scholar]
- Johnson, H.W.; Robinson, H.; Comstock, R.F. Estimates of Genetic and Environmental Variability in Soybean. Agron. J. 1955, 47, 314–318. [Google Scholar] [CrossRef]
- Olayiwola, M.O.; Soremi, P.A.S. Variability for dry fodder yield and component traits in cowpea [Vigna unguiculata (L.) Walp]. Electron. J. Plant Breed. 2014, 5, 58–62. [Google Scholar]
- Ogunniyan, D.J.; Olakojo, S.A. Genetic variation, heritability, genetic advance and agronomic character association of yellow elite inbred lines of maize (Zea mays L.). Niger. J. Genet. 2015, 28, 24–28. [Google Scholar] [CrossRef] [Green Version]
- Mostafavi, K.; Shoahosseini, M.; Geive, G.S. Multivariate analysis of variation among traits of corn hybrids traits under drought stress. Int. J. Agric. Sci. 2011, 1, 416–422. [Google Scholar]
- Oyetunde, O.A.; Badu-Apraku, B.; Ariyo, O.J.; Alake, C.O. Efficiencies of Heterotic Grouping Methods for Classifying Early Maturing Maize Inbred Lines. Agronomy 2020, 10, 1198. [Google Scholar] [CrossRef]
- Badu-Apraku, B.; Fakorede, M.A.B.; Gedil, M.; Annor, B.; Talabi, A.O.; Akaogu, I.C.; Oyekunle, M.; Akinwale, R.O.; Fasanmade, T.Y. Heterotic patterns of IITA and CIMMYT Early maturing Yellow Maize Inbreds under Contrasting Environments. Agron. J. 2016, 108, 1321–1336. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, K. Nutrient Seed Priming Improves Abiotic Stress Tolerance in Zea mays L. and Glycine max L. Ph.D. Thesis, University of Hohenheim, Stuttgart, Germany, 2012. [Google Scholar]
- Martínez-Ballesta, M.C.; Egea-Gilabert, C.; Conesa, E.; Ochoa, J.; Vicente, M.J.; Franco, J.A.; Bañon, S.; Martínez, J.J.; Fernández, J.A. The Importance of Ion Homeostasis and Nutrient Status in Seed Development and Germination. Agronomy 2020, 10, 504. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).