Abstract
Rhizospheres harbor many beneficial microorganisms interacting with the plant ecosystem. However, in agriculture, there is a tendency to remove any plant that is different from that being cultivated. This work aimed to display the root microbial communities of native vegetation growing wild in agricultural soil. Thus, high-throughput sequencing of culture-independent marker genes was performed for bacteria and fungi from these habitats after a period of high environmental temperatures. With respect to bacteria, results revealed a number of Operational Taxonomic Units (OTUs) ranging from 3210 to 3266. With respect to fungi, the results revealed a number of OTUs ranging from 963 to 973. Information on the composition of the rhizosphere microbial communities favours the understanding of their potential functions and their beneficial effects on the sustainability of the agrosystems.
1. Introduction
In the field, plant roots are considered vital areas of interaction with the soil microbiome [1]. Rhizospheres harbor a great diversity of microorganisms, many of which have a role as crucial regulators of substrate and energy affecting soil fertility and plant nutrient availability [2,3]. The absorption of ions and water taking place in the roots, along with the production of carbon-rich compounds, commonly leads to active plant–soil interaction areas, where rhizosphere microorganisms can be favouring plant nutrient acquisition and/or contributing to the recycling of nitrogen, phosphorous, organic compounds and other nutrients, and/or taking part in beneficial symbiotic associations, and synthesis of plant growth-promoting compounds and vegetative production [1,2,3]. This is of great importance for soil and plant health, plant development and sustainability, and efficient nutrient cycling, and therefore, it provides global benefits to plant ecosystems.
However, one of the most frequently applied agricultural soil management techniques consists of removing any plant different from that of the crop species being cultivated. In general, this strategy, called tillage, may cause soil disturbance, leading to higher potential for soil erosion and compaction, and loss of microbiological biodiversity, leading to an alteration of the processes taking place in the ecosystems because of changes in the cycles of energy and matter [4]. Under stressful conditions, the presence of osmolytes like sugars, amino acids, and polyols can help plants to maintain the correct cellular tonicity, positively influencing plant and soil health [5]. To a great extent, many of the mechanisms that contribute to advantageous biodiversity-ecosystem functional interactions take place belowground, enhancing the productivity and the resilience to extreme climatic events [2,6].
In order to find out the microbial diversity associated with the rhizosphere of native plants growing wild in soil devoted to agricultural crops, this work addressed high-throughput DNA sequencing to display the fungal and bacterial communities present in them, in environmental conditions of prolonged high temperatures and drought.
2. Materials and Methods
2.1. Rhizosphere Sampling and Processing
Composite plant samples were taken from four sampling sites within the same agricultural rainfed area in the Community of Madrid (Spain), after a summer period of prolonged drought and environmental high temperatures. The sites were selected based on the challenging conditions that were faced by plants and microorganisms. From each area, at least 10 whole plants from the wildly growing population were randomly collected, not necessarily of the same identity. They were kept intact without contact between the roots and their aerial parts until subsequent processing. Sampling from several areas, as well as processing, were performed at the same time, for comparative purposes. After cutting the roots under aseptic conditions, 3 g of each composite root sample were added to phosphate-buffered saline (PBS) buffer [pH 7.4; 137 mM NaCl, 10 mM Na2HPO4, 2.7 mM KCl, 1.9 mM KH2PO4] in 1:10 (w/v), and then shaken at 200 r.p.m. during 30 min, similarly to [4]. Immediately after shaking, suspensions were collected and kept at −25 °C in glycerol 25% (v/v) to maintain sample integrity.
2.2. DNA Extraction and Purification
Aliquots of 1 mL were taken from each of the suspensions and centrifuged at 14,000 r.p.m. for 10 min. The supernatants were discarded, and the pellets resuspended in the lysis buffer, thus beginning the extraction process. DNA extractions were performed with the commercial DNeasy® Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Purified DNAs were kept at −25 °C until further use.
2.3. High-Throughput DNA Sequencing
Libraries were prepared for bacteria and fungi by partial amplification of the 16S rRNA gene and the ITS genomic region, respectively. The primers included the Illumina sequencing primer sequences attached [7]. The oligonucleotide indices for multiplexing different libraries were subsequently merged. Negative controls with no DNA (BPCR) were considered in the PCRs to verify the absence of contamination during library preparation. The libraries were checked on 2% agarose gels and visualised under UV light to verify their size. They were purified using the Mag-Bind RXNPure Plus magnetic beads (Omega Bio-tek Inc, Norcross, GA, USA). Then, they were pooled in equimolar amounts according to quantification data by the Qubit dsDNA HS Assay (Thermo Fisher Scientific, Waltham, MA, USA), and the pool was sequenced in 2 GB of a NovaSeq PE250 run (Illumina Inc, San Diego, CA, USA) [7].
The quality of the FASTQ files was checked using the software FastQC version 0.11.9 and summarised using MultiQC. The obtained 16S and ITS amplicon reads were processed using QIIME2 [8], particularly the tool DADA2. The whole dataset was trimmed and filtered to avoid low-quality scores. The resulting sequences were clustered into amplicon sequence variants (ASVs). The number of times that each ASV was observed in each sample was established.
The taxonomy was assigned to bacterial and fungal ASVs using pre-trained classifiers of the SILVA and UNITE reference databases, respectively. Different filters were applied. Singletons, as well as ASVs occurring below 0.01% in each sample, sequences assigned only at the kingdom level (“Bacteria” or “Fungi”), and unassigned ASVs were excluded. Non-bacterial ASVs, such as eukaryotic sequences of chloroplast and mitochondrial origin, were removed. Final filtered ASV tables for bacteria and fungi data were converted into Biological Observation Matrix files (.biom), which were imported into R 3.6.1 [9] using the package phyloseq 1.24.2 to plot the results of the analyses. The representative sequences were extracted.
Stacked bar plots at each taxonomic level were exported as QIIME zipped visualisation (.qzv) files to be opened in the QIIME 2 View interface https://view.qiime2.org (accessed on 18 April 2025).
3. Results
Sample sizes of n = 10 native plants were sufficient for the production of diverse libraries due to the richness of microorganisms usually found in the rhizosphere. The obtained DNA concentrations ranged from 0.10 to 0.30 ng/µL.
3.1. Bacterial Diversity from the Rhizosphere of Native Plants
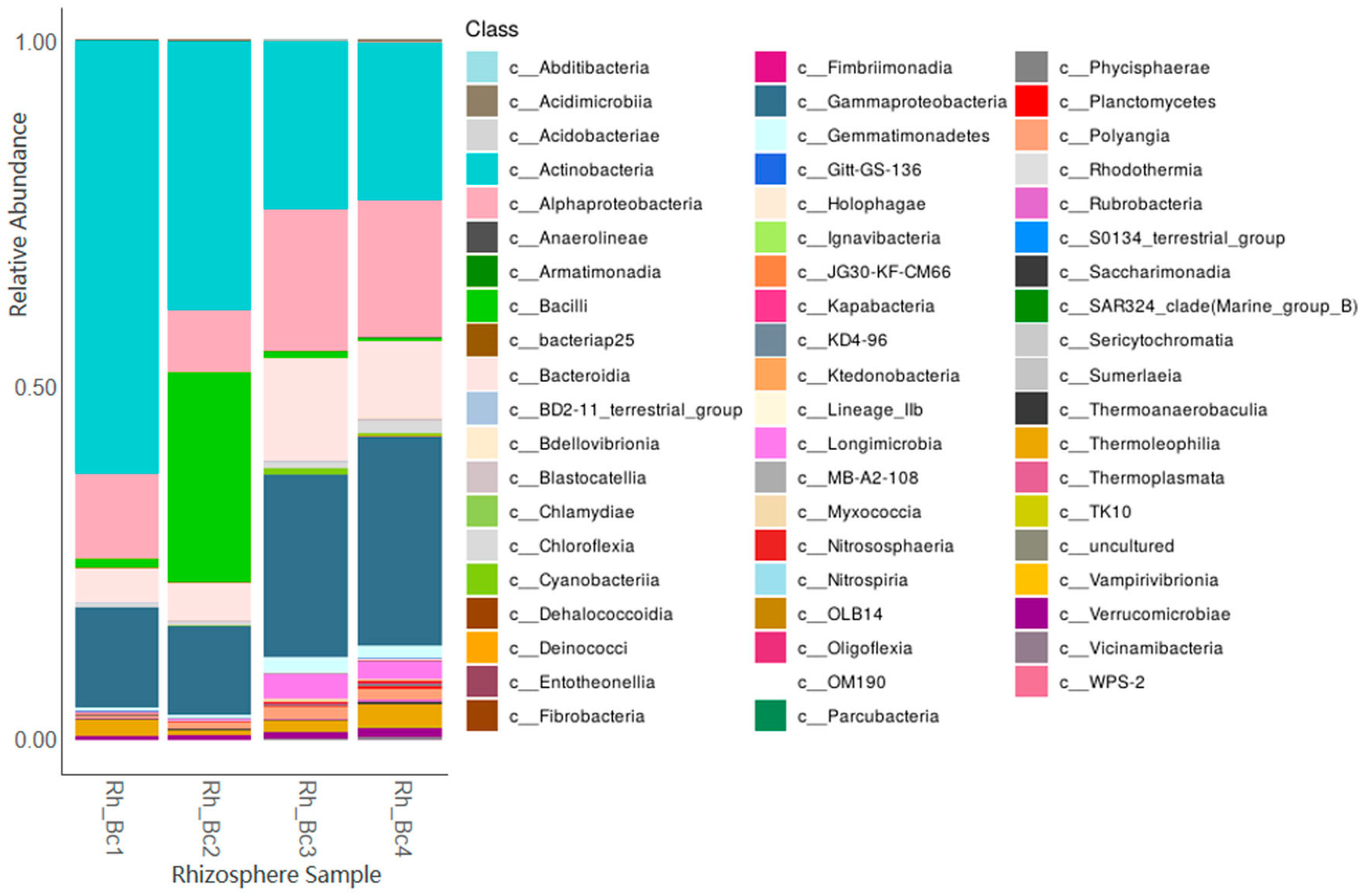
With respect to bacteria, results revealed a number of Operational Taxonomic Units (OTUs) ranging from 3210 to 3266, with the relatively most abundant identified families and/or genera being Bacillus, Caulobacter, Domibacillus, Erwiniaceae, Glycomyces, Lechevalieria, Massilia, Micrococcaceae, Paenarthrobacter, Promicromonospora, Pseudomonas, Rhizobiaceae, Sphingomonas, Streptomyces, and Terribacillus. Figure 1 displays the obtained data at the Class taxonomic level.
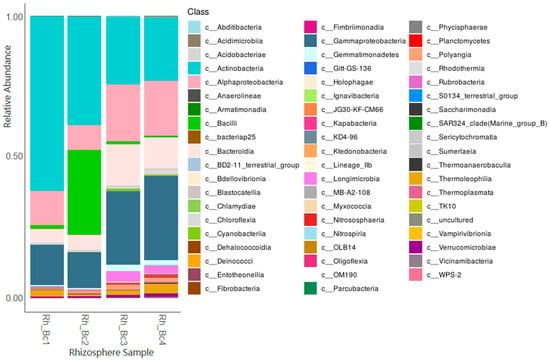
Figure 1.
Relative bacterial abundance in the DNAs of the analysed rhizospheres at the Class level.
3.2. Fungal Diversity from the Rhizosphere of Native Plants
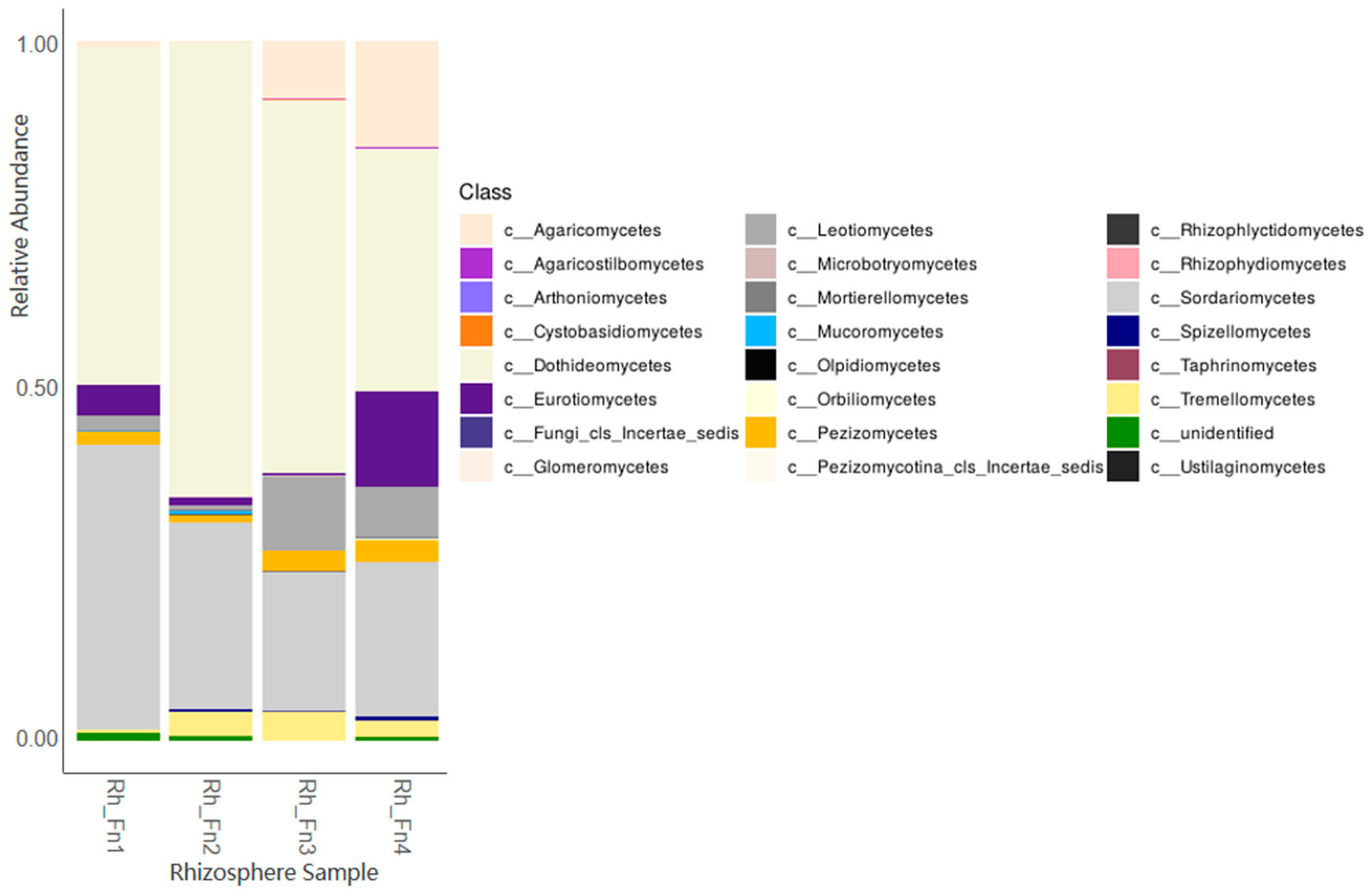
With respect to fungi, the results revealed a number of OTUs ranging from 963 to 973, with the relatively most abundant identified families and/or genera being Alternaria, Aspergillus, Aureobasidium, Chaetomium, Cladosporium, Coniothyrium, Didymellaceae, Entoloma, Fusarium, Macrophomina, Monosporascus, Poculum, and Sclerostagonospora. Figure 2 displays the obtained data at the Class taxonomic level.
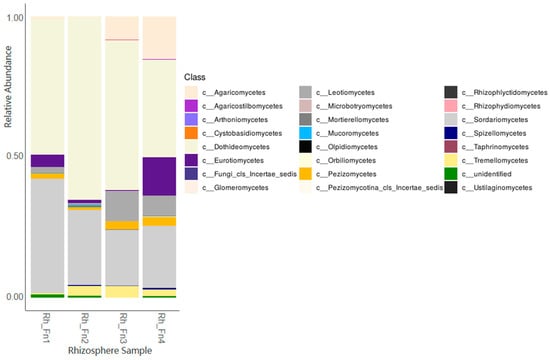
Figure 2.
Relative fungal abundance in the DNAs of the analysed rhizospheres at the Class level.
4. Discussion
A high number of bacterial and fungal OTUs was observed in the rhizosphere samples, similar to [2,10,11]. It is worth noting the great variety of these microorganisms that have been found in wild plants growing spontaneously in agricultural soil, which are usually eliminated, and especially because they were under unfavourable conditions, with prolonged low environmental humidity, absence of irrigation, and high temperatures. All of them, the majority along with the totality of the substantial minority species, can contribute to the sustainability of the agricultural field. Soil bacteria and fungi mainly participate in the regulation of the dynamics of numerous physiological processes, such as formation of soil structure, stabilization of soil organic matter and decomposition of residues, compound transformation, nitrogen fixation, biofertilization, hormone production, biocontrol of diseases caused by root pathogens, shifts in habitats for other microorganisms, and protection against drought [12,13]. From the bacterial and fungal families and/or genera detected in this work as being more abundant, the most frequently reported taking part in nutrient cycling and availability would be Aspergillus, Bacillus, Pseudomonas, Streptomyces, and Rhizobiaceae, whereas those having a role in protecting plants from pathogens and promoting sustainable agricultural practices would be Aureobasidium, Bacillus, Pseudomonas, and Streptomyces [12,13,14]. Information on the composition of the rhizosphere bacterial and fungal communities allows for a deeper understanding of their potential functions in soil processes of agricultural ecosystems and, eventually, of their beneficial effects on plant growth [10,11,12,13]. This knowledge can be used to identify strategies to control or adjust the rhizosphere microbiome for microbial benefits to efficient nutrient cycling and soil health [11,13], and therefore, to guide and monitor crop and/or land management under challenging environmental conditions [15].
Moreover, this work reminds us of the importance of the often overlooked microbial communities that thrive in challenging agricultural environments and the need to embrace the natural ecosystems that coexist with cultivated crops.
Author Contributions
Conceptualization, B.Á.; methodology, M.A.-M., E.M.-G., L.T.-H., S.B.-L., A.R.-C. and M.M.-I.; analysis, M.A.-M., E.M.-G., L.T.-H. and B.Á.; writing, B.Á.; funding acquisition, B.Á. All authors have read and agreed to the published version of the manuscript.
Funding
The authors thank funding from IMIDRA Plant Protection Laboratory.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Acknowledgments
A.R.-C. was the recipient of a grant from Programa de Garantía Juvenil of the Consejería de Educación, Universidades, Ciencia y Portavocía of the Community of Madrid (Spain) and the Fondo Social Europeo of the European Union (EU). DNA metabarcoding analyses were carried out by All Genetics and Biology S.L. (www.allgenetics.eu).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Molefe, R.R.; Amoo, A.E.; Babalola, O.O. Communication Between Plant Roots and the Soil Microbiome; Involvement in Plant Growth and Development. Symbiosis 2023, 90, 231–239. [Google Scholar] [CrossRef]
- Maheshwari, D.; Agarwal, M.; Dheeman, S. Trends and Prospects of Microbial Diversity in Rhizosphere. In Bacterial Diversity in Sustainable Agriculture. Sustainable Development and Biodiversity; Maheshwari, D., Ed.; Springer: Cham, Switzerland, 2014; Volume 1, pp. 1–22. [Google Scholar] [CrossRef]
- Schmidt, J.E.; Kent, A.D.; Brisson, V.L.; Gaudin, A.C.M. Agricultural Management and Plant Selection Interactively Affect Rhizosphere Microbial Community Structure and Nitrogen Cycling. Microbiome 2019, 7, 146. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, B.; Tihomirova-Hristova, L.; Bielsa-Lozoya, S.; Morate-Gutiérrez, E.; Antón-Iruela, O.; Bienes, R.; García-Díaz, A.; Sastre, B. Effect of Olive Grove Management by Groundcovers on Soil Microbiological Biomass. Modern Agric. Sci. Technol. 2021, 7, 7–12. [Google Scholar] [CrossRef]
- Wijayasinghe, Y.S.; Tyagi, A.; Poddar, N.K. Regulation of Cell Volume by Osmolytes. In Cellular Osmolytes; Singh, L.R., Dar, T.A., Eds.; SpringerNature: Singapore, 2017; Chapter 9; pp. 195–228. [Google Scholar] [CrossRef]
- Chauhan, P.; Sharma, N.; Tapwal, A.; Kumar, A.; Verma, G.S.; Meena, M.; Seth, C.S.; Swapnil, P. Soil Microbiome: Diversity, Benefits and Interactions with Plants. Sustainability 2023, 15, 14643. [Google Scholar] [CrossRef]
- Illumina, I. Effects of Index Misassignment on Multiplexing and Downstream Analysis. 2017. Available online: https://www.illumina.com (accessed on 1 December 2024).
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- R-Team-Core. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 1 December 2024).
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The Rhizosphere Microbiome: Significance of Plant Beneficial, Plant Pathogenic, and Human Pathogenic Microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Ling, N.; Wang, T.; Kuzyakov, Y. Rhizosphere Bacteriome Structure and Functions. Nat. Commun. 2022, 13, 836. [Google Scholar] [CrossRef] [PubMed]
- Frąc, M.; Hannula, S.E.; Bełka, M.; Jędryczka, M. Fungal Biodiversity and Their Role in Soil Health. Front. Microbiol. 2018, 9, 707. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Song, Y.; An, Y.; Lu, Y.; Zhong, G. Soil Microorganisms: Their Role in Enhancing Crop Nutrition and Health. Diversity 2024, 16, 734. [Google Scholar] [CrossRef]
- Meza-Manzaneque, B.; Pérez-Díaz, M.; Biosca, E.G.; Álvarez, B. Isolation and Characterization of Agricultural Soil Bacteria with Biotechnological and Biological Control Potential Applications. Biol. Life Sci. Forum 2024, 31, 28. [Google Scholar] [CrossRef]
- Dar, Z.A.; Bhat, R.A.; Bhat, J.I.A.; Mir, S.A.; Amin, A.; Rashid, A.; Rifat, B.; Lone, R. Microbial Diversity and Their Role in Plant and Soil Health Under Stress Conditions. In In Vitro Plant Breeding Towards Novel Agronomic Traits; Kumar, M., Muthusamy, A., Kumar, V., Bhalla-Sarin, N., Eds.; SpringerNature: Singapore, 2019; Chapter 9; pp. 149–166. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).