Abstract
The intestinal microbiota plays an important role in the homeostasis of the intestinal tract, and the fact that exercise could have an impact on its functionality is currently the subject of various studies. Exercise is used as a strong preventive and treatment strategy in numerous chronic diseases, influencing metabolism, immunity, and physiological functions. Some recent studies provide information on exercise as a factor that could favor the growth of beneficial species and increase the diversity of the intestinal microbiota. However, the difficulty in determining the true effect of exercise is only due to the confounding influence of lifestyle and dietary habits. Regardless, there is still interest in the implications of the gut microbiota for elite sports because the details are considered crucial for success in different competitions. This narrative review tries to collect the information available in the literature on the role that exercise plays in the modulation of the balance of the intestinal microbiota.
1. Gut Microbiota
The main objective of the narrative is to analyze the association between the intestinal microbiota and exercise and to conclude if there is sufficient evidence to consider new approaches for intestinal recovery and protection as a new important factor in performance.
Joshua Lederberg, an American biologist who received the Nobel Prize in Medicine in 1958, stated that the microorganisms found in humans protect us symbiotically in our own body. These communities of microorganisms, known today under the term microbiota, consist of a variety of microorganisms that includes eukaryotes, archaea, bacteria, and viruses, constituting about 1.3% of the body mass, and are essential to maintain optimal health. These microorganisms inhabit different parts of the body, colonizing the mucosa within a given anatomical niche, such as the respiratory system, the walls of the digestive system or also the urogenital system, among other surfaces []. More diversity of the microbiota is found in the intestinal tract and mouth, while the skin contains less diversity, followed by the vagina, although a consensus has not yet been reached on the number of species that inhabit it [].
The microbiota is known as the set of microorganisms residing in each ecosystem, with a symbiotic relationship and with adaptive properties and rapid renewal, forming a large metabolic unit. The intestinal microbiota is a set of microorganisms made up of approximately 100,000 million bacteria that live in our intestine. The microbiota is responsible, among other functions, for maintaining the well-being of the intestinal mucosa, helping us digest food and converting harmful elements into less toxic substances.
In 2014, the term “intestinal flora” was coined to describe the intestinal microbiota, although later, it was changed to gut microbiota [].
Some gut microbiota bacteria attach to the cells’ external wall and to specific receptors called adhesins. They can adapt to the specific conditions of the environment, such as humidity, temperature, or pH, being able to activate defense mechanisms in case of interaction, for example, with a harmful virus. The homeostasis of the intestinal tract depends on the balance between the microbiota, intestinal permeability, and local immunity; in the event of any non-adaptive alteration, it can produce negative effects on the host itself. When these alterations occur, it is known as intestinal dysbiosis, which can affect digestion, nutrient absorption, vitamin production, and the control of harmful microorganisms [].
Most of the bacteria that make up the gut microbiota live in the colon []. The colon is described as a potentially active organ due to the extensive activity of the gut microbiota and has been compared to the liver due to its high metabolic capacity []. The microbiota is dominated by four main phyla: Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria []. Although there are also bacteria in smaller proportions that are vital for the function of the microbiota, Firmicutes and Bacteriodetes stand out due to their proportion throughout the intestinal tract, as they represent 60% and 20%, respectively, of all the gut microbiota, while the Proteobacteria phylum covers 5% and Actinobacteria 3%, approximately [].
2. The Gut Microbiota Function
The functions of the gut microbiota include the following:
2.1. Protection Function in the Intestinal Barrier
The gut microbiota plays an important role in helping to maintain the integrity of the intestinal barrier, thus protecting the host from pathogens. In turn, the microbiota develops defense mechanisms together with the epithelial cells, which are responsible for establishing an effective and harmonious intestinal barrier, preventing the passage of harmful substances, antigens, toxins, and microbial products and, on the other hand, the absorption of nutrients benign [].
The main elements that form the intestinal barrier are the mucus, the tight junctions between enterocytes, antimicrobial peptides, and immunoglobulin A (IgA) secretion []. The intercellular junctions of the intestinal barrier separate the internal environment from the external environment, making it impermeable to certain pathogenic substances and permeable to nutrients [].
The mucosa interacts with the microbiota to maintain intestinal homeostasis; in addition, the intestinal microbiota induces the biosynthesis of antimicrobial peptides by mucosal Paneth cells, such as cathelicidins, lectins, and defensins, by activating their membrane receptors. Bacteriodetes thetaiotaomicron and Lactobacillus innocua are two of the main agents that stimulate Paneth cells [].
Another reinforcement of this barrier function by the microbiota of the intestinal lumen is the production of lactic acid by Lactobacillus, which reinforces the ability of human lysozyme released in the lumen to destroy peptidoglycan []. Short-chain fatty acids (SCFAs) released by bacterial fermentation also stimulate the production of the antimicrobial peptide cathelicidin. These SCFAs acidify the intestinal lumen and prevent colonization by pathogens sensitive to this pH, such as Salmonella, Escherichia, and Clostridium difficile [].
The homeostasis of the barrier function would result in an optimal symbiosis; otherwise, if the function of the intestinal barrier does not favor an adequate mechanism, it will have an exaggerated or abnormal immune response that would lead to poor control of intestinal permeability, leading to what we know as intestinal dysbiosis [,].
2.2. Function on the Development of the Immune System
The gut microbiota contributes to gut immunomodulation in conjunction with the innate and adaptive immune systems []. In line with the previous protection function, the immune system has co-evolved to maintain a symbiotic relationship with the commensal species of the microbiota while keeping other species considered pathogenic in this intestinal ecosystem under control. About 70% of the lymphocytes in the human body live in the digestive system, which gives us an idea of the importance of this function in the development [].
Components and cell types of the immune system involved in the immunomodulatory process include gut-associated lymphoid tissues (GALT), effector and regulatory T cells, IgA-producing B (plasma) cells, Group 3 innate lymphoid cells, resident macrophages, and dendritic cells in the lamina itself [].
The role of the gut microbiota in forming a functional GALT is implicit in the poor development of Peyer’s patches and isolated lymphoid follicles that are marked by an abundance of IgE B cells rather than IgA B cells []. The absence of the intestinal microbiota causes the plasma cell populations of the intestinal mucosa to differentiate mainly towards greater production of IgE, instead of IgA, which is associated with an increased risk of allergies [].
Effector T cells are regulated in the mucosa by Th17 cells. Some bacteria, such as Bacillus fragilis, belonging to the Bifidobacterium or Clostridium genera, activate Treg lymphocytes via Toll-like receptors (TLR) that are part of the innate immune system, inducing systemic anti-inflammatory effects, which helps to develop tolerance to antigens present in the diet and some bacteria in the lumen [].
This cross-communication between the microbiota and the epithelial cells is also responsible for the release of IgA and for stimulating the production of glycoproteins to form more mucus. The main stimulators of epithelial IgA production are Gram-negative species such as Bacteroides, which activate dendritic cells in the mucosa so that they induce IgA production by plasma cells (activated lymphocytes) in the submucosa. This IgA prevents microorganisms from activating pro-inflammatory pathways, as they cannot bind when coated by these antibodies, to receptors such as TLRs. In turn, IgA collaborates in capturing antigens and prevents their penetration through the intestinal barrier, limiting the movement of pathogens and neutralizing toxins throughout the lumen []. The presence of high populations of Suturella, on the other hand, reduces IgA production in the lumen, since this bacterium releases IgA-degrading proteases []. The complexity of the intestinal microbiota, per se, is a defense against the colonization of this ecosystem by other types of microorganisms []. This is especially important in adults who have a much more stable intestinal microbiota than children or subjects with intestinal dysbiosis. The production of bacteriocins by the resistant microbiota is responsible for this resistance to colonization of the lumen by other bacteria. Similarly, the bacteriophages in the intestinal lumen can prevent the establishment of susceptible bacterial species. All these mechanisms prevent the bacterial translocation factor through the intestinal epithelium and therefore prevent a systemic immune response to a possible infection [].
2.3. The Intestinal Microbiota Performs a Metabolic Function
Contributing to the digestion of nutrients from the diet []. The gut microbiota obtains its nutrients mainly from carbohydrates in the diet. Fermentation of carbohydrates that escape proximal digestion and indigestible oligosaccharides are metabolized by bacteria such as Bacteroides, Roseburia, Bifidobacterium, Faecalibacterium, and Enterobacteria, resulting in the synthesis of SCFAs such as butyrate, propionate, and acetate, which are rich sources of energy for the host [].
This metabolic process is favored by the more acidic pH of the right colon. In contrast, in the left colon, microorganisms grow more slowly due to the low arrival of food with a more neutral pH [].
Members of the genus Bacteroides are the predominant organisms involved in carbohydrate metabolism, achieved by expressing enzymes such as glycosyltransferases, glycoside hydrolases, and polysaccharide lyases. The best example among these organisms is Bacteroides thetaiotaomicron, which is endowed with a genome encoding more than 260 hydrolases [].
The gut microbiota also has an efficient protein metabolizing machinery that functions through microbial proteinases and peptidases in conjunction with human proteinases. Various amino acid transporters in the bacterial cell wall facilitate the entry of amino acids from the intestinal lumen into the bacteria, where they convert the amino acids into small signaling molecules and antimicrobial peptides (bacteriocins) [].
Another metabolic function of great importance is described in the literature, such as the synthesis of vitamins such as vitamin K and vitamin B, and the function of lipid regulation. In addition, members of the genus Bacteroides synthesize conjugated linoleic acid (CLA), which has antidiabetic, antiatherogenic, antiobesogenic, hypolipidemic, and immunomodulatory properties []. The gut microbiota has also been shown to positively impact lipid metabolism by suppressing the inhibition of lipoprotein lipase (LPL) activity in adipocytes [].
3. Factors That Modify the Gut Microbiota
Many factors could modify the gut microbiota. The ones that stand out are diet or eating habits, probiotics, prebiotics and polyphenols, pharmacological treatments, and physical exercise [].
Other factors involved, apart from those mentioned above, are stress, genetics, age, types of birth, and exercise [], (Figure 1).
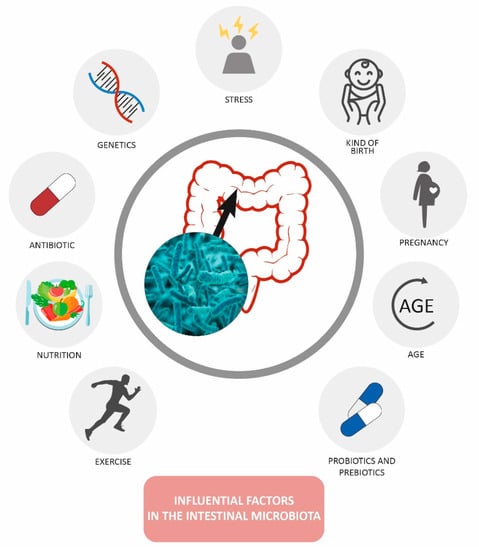
Figure 1.
Factors modifying the gut microbiota [].
Exercise and Gut Microbiota
There are several potential mechanisms by which exercise could modify the gut microbiota. Several studies by Hoffman Goetz and colleagues [,] have reported that exercise alters intraepithelial lymphocyte gene expression, downregulating proinflammatory cytokines and up-regulating anti-inflammatory cytokines and antioxidant enzymes. These immune cells reside close to microbial communities and produce antimicrobial factors that are essential for mediating host-microbial homeostasis [].
Similarly, exercise can affect the integrity of the intestinal mucus layer, which plays an important role in preventing pathogens from adhering to the intestinal epithelium and serves as an important substrate for certain mucosa-associated bacteria, such as A. muciniphila. Exercise raises core temperature and causes heat stress, particularly when performed for prolonged periods or in a hot environment []. Exercise can also reduce intestinal blood flow by more than 50%, with significant intestinal ischemia occurring within 10 min of high-intensity exercise []. At rest, the splanchnic bed undergoes rapid reperfusion. Although the intestine is an anaerobic environment, intestinal epithelial cells primarily use oxidative metabolism, and high-intensity exercise is known to transiently impair intestinal barrier function []. Therefore, exercise-induced heat stress and ischemia may produce interactions between the intestinal mucosal immune system and the microbiota residing in the intestinal lumen.
Evidence for the role of exercise in modifying the human gut microbiota first emerged from cross-sectional studies in 2014 []. Clarke et al. found that the gut microbiota of professional rugby players had a higher alpha diversity and a higher relative abundance of 40 different bacterial taxa than the gut microbiota of the control group, being sedentary subjects []. More recently, Bressa et al. compared active women with sedentary controls and found that women who engaged in at least 3 h of exercise per week had increased levels of Faecalibacterium prausnitzii, Roseburia hominis, and Akkermansia muciniphila [], which are producers of butyrate [], while A. muciniphila has been associated with a lean body mass index (BMI) and better metabolic health [].
Several studies have also attempted to correlate the composition and metabolic capacity of the microbiota with cardiorespiratory fitness, with Durk et al. showing a higher ratio of Firmicutes to Bacteroidetes in subjects with better metabolic health, and was significantly correlated with maximal oxygen consumption (VO2max) []. Estaki et al. found in younger adults that microbial diversity and abundance of butyrate-producing bacterial taxa were positively correlated with cardiorespiratory fitness [], while Barton et al. found, through metagenomic analyses differences in fecal microbiota between athletes and sedentary controls. Athletes showed relative increases in different metabolic pathways, such as amino acid biosynthesis and carbohydrate metabolism, as well as increased production of SCFAs, such as acetate, propionate, and butyrate produced by the microbiota [].
However, all these studies were limited by their cross-sectional design and their difficulty in controlling diet effects on the gut microbiota. There is considerable inter-individual variability in the microbiota composition, and active individuals tend to eat differently from sedentary ones []. For example, Clarke and colleagues found that increased protein intake by elite rugby players explained many of the observed differences in gut microbiota []. These limitations suggested the need to analyze longitudinal studies to determine whether exercise independently alters the gut microbiota in humans.
Recently, Allen and colleagues obtained results in the first controlled longitudinal study with a strict diet to evaluate the effects of exercise on the gut microbiota. Various bacterial taxa were differentially altered by exercise depending on the subjects’ BMI. For example, exercise increased Faecalibacterium species in lean subjects but reduced their abundance in obese subjects, Bacteroides species decreased in lean subjects and increased in obese subjects. Six weeks of exercise also increased the abundance of butyrate-producing taxa and fecal acetate concentrations, but only in lean subjects. Interestingly, most bacterial taxa and SCFAs that increased with exercise subsequently decreased during the 6-week sedentary rest period, indicating that the effects of exercise on the microbiota were transient and reversible [].
Similarly, Cronin and colleagues sought to determine whether a short-term exercise regimen, with or without whey protein supplementation, could alter gut microbial composition and function in predominantly overweight or obese adult men and women. The post-intervention evaluation revealed no significant changes in taxonomic composition or metabolic pathways in either exercise group compared to baseline. However, a trend for increased bacterial diversity was observed in the exercise and exercise plus whey protein groups compared to the group receiving whey protein alone. Metagenomic and metabolomic analyzes revealed only modest alterations in microbial metabolism. Although the study had a large sample size, the authors note that self-reported maintenance of usual dietary intake and a wide BMI range may have prevented the detection of more significant changes [].
Munukka and colleagues conducted a similar study to determine whether resistance exercise could affect the gut microbiota in previously sedentary overweight women. Six weeks of light-to-moderate intensity exercise cycles increased the relative abundance of A. muciniphila and a decrease in the phylum Proteobacteria. What is most interesting is that only the microbiota of about half of the subjects responded significantly to exercise. Metagenomic analysis revealed that exercise training decreased the abundance of several genes related to fructose and amino acid metabolism [].
These findings suggest that exercise has independent effects on the gut microbiota, but training of longer duration or intensity may be necessary to induce significant taxonomic and metagenomic changes. In addition, the microbiota of lean people may respond differently to an exercise intervention than that of overweight or obese people (Figure 2).
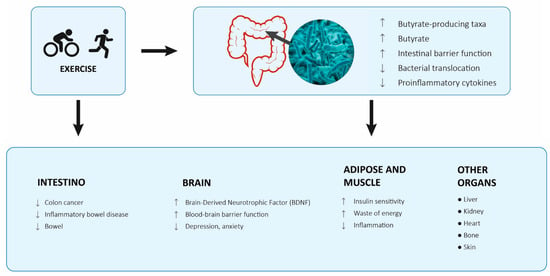
Figure 2.
Proposed model of how exercise modulates different organs and the gut microbiota and epithelium [].
Likewise, whenever there is a reduced immune function, there will be a lower capacity in muscle strength, coordination, or speed, so a decrease in mucosal immunity is associated with an alteration in the athlete’s efficiency. Although it occurs more commonly in endurance exercises than in team sports []. Altered gut motility or enteric nervous system activity is another mechanism by which exercise may influence the gut microbiota. Exercise reduces transit time in the large intestine and has been shown to speed the movement of gas through the gastrointestinal tract [,].
Exercise impacts the autonomic nervous system by increasing general vagal and sympathetic tone []. However, regional, or global changes in gastrointestinal transit are likely to deeply affect the intestinal pH, mucus secretion, biofilm formation, and nutrient availability to the microbiota. Mechanical forces increase in the abdomen during most types of aerobic exercise, which could potentially influence intestinal motility or increase the mixing of intestinal contents [].
Physical training can also alter the enterohepatic circulation of bile acids []. Bile acids are potent regulators of gut microbiota community structure, and the absence of these molecules is associated with significant alterations in gut microbial communities [].
We have seen how physical exercise can modulate the intestinal microbiota, we also see that adaptations to exercise can also be influenced by the intestinal microbiota, playing an important role in the production, storage, and expenditure of energy obtained from the diet, as well as in inflammation, oxidative reactions, and hydration status [].
The intestinal microbiota promotes the digestion and absorption of food to produce energy by the host, in this process the colon metabolizes complex carbohydrates and later they are fermented into SCFA such as butyrate, acetate and propionate []. Propionate and acetate are transported in the bloodstream to different organs in the human body where they are used as substrates for energy metabolism, particularly by hepatocyte cells, which use propionate for gluconeogenesis []. Given the energy requirements during physical exercise, the intestinal microbiota exerts positive effects on the performance of athletes by providing energy substrate to the muscle through the production of SCFA [,].
One of the main physiological adaptations to physical exercise is the modulation of oxidative and nitrosativo stress to avoid tissue damage from intestinal permeability and bacterial translocation []. It has been described that endurance athletes have a high prevalence of upper respiratory tract infections and gastrointestinal problems, including increased permeability of the gastrointestinal epithelial wall, also called “leaky gut”, altered mucosal thickness, and higher bacterial translocation rates. Athletes exposed to high-intensity exercise show a higher incidence of gastrointestinal problems, symptoms such as diarrhea, bloating, nausea, and even bleeding [].
Ismaeel et al. recently described that ROS can be decisive for the cell adaptation process, acting as redox signaling molecules. The intestinal microbiota regulates the maintenance of the gastrointestinal redox state and mitochondrial morphology, through the production of SCFAs and secondary bile acids, which will improve the functioning of the intestinal barrier and the immune response of the mucosa [].
Recently, the intestinal microbiota has been related to the maintenance of adequate hydration during exercise. The phylum Bacteroidetes has been shown to reduce plasma sodium levels, while the Clostridium genus reduced plasma osmolality levels in healthy young individuals. These results suggested that the microbiota influences cellular solute transport across the intestinal mucosa and contributes to hydration status, while reducing plasma osmolality. In the same experiment, high numbers of Bifidobacterium influenced T cell levels, reflecting interaction with the host’s immune response. Because good hydration status and a properly functioning protective gut barrier are essential for athletic performance, and because some athletes do not meet their fluid needs during exercise, it is important to understand the role of the microbiota in the water transport and changes associated with the intestinal mucosa [].
4. Methods Used to Study the Microbiota
Currently, the analysis of the intestinal microbiota is carried out with the stool samples of the subjects participating in the studies, before DNA extraction, it can be considered to lyophilize the samples, collected in the sterile container, in which, in this way, the percentage of water can be obtained. Next, the most widely used standardized technique is DNA extraction using the IHMS (International Human Microbiota Standards) standard operating protocol. The standardized IHMS “Q” protocol is the one recommended by the International Microbiome Consortium []. The homogenizer (Bullet Blender Storm, Next Advance, NY, USA) was used using glass balls. DNA quantification was performed with the PicoGreen dsDNA Quant iT kit (ThermoFisher Scientific, Waltham, MA, USA) and the FP 8300 spectrofluorometer (Jasco, Tokyo, Japan) and DNA purity estimation was performed with a spectrophotometer, using Nanodrop 1100 (Thermofisher Scientific, Waltham, MA, USA).
The product of the extraction is used for massive sequencing. For sequencing, libraries are generated by amplifying the V3 and V4 hypervariable regions of the 16S rRNA gene, which are present in all bacterial species.
The primers used for library generation included the double index sequence needed for multiplexing along with heterogeneity spacers to increase library complexity. The product was a 459 bp amplification which is visualized on a Gelred stained 0.8% agarose gel (Merck KGaA, Darmstadt, Germany). DNA amplifications are typically sequenced on a MiSeq Illumina platform (Illumina, San Diego, CA). Sequence results can be analyzed using Quantitative Insights into Microbial Ecology (QIIME2), v. 18.2, [].
Reads are assigned to an Organizational Taxonomic Unit (OTU) when sequence identity was greater than or equal to 97%. Sequences that did not match any reference are discarded. The analysis of beta diversity allows us to know the distribution of each microbiota sample in space, considering the distance matrix that exists between the different OTUs that make up the community, so that the microbiota samples that are close in space they are more like each other. Four indices are commonly used to determine beta diversity:
- Bray Curtis: this index is not affected by null values (when a species is not detected, or its value is 0). It is a quantitative index because it considers the abundance of species and does not consider phylogenetic distances. This index is between 0 and 1.
- Jaccard: measures the degree of similarity, its range is from 0 to 1, it does not consider the abundance of the species but only their presence or absence, so it is affected by null values.
- Weighted Unifrac and Unweighted Unifrac: Unifrac methods consider information about the abundance of species and their phylogenetic distance. The Unweighted parameter is qualitative because it only considers the presence or absence of individuals, while the Weighted parameter considers the abundances.
The analysis of alpha diversity allows us to know quantitatively, how diverse the microbiota is, how many different species there are. Four indices are commonly used to terminate alpha diversity:
- Observed OTUs: this index allows us to know at a quantitative level the number of OTUs in each sample.
- Pielou index (evenness): this index is indicative of how the species are distributed, of their uniformity in the microbiota, if there are a few species that dominate the microbiota (low evenness) or the abundance of the species is similar (high evenness).
- Shannon index: the Shannon index is an index that considers the richness of species (richness) and the scale considering the distribution of abundance of each species (evenness).
- Faith’s index: it is an index that takes taxonomy into account, that is, it is a phylogenetic index that measures the distances of the branches that separate one OTU from another, the more different the 2 OTUS are phylogenetically, the higher the Faith’s index, the greater the Phylogenetic separation of all OTUS within Faith’s largest index sample.
5. Conclusions
The key component that determines the correct function of the intestinal barrier is the gut microbiota. However, more studies are needed to understand how the gut microbiota composition can influence physical performance. Given that details and small gains are recognized as crucial for competitive success in athletes, it is reasonable to consider the gut microbiota as a potential factor that could modulate physical efficiency and would need further study to determine its importance and to find strategies that can mediate the effects of exercise.
Author Contributions
D.D.-B.: writing and review of the drafts of the manuscript and acquisition, analysis, and interpretation of data. G.G.-P.-d.-S.: design and conceptualization of the study, writing and review of the drafts of the manuscript, and supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no potential conflict of interest with respect to the research, authorship, and/or publication of this article.
References
- Brüls, T.; Weissenbach, J. The human metagenome: Our other genome? Hum. Mol. Genet. 2011, 20, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Prats, G. Microbiologia y Parasitología Médica; Médica Panamericana: Madrid, Spain, 2012; ISBN 9788498354294. [Google Scholar]
- Sebastián-Domingo, J.J.; Sánchez-Sánchez, C. From the intestinal flora to the microbiome. Rev. Esp. Enferm. Dig. 2018, 110, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Richmond, C.R. ICRP report of the task group on reference man. Int. J. Nucl. Med. Biol. 1985, 12, 251. [Google Scholar] [CrossRef]
- Hopkins, M.J.; Macfarlane, G.T.; Furrie, E.; Fite, A.; Macfarlane, S. Characterisation of intestinal bacteria in infant stools using real-time PCR and northern hybridisation analyses. FEMS Microbiol. Ecol. 2005, 54, 77–85. [Google Scholar] [CrossRef]
- Stephens, R.W.; Arhire, L.; Covasa, M. Gut Microbiota: From Microorganisms to Metabolic Organ Influencing Obesity. Obesity 2018, 26, 801–809. [Google Scholar] [CrossRef]
- Sun, J.; Kato, I. Gut microbiota, inflammation and colorectal cancer. Genes Dis. 2016, 3, 130–143. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef]
- Jandhyala, S.M. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787. [Google Scholar] [CrossRef]
- Rakoff-Nahoum, S.; Paglino, J.; Eslami-Varzaneh, F.; Edberg, S.; Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004, 118, 229–241. [Google Scholar] [CrossRef]
- Kim, D.; Yoo, S.A.; Kim, W.U. Gut microbiota in autoimmunity: Potential for clinical applications. Arch. Pharm. Res. 2016, 39, 1565–1576. [Google Scholar] [CrossRef]
- Ren, C.; Dokter-Fokkens, J.; Figueroa Lozano, S.; Zhang, Q.; de Haan, B.J.; Zhang, H.; Faas, M.M.; de Vos, P. Lactic Acid Bacteria May Impact Intestinal Barrier Function by Modulating Goblet Cells. Mol. Nutr. Food Res. 2018, 62, 1700572. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.R. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 2009, 9, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Vancamelbeke, M.; Vermeire, S. The intestinal barrier: A fundamental role in health and disease. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 821–834. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Zeng, M.Y.; Núñez, G. The interplay between host immune cells and gut microbiota in chronic inflammatory diseases. Exp. Mol. Med. 2017, 49, e339. [Google Scholar] [CrossRef]
- Honda, K.; Littman, D.R. The microbiota in adaptive immune homeostasis and disease. Nature 2016, 535, 75–84. [Google Scholar] [CrossRef]
- Durkin, H.G.; Bazin, H.; Waksman, B.H. Origin and fate of IgE-bearing lymphocytes: I. Peyer’s patches as differentiation site of cells simultaneously bearing IgA and IgE. J. Exp. Med. 1981, 154, 640–648. [Google Scholar] [CrossRef]
- Cani, P.D.; Van Hul, M.; Lefort, C.; Depommier, C.; Rastelli, M.; Everard, A. Microbial regulation of organismal energy homeostasis. Nat. Metab. 2019, 1, 34–46. [Google Scholar] [CrossRef]
- Theilmann, M.C.; Goh, Y.J.; Nielsen, K.F.; Klaenhammer, T.R.; Barrangou, R.; Hachem, M.A. Lactobacillus acidophilus metabolizes dietary plant glucosides and externalizes their bioactive phytochemicals. MBio 2017, 8, e01421-17. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Cummings, J.H.; Allison, C. Protein degradation by human intestinal bacteria. J. Gen. Microbiol. 1986, 132, 1647–1656. [Google Scholar] [CrossRef]
- Morales, P.; Brignardello, J.; Gotteland, M. La microbiota intestinal: Un nuevo actor en el desarrollo de la obesidad. Rev. Med. Chile 2010, 138, 1020–1027. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Lombard, V.; Henrissat, B. Complex carbohydrate utilization by the healthy human microbiome. PLoS ONE 2012, 7, e28742. [Google Scholar] [CrossRef] [PubMed]
- De Biase, D.; Pennacchietti, E. Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: Function, distribution and biomedical implications of the gadBC operon. Mol. Microbiol. 2012, 86, 770–786. [Google Scholar] [CrossRef] [PubMed]
- Churruca, I.; Fernández-Quintela, A.; Portillo, M.P. Conjugated linoleic acid isomers: Differences in metabolism and biological effects. BioFactors 2009, 35, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Devillard, E.; McIntosh, F.M.; Duncan, S.H.; Wallace, R.J. Metabolism of linoleic acid by human gut bacteria: Different routes for biosynthesis of conjugated linoleic acid. J. Bacteriol. 2007, 189, 2566–2570. [Google Scholar] [CrossRef]
- Cerdá, B.; Pérez, M.; Pérez-Santiago, J.D.; Tornero-Aguilera, J.F.; González-Soltero, R.; Larrosa, M. Gut microbiota modification: Another piece in the puzzle of the benefits of physical exercise in health? Front. Physiol. 2016, 7, 51. [Google Scholar] [CrossRef]
- Clarke, S.F.; Murphy, E.F.; O’Sullivan, O.; Lucey, A.J.; Humphreys, M.; Hogan, A.; Hayes, P.; O’Reilly, M.; Jeffery, I.B.; Wood-Martin, R.; et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 2014, 63, 1913–1920. [Google Scholar] [CrossRef]
- Bressa, C.; Bailén-Andrino, M.; Pérez-Santiago, J.; González-Soltero, R.; Pérez, M.; Montalvo-Lominchar, M.G.; Maté-Muñoz, J.L.; Domínguez, R.; Moreno, D.; Larrosa, M. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS ONE 2017, 12, e0171352. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef]
- Dao, M.C.; Everard, A.; Aron-Wisnewsky, J.; Sokolovska, N.; Prifti, E.; Verger, E.O.; Kayser, B.D.; Levenez, F.; Chilloux, J.; Hoyles, L.; et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: Relationship with gut microbiome richness and ecology. Gut 2016, 65, 426–436. [Google Scholar] [CrossRef]
- Durk, R.P.; Castillo, E.; Márquez-Magaña, L.; Grosicki, G.J.; Bolter, N.D.; Matthew Lee, C.; Bagley, J.R. Gut microbiota composition is related to cardiorespiratory fitness in healthy young adults. Int. J. Sport Nutr. Exerc. Metab. 2019, 29, 249–253. [Google Scholar] [CrossRef]
- Estaki, M.; Pither, J.; Baumeister, P.; Little, J.P.; Gill, S.K.; Ghosh, S.; Ahmadi-Vand, Z.; Marsden, K.R.; Gibson, D.L. Cardiorespiratory fitness as a predictor of intestinal microbial diversity and distinct metagenomic functions. Microbiome 2016, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Barton, W.; Penney, N.C.; Cronin, O.; Garcia-Perez, I.; Molloy, M.G.; Holmes, E.; Shanahan, F.; Cotter, P.D.; O’Sullivan, O. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 2018, 67, 625–633. [Google Scholar] [CrossRef]
- Aya, V.; Flórez, A.; Perez, L.; Ramírez, J.D. Association between physical activity and changes in intestinal microbiota composition: A systematic review. PLoS ONE 2021, 16, e0247039. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.M.; Mailing, L.J.; Niemiro, G.M.; Moore, R.; Cook, M.D.; White, B.A.; Holscher, H.D.; Woods, J.A. Exercise Alters Gut Microbiota Composition and Function in Lean and Obese Humans. Med. Sci. Sports Exerc. 2018, 50, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Cronin, O.; Barton, W.; Skuse, P.; Penney, N.C.; Garcia-Perez, I.; Murphy, E.F.; Woods, T.; Nugent, H.; Fanning, A.; Melgar, S.; et al. A Prospective Metagenomic and Metabolomic Analysis of the Impact of Exercise and/or Whey Protein Supplementation on the Gut Microbiome of Sedentary Adults. mSystems 2018, 3, e00044-18. [Google Scholar] [CrossRef] [PubMed]
- Munukka, E.; Ahtiainen, J.P.; Puigbó, P.; Jalkanen, S.; Pahkala, K.; Keskitalo, A.; Kujala, U.M.; Pietilä, S.; Hollmén, M.; Elo, L.; et al. Six-week endurance exercise alters gut metagenome that is not reflected in systemic metabolism in over-weight women. Front. Microbiol. 2018, 9, 2323. [Google Scholar] [CrossRef]
- Mailing, L.J.; Allen, J.M.; Buford, T.W.; Fields, C.J.; Woods, J.A. Exercise and the Gut Microbiome: A Review of the Evidence, Potential Mechanisms, and Implications for Human Health. Exerc. Sport Sci. Rev. 2019, 47, 75–85. [Google Scholar] [CrossRef]
- Nieman, D.C. Exercise immunology: Practical applications. Int. J. Sport. Med. Suppl. 1997, 18, S91–S100. [Google Scholar] [CrossRef]
- Song, B.K.; Cho, K.O.; Jo, Y.; Oh, J.W.; Kim, Y.S. Colon transit time according to physical activity level in adults. J. Neurogastroenterol. Motil. 2012, 18, 64–69. [Google Scholar] [CrossRef]
- Dainese, R.; Serra, J.; Azpiroz, F.; Malagelada, J.R. Effects of physical activity on intestinal gas transit and evacuation in healthy subjects. Am. J. Med. 2004, 116, 536–539. [Google Scholar] [CrossRef]
- Freeman, J.V.; Dewey, F.E.; Hadley, D.M.; Myers, J.; Froelicher, V.F. Autonomic Nervous System Interaction with the Cardiovascular System during Exercise. Prog. Cardiovasc. Dis. 2006, 48, 342–362. [Google Scholar] [CrossRef] [PubMed]
- Meissner, M.; Lombardo, E.; Havinga, R.; Tietge, U.J.F.; Kuipers, F.; Groen, A.K. Voluntary wheel running increases bile acid as well as cholesterol excretion and decreases atherosclerosis in hypercholesterolemic mice. Atherosclerosis 2011, 218, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Kakiyama, G.; Pandak, W.M.; Gillevet, P.M.; Hylemon, P.B.; Heuman, D.M.; Daita, K.; Takei, H.; Muto, A.; Nittono, H.; Ridlon, J.M.; et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J. Hepatol. 2013, 58, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Mach, N.; Fuster-Botella, D. Endurance exercise and gut microbiota: A review. J. Sport Health Sci. 2017, 6, 179–197. [Google Scholar] [CrossRef]
- Hsu, Y.J.; Chiu, C.C.; Li, Y.P.; Huang, W.C.; Huang, Y.T.; Huang, C.C.; Chuang, H.L. Effect of intestinal microbiota on exercise performance in mice. J. Strength Cond. Res. 2015, 29, 552–558. [Google Scholar] [CrossRef]
- Samuel, B.S.; Shaito, A.; Motoike, T.; Rey, F.E.; Backhed, F.; Manchester, J.K.; Hammer, R.E.; Williams, S.C.; Crowley, J.; Yanagisawa, M.; et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. USA 2008, 105, 16767–16772. [Google Scholar] [CrossRef]
- Lambert, J.E.; Myslicki, J.P.; Bomhof, M.R.; Belke, D.D.; Shearer, J.; Reimer, R.A. Exercise training modifies gut microbiota in normal and diabetic mice. Appl. Physiol. Nutr. Metab. 2015, 40, 749–752. [Google Scholar] [CrossRef]
- Bessa, A.L.; Oliveira, V.N.; Agostini, G.G.; Oliveira, R.J.S.; Oliveira, A.C.S.; White, G.E.; Wells, G.D.; Teixeira, D.N.S.; Espindola, F.S. EXERCISE intensity and recovery: Biomarkers of injury, inflammation, and oxidative stress. J. Strength Cond. Res. 2016, 30, 311–319. [Google Scholar] [CrossRef]
- Lamprecht, M.; Frauwallner, A. Exercise, Intestinal Barrier Dysfunction and Probiotic Supplementation. Acute Top. Sport Nutr. 2012, 59, 47–56. [Google Scholar]
- Rehrer, N.J.; Brouns, F.; Beckers, E.J.; Frey, W.O.; Villiger, B.; Riddoch, C.J.; Menheere, P.P.; Saris, W.H. Physiological changes and gastro-intestinal symptoms as a result of ultra-endurance running. Eur. J. Appl. Physiol. Occup. Physiol. 1992, 64, 1–8. [Google Scholar] [CrossRef]
- Ismaeel, A.; Holmes, M.; Papoutsi, E.; Panton, L.; Koutakis, P. Resistance Training, Antioxidant Status, and Antioxidant Supplementation. Int. J. Sport Nutr. Exerc. Metab. 2019, 29, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.; Mach, N. The crosstalk between the gut microbiota and mitochondria during exercise. Front. Physiol. 2017, 8, 319. [Google Scholar] [CrossRef] [PubMed]
- Useros, N.R.; Gheorghe, A.; Labajos, R.S.; Rebato, E.N.; Sanchez, A.M. HYDRAGUT study: Influence of HYDRAtion status on the GUT microbiota and their impact on the immune system. FASEB J. 2015, 29, 593-1. [Google Scholar] [CrossRef]
- Fiedorová, K.; Radvanský, M.; Němcová, E.; Grombiříková, H.; Bosák, J.; Černochová, M.; Lexa, M.; Šmajs, D.; Freiberger, T. The impact of DNA extraction methods on stool bacterial and fungal microbiota community recovery. Front. Physiol. 2019, 10, 821. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Dillon, M.R.; Zhang, Y.; Rideout, J.R.; Bolyen, E.; Li, H.; Albert, P.S.; Caporaso, J.G. q2-longitudinal: Longitudinal and Paired-Sample Analyses of Microbiome Data. mSystems 2018, 3, e00219-18. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).