Fipexide Rapidly Induces Callus Formation in Medicago sativa by Regulating Small Auxin Upregulated RNA (SAUR) Family Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Callus Induced by Different PGRs
2.3. Paraffin Observation of FPX-Treated and Auxin- and Cytokinin-Treated Callus
2.4. RNA-seq Analysis of FPX-Treated Callus
2.5. Quantitative Real-Time PCR
2.6. Statistical Analysis
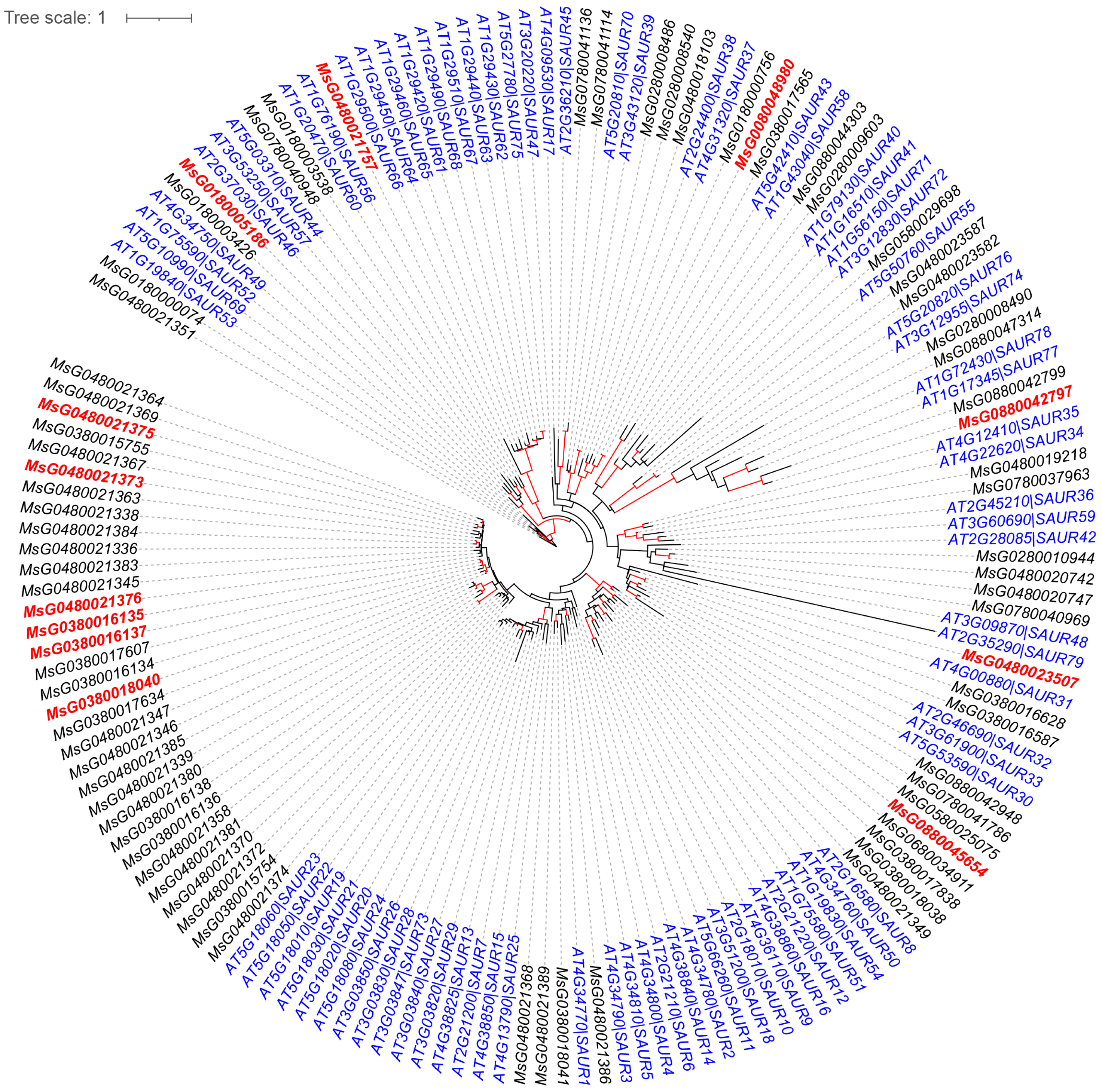
2.7. Phylogenetic Analysis
3. Results
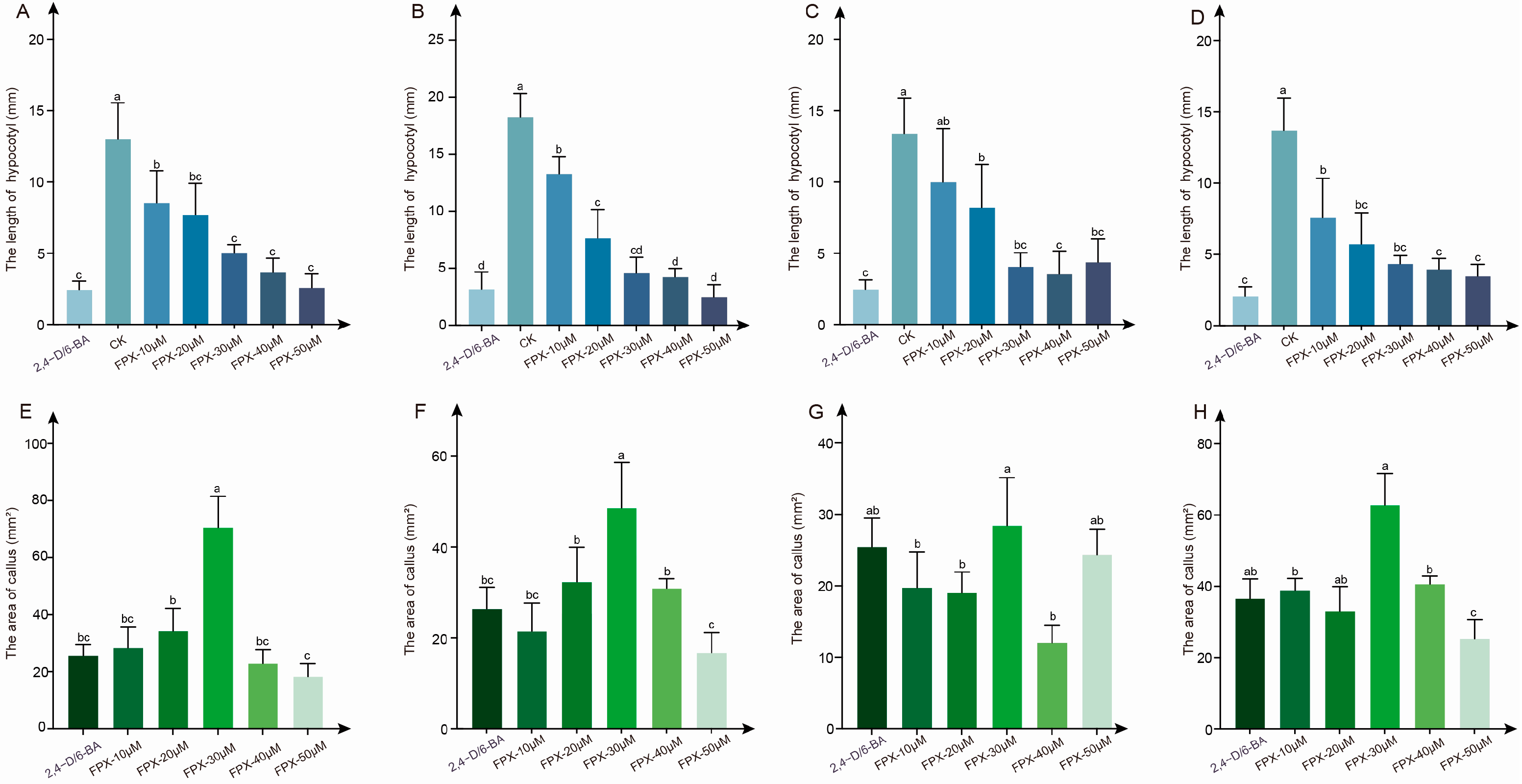
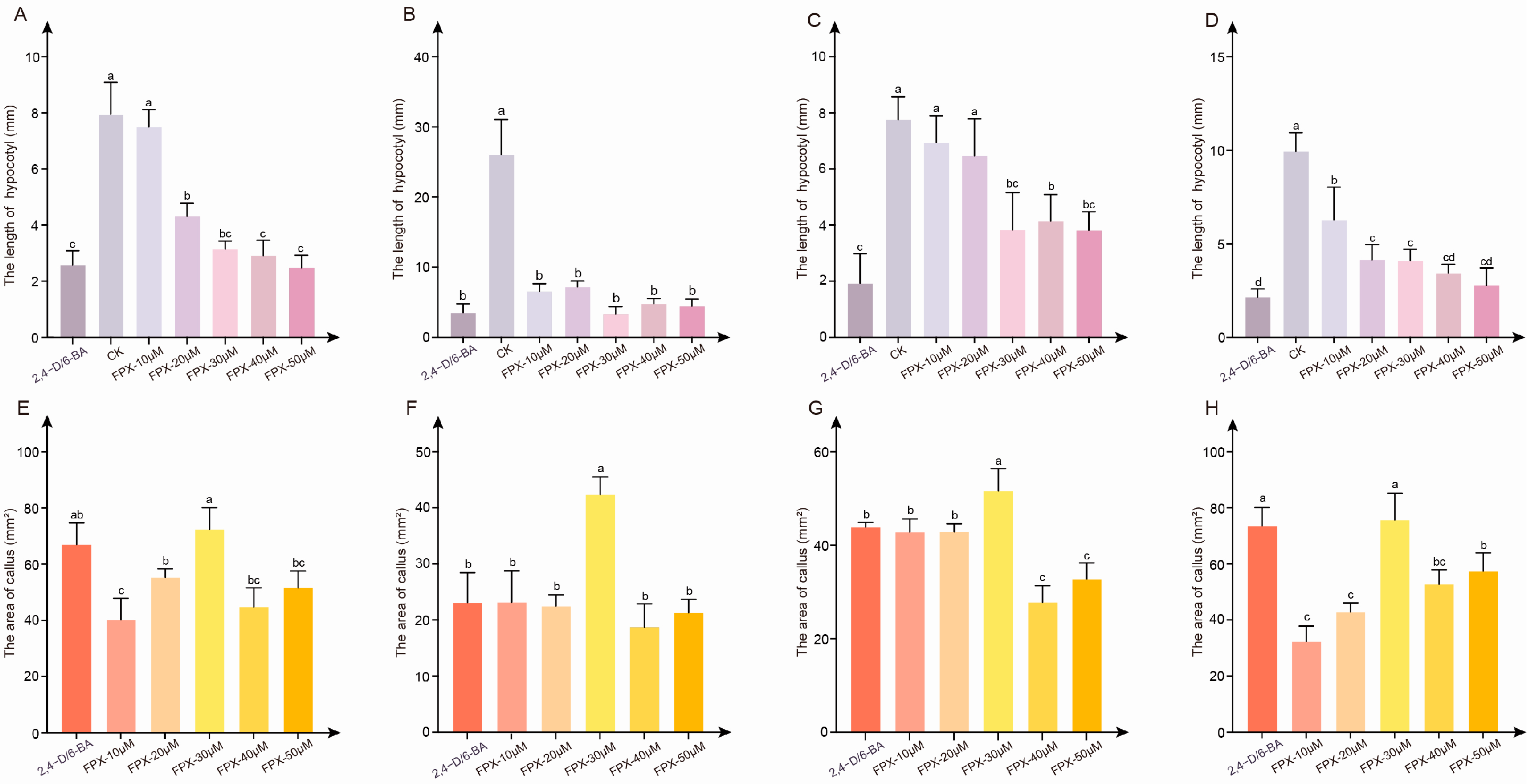
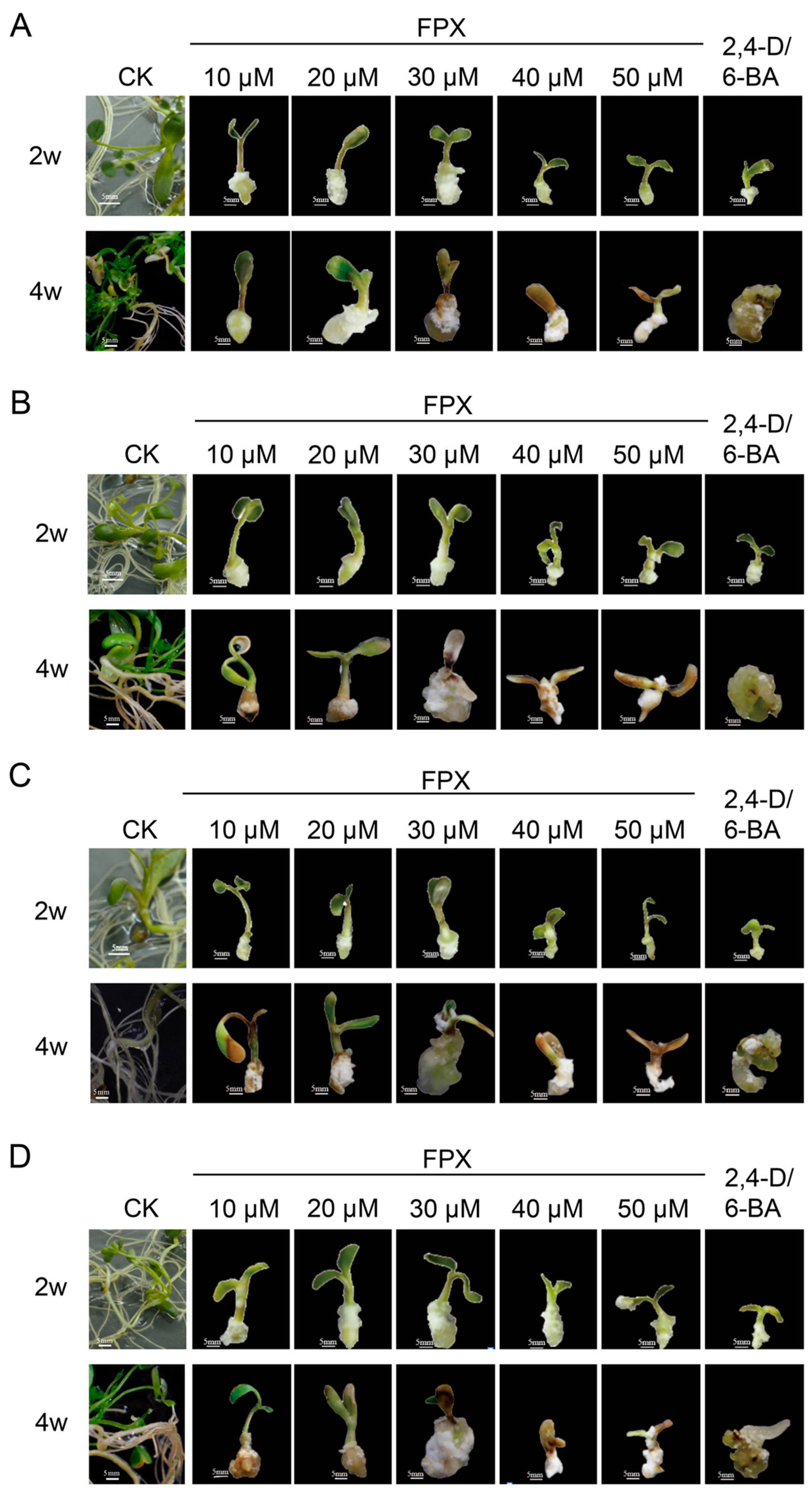
3.1. FPX Treatment Rapidly Induced Callus Formation
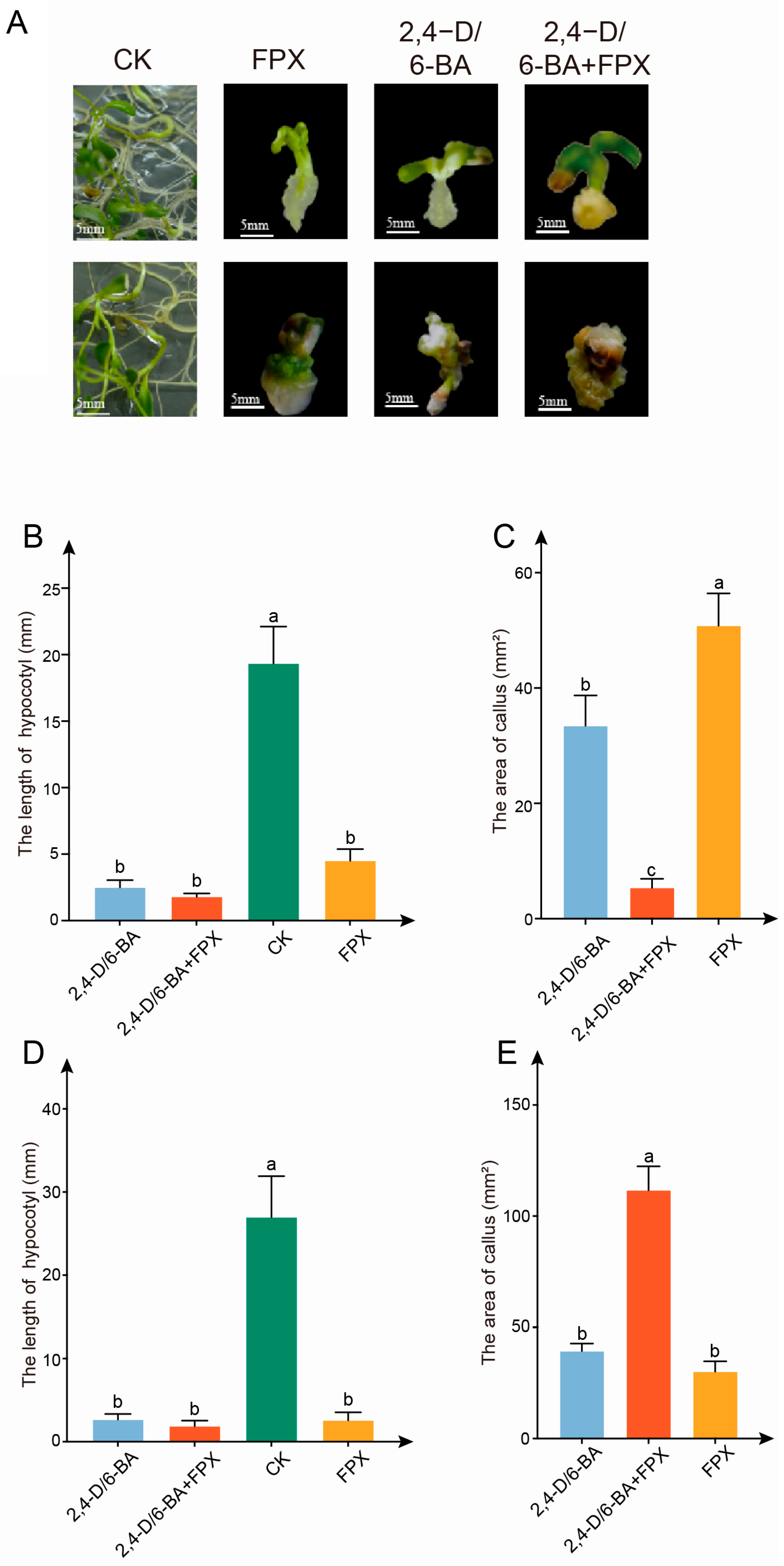
3.2. The Effect of FPX-Auxin/Cytokinin Combined Treatment Is Better than Single Growth Regulator Treatment
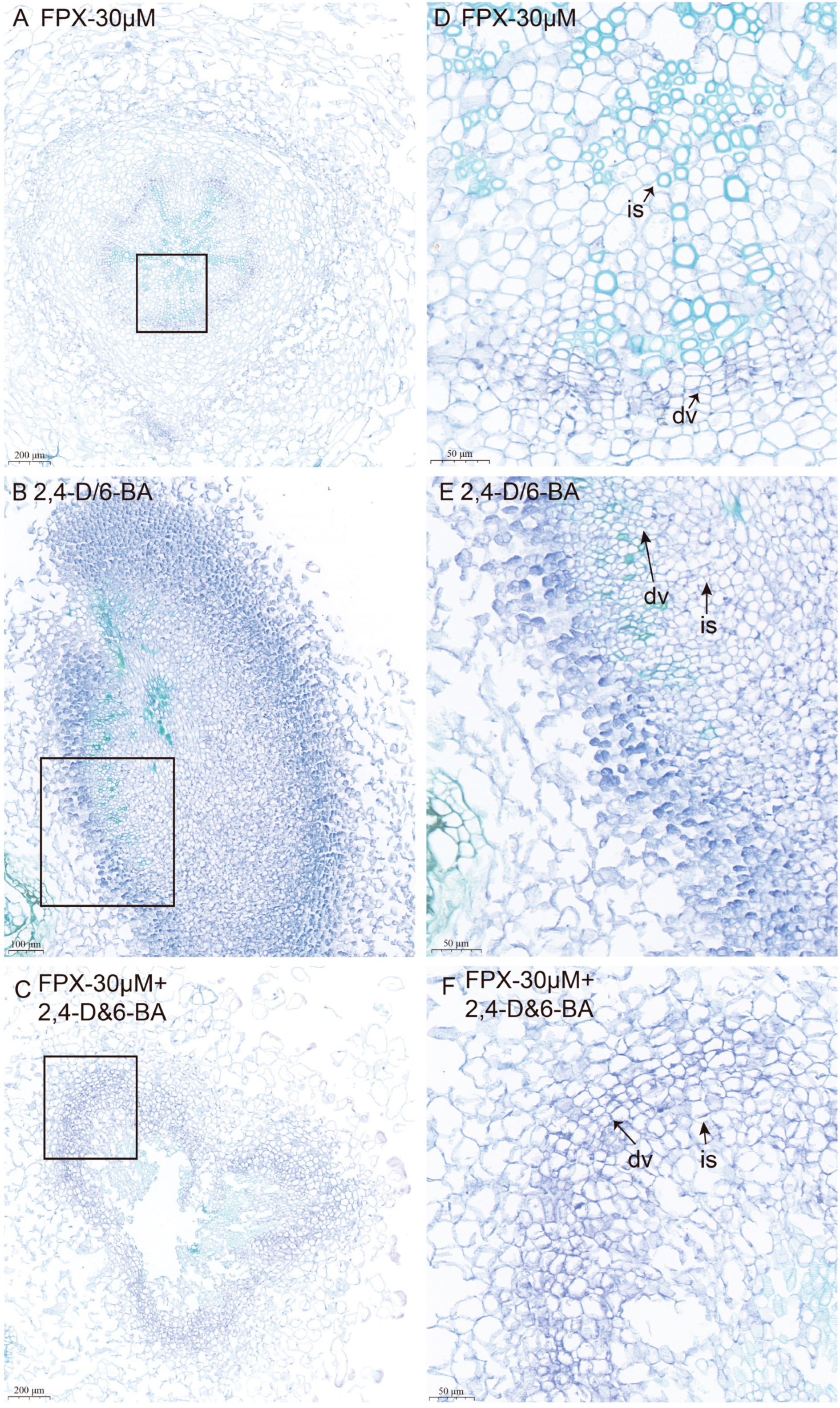
3.3. The Cellular Organization of Callus of Seeds or Hypocotyl Differs Between FPX and Auxin/Cytokinin Treatments
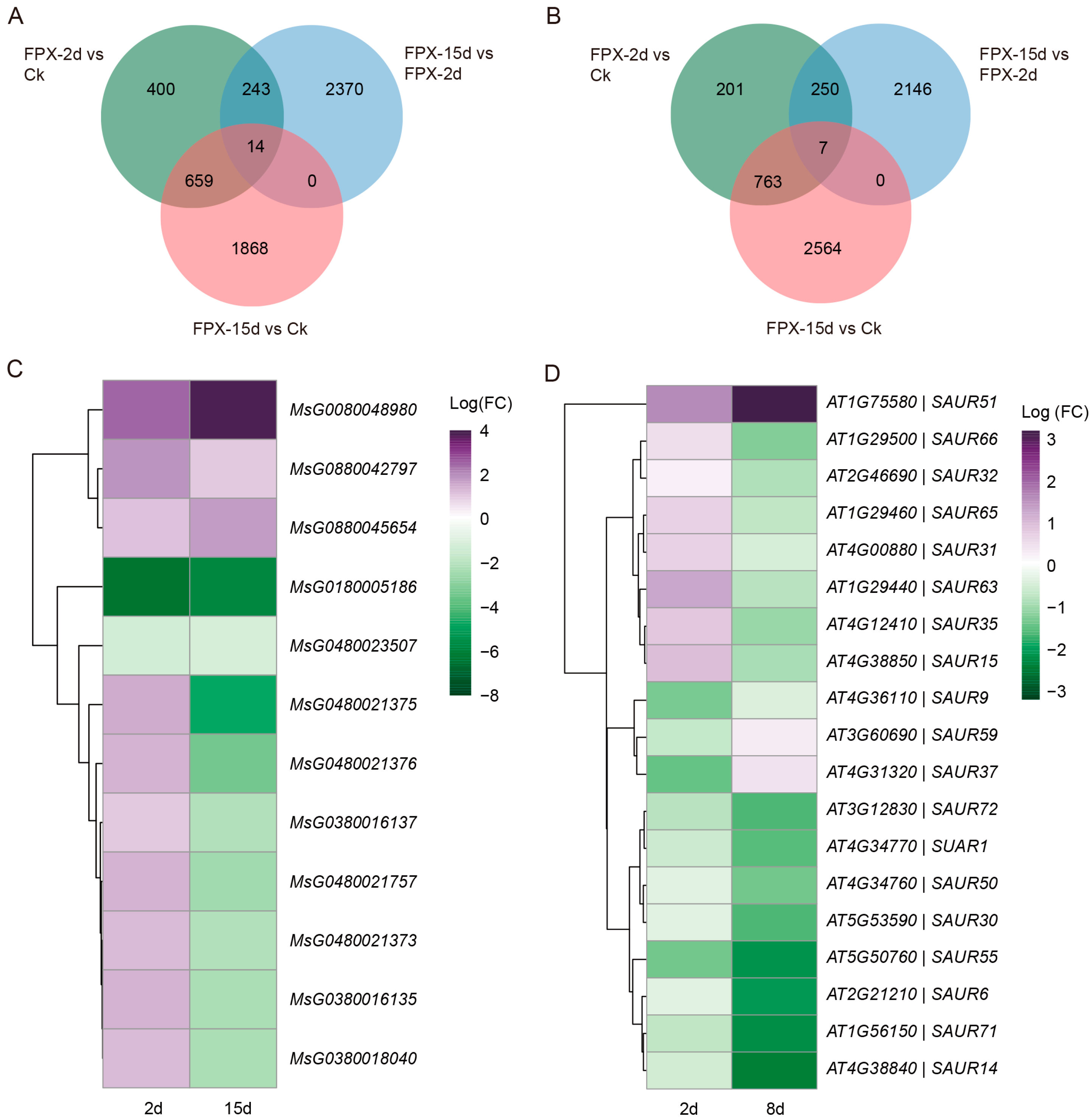
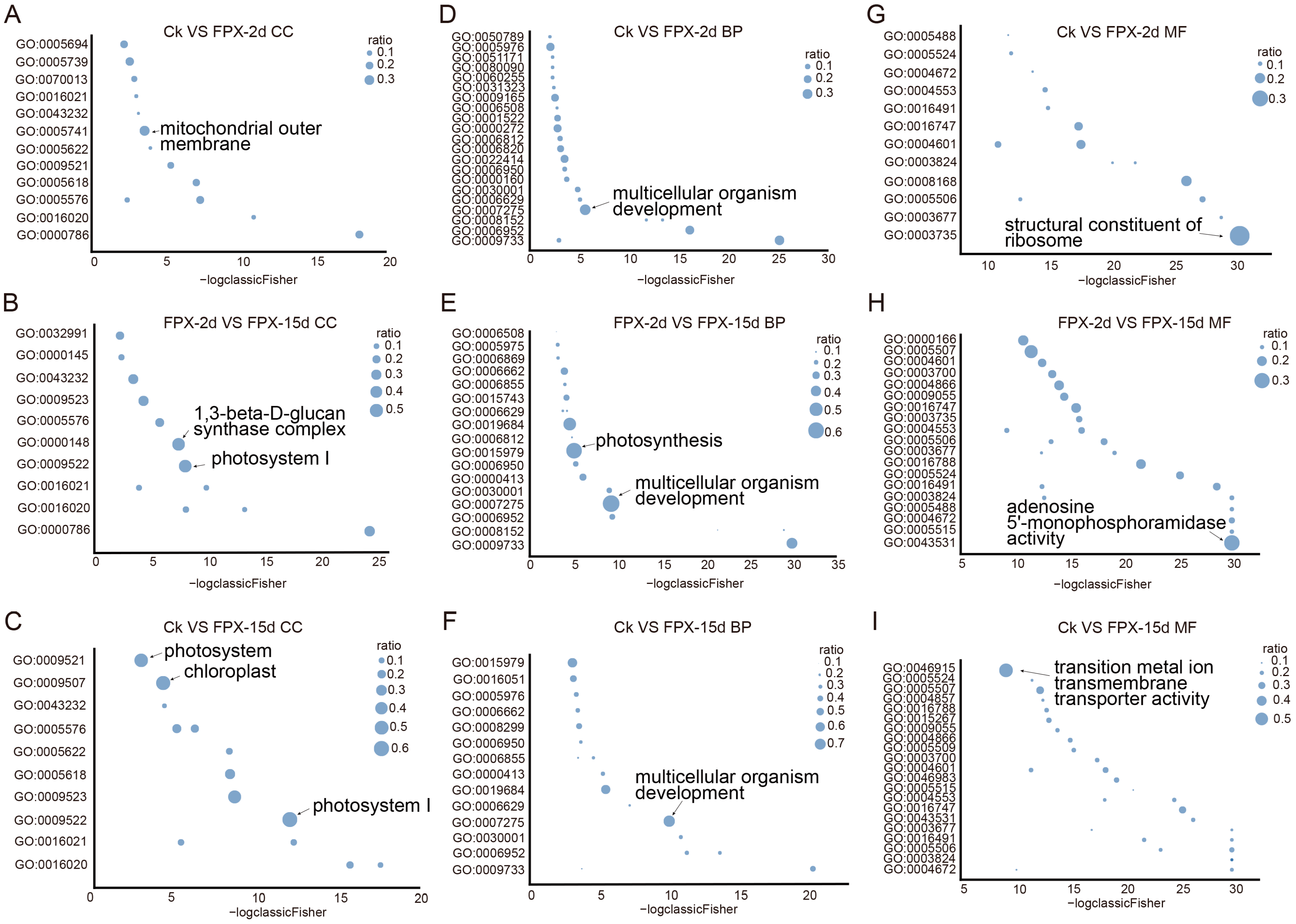
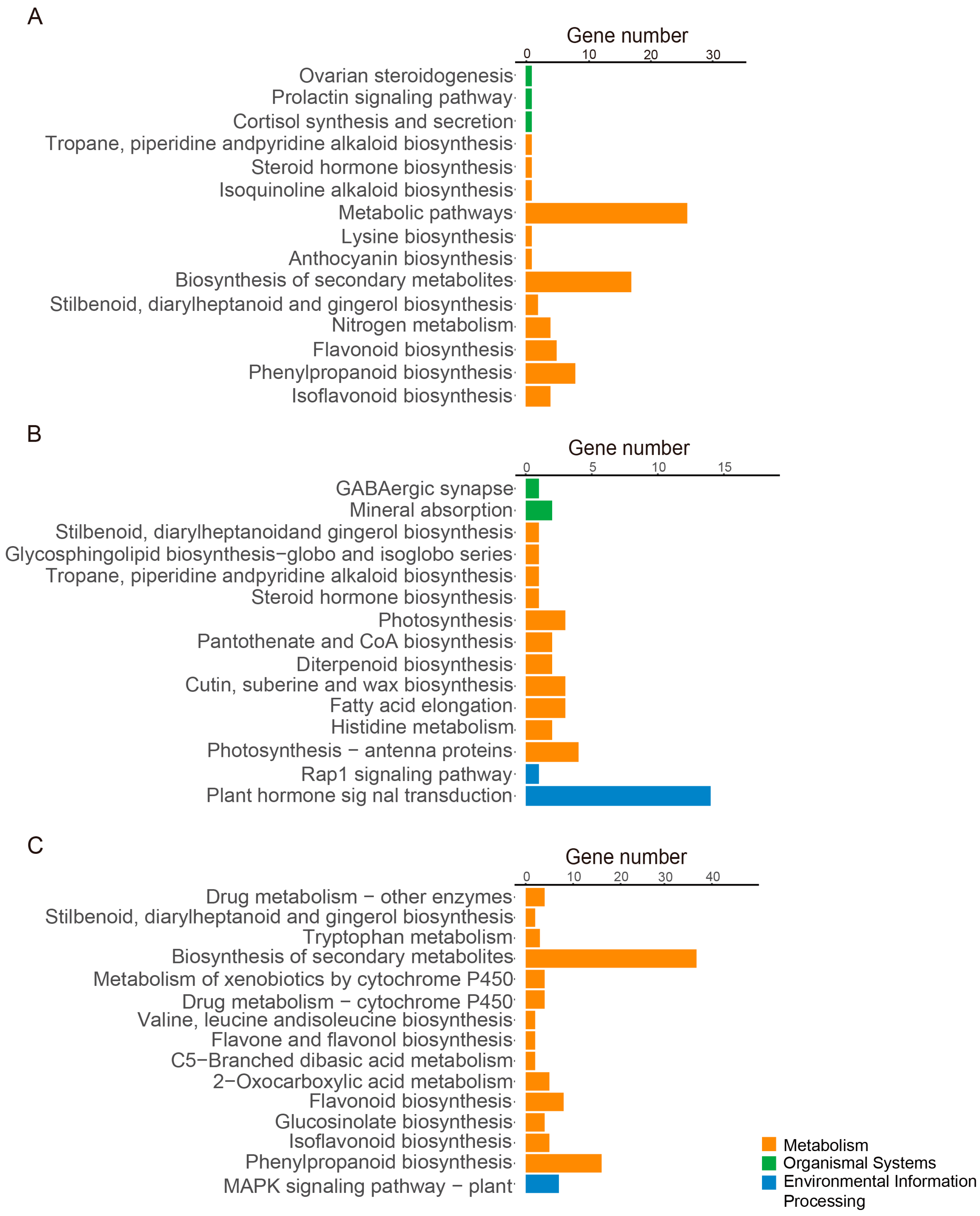
3.4. FPX Regulates Auxin-Related Genes and Plant Metabolism Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviation
| FPX | Fipexide |
References
- Horsch, R.B.; Fraley, R.T.; Rogers, S.G.; Sanders, P.R.; Lloyd, A.; Hoffmann, N. Inheritance of functional foreign genes in plants. Science 1984, 223, 496–498. [Google Scholar] [CrossRef] [PubMed]
- Anjanappa, R.B.; Gruissem, W. Current progress and challenges in crop genetic transformation. J. Plant Physiol. 2021, 261, 153411. [Google Scholar] [CrossRef] [PubMed]
- Lesh, M. Splice of Life: The Case for GMOs and Gene Editing; Adam Smith Institute: London, UK, 2021. [Google Scholar]
- Yin, K.; Gao, C.; Qiu, J.L. Progress and prospects in plant genome editing. Nat. Plants 2017, 6, 17107. [Google Scholar] [CrossRef] [PubMed]
- Gosal, S.S.; Kang, M.S. Plant Tissue Culture and Genetic Transformation for Crop Improvement. In Improving Crop Resistance to Abiotic Stress; Tuteja, N., Gill, S.S., Tiburcio, A.F., Tuteja, R., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2012; pp. 357–397. [Google Scholar] [CrossRef]
- Gautheret, R.J. Sur la possibilité de réaliser la culture indéfinie des tissus de tubercules de carotte. C. R. Hebd. Seances Acad. Sci. 1939, 208, 118–120. [Google Scholar]
- Nobécourt, P. Sur la pérennité et l’augmentation de volume des cultures de tissues végétaux. C. R. Seances Soc. Biol. 1939, 130, 1270–1271. [Google Scholar]
- White, P.R. Potentially unlimited growth of excised plant callus in an artificial nutrient. Am. J. Bot. 1939, 26, 59–64. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Sugimoto, K.; Iwase, A. Plant callus: Mechanisms of induction and repression. Plant Cell 2013, 25, 3159–3173. [Google Scholar] [CrossRef]
- Skoog, F.; Miller, C.O. Chemical regulation of growth and organ formation in plant tissue cultured in vitro. Symp. Soc. Exp. Biol. 1957, 11, 118–131. [Google Scholar]
- Nakano, T.; Tanaka, S.; Ohtani, M.; Yamagami, A.; Takeno, S.; Hara, N.; Mori, A.; Nakano, A.; Hirose, S.; Himuro, Y.; et al. FPX is a novel chemical inducer that promotes callus formation and shoot regeneration in plants. Plant Cell Physiol. 2018, 59, 1555–1567. [Google Scholar] [CrossRef]
- Tanahara, Y.; Yamanaka, K.; Kawai, K.; Ando, Y.; Nakatsuka, T. Establishment of an efficient transformation method of garden stock (Matthiola incana) using a callus formation chemical inducer. Plant Biotechnol. 2022, 39, 273–280. [Google Scholar] [CrossRef]
- Li, X.; Brummer, E.C. Applied genetics and genomics in alfalfa breeding. Agronomy 2012, 2, 40–61. [Google Scholar] [CrossRef]
- Kumar, S. Biotechnological advancements in alfalfa improvement. J. Appl. Genet. 2011, 52, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Fu, C.; Wang, Z.Y. A unified Agrobacterium-mediated transformation protocol for alfalfa (Medicago sativa L.) and Medicago truncatula. In Transgenic Plants: Methods and Protocols; Kumar, S., Barone, P., Smith, M., Eds.; Humana Press: New York, NY, USA, 2019; pp. 153–163. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Plant Physiol. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Pertea, G.; Pertea, M. GFF utilities: GffRead and GffCompare. F1000Research 2020, 9, 304. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate—A practical and powerful approach to Multiple testing. J. R. Statist. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Lyu, Y.; Zhu, S.; Zhi, X.; Ji, Y.; Fan, Y.; Dong, F. Improving subseasonal-to-seasonal prediction of summer extreme precipitation over southern China based on a deep learning method. Geophys. Res. Lett. 2023, 50, e2023GL106245. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Shen, C.; Du, H.; Chen, Z.; Lu, H.; Zhu, F.; Chen, H.; Meng, X.; Liu, Q.; Liu, P.; Zheng, L.; et al. The chromosome-level genome sequence of the autotetraploid alfalfa and resequencing of core germplasms provide genomic resources for alfalfa research. Mol. Plant 2020, 13, 1250–1261. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34 (Suppl. S2), W609–W612. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Stortenbeker, N.; Bemer, M. The SAUR gene family: The plant’s toolbox for adaptation of growth and development. J. Exp. Bot. 2019, 70, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Spartz, A.K.; Ren, H.; Park, M.Y.; Grandt, K.N.; Lee, S.H.; Murphy, A.S.; Sussman, M.R.; Overvoorde, P.J.; Gray, W.M. SAUR Inhibition of PP2C-D Phosphatases Activates Plasma Membrane H+-ATPases to Promote Cell Expansion in Arabidopsis. Plant Cell 2014, 26, 2129–2142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yu, Z.; Yao, X.; Chen, J.; Chen, X.; Zhou, H.; Lou, Y.; Ming, F.; Jin, Y. Genome-wide identification and characterization of small auxin-up RNA (SAUR) gene family in plants: Evolution and expression profiles during normal growth and stress response. BMC Plant Biol. 2021, 21, 4. [Google Scholar] [CrossRef]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inzé, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.J.; Gray, W.M. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef]
- Kant, S.; Bi, Y.M.; Zhu, T.; Rothstein, S.J. SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol. 2009, 151, 691–701. [Google Scholar] [CrossRef]
- Ren, H.; Gray, W.M. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 2006, 88, 360–371. [Google Scholar] [CrossRef]
- Chen, Y.; Hao, X.; Cao, J. Small auxin upregulated RNA (SAUR) gene family in maize: Identification, evolution, and its phylogenetic comparison with Arabidopsis, rice, and sorghum. J. Integr. Plant Biol. 2013, 56, 133–150. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, W.; Li, S.; Lan, B.; Gai, Y.; Du, F.K.; Yin, K. Fipexide Rapidly Induces Callus Formation in Medicago sativa by Regulating Small Auxin Upregulated RNA (SAUR) Family Genes. Crops 2025, 5, 28. https://doi.org/10.3390/crops5030028
Zhao W, Li S, Lan B, Gai Y, Du FK, Yin K. Fipexide Rapidly Induces Callus Formation in Medicago sativa by Regulating Small Auxin Upregulated RNA (SAUR) Family Genes. Crops. 2025; 5(3):28. https://doi.org/10.3390/crops5030028
Chicago/Turabian StyleZhao, Wenxuan, Siyang Li, Bo Lan, Yunpeng Gai, Fang K. Du, and Kangquan Yin. 2025. "Fipexide Rapidly Induces Callus Formation in Medicago sativa by Regulating Small Auxin Upregulated RNA (SAUR) Family Genes" Crops 5, no. 3: 28. https://doi.org/10.3390/crops5030028
APA StyleZhao, W., Li, S., Lan, B., Gai, Y., Du, F. K., & Yin, K. (2025). Fipexide Rapidly Induces Callus Formation in Medicago sativa by Regulating Small Auxin Upregulated RNA (SAUR) Family Genes. Crops, 5(3), 28. https://doi.org/10.3390/crops5030028





