Abstract
The accumulation of ions due to increased salinity in the soil is one of the major abiotic stressors of cultivated plants that negatively affect their productivity. The model plant, Medicago truncatula, is the only Medicago species that has been extensively studied, whereas research into increased salinity adaptation of two important forage legumes, M. sativa and M. arborea, has been limited. In the present study, the expression of six genes, namely SOS1, SOS3, NHX2, AKT, AVP and HKT1 was monitored to investigate the manner in which sodium ions are blocked and transferred to the various plant parts. In addition, in silico miRNA analysis was performed to identify miRNAs that possibly control the expression of the genes studied. The following treatments were applied: (1) salt stress, with initial treatment of 50 mM NaCl and gradual acclimatization every 10 days, (2) salt shock, with continuous application of 100 mM NaCl concentration and (3) no application of NaCl. Results showed that M. arborea appeared to overexpress and activate all available mechanisms of resistance in conditions of increased salinity, while M. sativa acted in a more targeted way, overexpressing the HKT1 and AKT genes that contribute to the accumulation of sodium ions, particularly in the root. Regarding miRNA in silico analysis, five miRNAs with significant complementarity to putative target genes, AKT1, AVP and SOS3 were identified and served as a first step in investigating miRNA regulatory networks. Further miRNA expression studies will validate these results. Our findings contribute to the understanding of the molecular mechanisms underlying salt-responsiveness in Medicago and could be used in the future for generating salt-tolerant genotypes in crop improvement programs.
Keywords:
salinity; gene expression; resilient mechanisms; ion transportation; Medicago sp.; miRNA; MiR156 1. Introduction
Alfalfa (Medicago sativa L.) is the most extensively cultivated perennial forage legume with high ecological importance [1]. Tree medick (Medicago arborea L.) is a Mediterranean stress-tolerant leguminous fodder shrub [2] that could play an important role as a genetic resource for the improvement of Medicago sp. salt tolerance [3]. Salinity is a major environmental abiotic stress factor in crop productivity worldwide that impacts plant development and yield via two main mechanisms: hyperosmotic pressure, which is caused by low water availability, and ion toxicity (especially Na+), arising from solute imbalances [4]. However, little is known regarding the molecular mechanisms involved in the responses of Medicago sp. to salinity except Medicago truncatula.
Salt tolerance in higher plants is a quantitative trait that is regulated by polygenes and numerous signaling pathways, and a complex interaction system exists between the genes and the signaling pathways [5,6]. Genes that contribute to salt tolerance in plants can be categorized into three functional groups: the first contains genes that regulate salt uptake and transport, the second contains genes with osmotic function, and the third contains genes that regulate plant growth under saline soil [3]. The salt overly sensitive (SOS) signal transduction pathway is pivotal for discharging Na+ and maintaining ionic balance on a cellular level and confers improved salt tolerance in plants [7]. Several studies reported that the overexpression of Arabidopsis thaliana SOS1, SOS2 and SOS3 genes enhances salt tolerance in a variety of plant species including M. sativa [1]. Previous studies have also indicated that overexpression of the stress-associated gene encoding the compatible solute, the ion transporter SeNHX1, results in enhanced tolerance to salinity stress in alfalfa [8]. Na+/H+ antiporters (NHXs), which are located on plasma membranes and tonoplast, participate in the maintenance of Na+ homeostasis by transporting Na+ from the cytoplasm into either the extracellular spaces or vacuoles [9]. AKT and HKT1 genes play a major role in the transport of Na+ from shoots to roots, thus preventing leaf damage [10]. AVP gene generates a proton gradient for the driving force for Na+/H+ antiporter and other transporters on the vacuolar membranes, leading to increased salt tolerance and osmotic adjustment in plants [11].
A plethora of miRNAs have been reported to contribute to plant stress responses [12,13] and stimulate interest due to their ubiquitous interaction with target genes related to stress tolerance [14,15]. Recent studies have confirmed the existence of highly conserved, yet not functionally characterized miRNAs, that are induced upon several abiotic stressors [16,17]. Research into miRNAs involved in alkali stress is relatively rare. Regardless of the species, the miR156, miR319, miR398, miR172, miR408, miR169 and miR528 families were most frequently regulated by salt/alkali stress responses [16,18]. Regarding the Medicago species, further elucidation of the molecular mechanism’s underlying responses to salt/alkali stress can be accomplished by identifying miRNAs and their putative target genes.
The present work was carried out following the findings of a previous study [3] concerning the species M. arborea and M. sativa. An effort has been made to further clarify those findings, suggesting the molecular mechanisms that the parents of the Alborea hybrid utilize to efficiently respond against salinity stress. To the best of our knowledge, there are no reports regarding the response of the Alborea hybrid to salinity, except from our previous studies [3,19] that clearly demonstrated Alborea’s sensitivity to this stressor. To that end, the present study focused on salinity-induced changes in six genes, namely SOS1, SOS3, NHX2, AKT, AVP and HKT1, to investigate more extensively the way sodium ions are blocked and transferred to the various plant parts, preventing ion toxicity and ensuring Na+ homeostasis. Additionally, an in-silico analysis was conducted to identify miRNAs that may interact with the genes under study.
2. Results and Discussion
2.1. Gene Expression Levels
Figure 1 displays the expression levels of six salt stress-related genes in M. sativa and M. arborea. Information and additional data regarding phenotypical and physiological differences among the two species have already been published in previous works [3,19].
From the SOS gene family, we studied two genes, namely SOS1 and SOS3, which are directly involved in the regulation of salt tolerance by eliminating Na+ from plant roots. According to our results, M. arborea showed generally higher gene expression values than M. sativa for all treatments, for both shoots and roots, while M. sativa overexpressed the genes more targeted focusing on roots. However, the highest SOS1 and SOS3 gene expression values appeared in plants (both roots and shoots) that were subjected to 100 mM NaCl. (Figure 1(a1,b2)). According to Mäser et al. [20], overexpression of SOS genes in M. falcata and M. truncatula shows that Na+ release from cells is an important factor in plant survival under saline conditions, and activation of this mechanism can improve plant resistance.
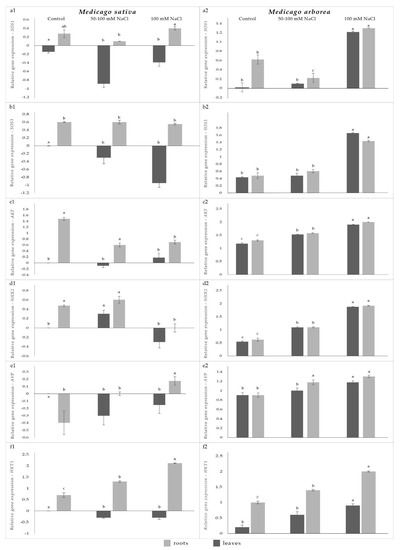
Figure 1.
Mean expression values of roots (light grey) and leaves (dark grey) of: (a1) SOS1 gene in M. sativa, (a2) SOS1 gene in M. arborea, (b1) SOS3 gene in M. sativa, (b2) SOS3 gene in M. arborea, (c1) AKT gene in M. sativa, (c2) AKT gene in M. arborea, (d1) NHX2 gene in M. sativa, (d2) NHX2 gene in M. arborea, (e1) AVP gene in M. sativa, (e2) AVP gene expression in M. arborea, (f1) HKT1 gene in M. sativa, (f2) HKT1 gene in M. arborea in three different salinity treatments. Comparisons between genotypes differ significantly at p < 0.05. Means followed by a different letter differ significantly (p < 0.05) based on the Tukey analysis. Relative expression ratios were calculated using the 2−∆∆CT method [21]. The first two bars are for control treatment, the next two bars are for the 50–100 mM NaCl treatment and the final two bars are for the 100 mM NaCl treatment.
They also argue that Na+/H+ antiporter SOS genes had a role in counteracting root to shoot Na+ transport. SOS genes are expressed in the root and leaf vasculature, predominately in the root tips, and have been considered to re-extract Na+ from the root xylem under salt stress via a proposed reversal of the SOS transporter. Sandhu [10] highlighted the major role that AKT and HKT1 genes play in the transport of Na+ from shoots to roots. In this study, the two genes were overexpressed in the roots of M. sativa while the expression values for both genes in leaves were very low for all treatments (Figure 1(c1,f1)). Specifically for the HKT1 gene, the most elevated expression level was observed in the roots of plants subjected to 100 mM NaCl. Similarly, ref. [20] highlighted that in Arabidopsis the HKT1 gene was strongly expressed in roots. Inversely, the overexpression of these two genes in M. arborea plants was very profound for both leaves and roots of the stressed plants, especially in plants which were subjected to salt shock (Figure 1(c2,f2)). Additionally, it was [10] demonstrated by testing 12 genotypes of M. sativa that the two genotypes of highest salinity resistance overexpressed the AKT and HKT1 genes, mainly in roots compared to leaves, suggesting the contribution of these genes to salinity tolerance. Mäser et al. [21] also showed that in the Arabidopsis thaliana species, K+ starvation induced HKT1 expression in root cortex cells and enhanced inward Na+ currents that included HKT1-like properties in root cortex cells; therefore, disruption of HKT1 facilitates the net transfer of Na+ to shoots. Furthermore, HKT1 could counteract shoot-ward Na+ transport either by sequestering Na+ in the roots and/or preventing it from being loaded into the xylem by re-extracting Na+ that is being loaded into the xylem, or by removing Na+ from leaves and transporting Na+ to roots via the phloem.
The NHX2 and AVP genes are known to play an important role in isolating sodium in the vacuole to protect the cytoplasm from toxic effects. Overexpression of NHX2 gene in alfalfa transgenic plants [8,22] and the AVP gene in Arabidopsis transgenic plants [23,24] improved salinity tolerance by improving intracellular potassium homeostasis. Additionally, several studies in cultivated crops highlighted the importance of AVP overexpression for providing increased plant yield under salinity and drought stress conditions [25,26,27]. In this study, M. sativa demonstrated the highest gene expression value for NHX2 in both roots and leaves for plants subjected to gradually increased salt concentrations (Figure 1(d1)). On the contrary, regarding AVP expression profile in M. sativa, the highest up-regulation was observed in the roots of plants which were treated with 100 mM NaCl (Figure 1(e1)). In contrast to M. sativa, gene expression profiles for all M. arborea salt treatments were clearly elevated (Figure 1(d2,e2)).
Considering all gene expression values under increased salinity conditions for both M. sativa and M. arborea, we can conclude that M. arborea seems to overexpress all examined genes in both shoots and roots, while M. sativa acts in a more targeted manner, overexpressing specific genes (HKT1 and AKT), which help the accumulation of Na+ more specifically in roots. The same trend was observed in another study [3] in which gene expression related to ion transport was carried out under salinity stress. Both studies agree that M. sativa uses a more cost-efficient strategy to respond to salinity by activating specific genes.
2.2. miRNA In Silico Analysis
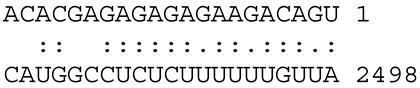
Given the importance of miRNAs in the regulation of gene expression during plant development and in response to abiotic stresses, we performed in silico analysis by use of the ‘psRNAΤarget’ tool, to uncover miRNAs that may target the genes under study (Table 1). The analysis used miRNA and mRNA sequences from M. truncatula since most Medicago miRNAs sequences in the miRNA databases have been derived from this model leguminous plant, as well as due to its high genetic relationship with related species of the Medicago genus, such as M. sativa and M. arborea. Significant complementarity was detected between the conserved miRNA mtr-miR156a and the mRNA of the gene MtAVP (Pyrophospate-energized vacuolar membrane proton pump) in 15 consecutive positions (1–15 bp) from the 5′ end of the miRNA (Table 1). G-U wobbles are allowed in miRNA-target complementarities, and in plants, it seems that complementarity in at least 1–13 consecutive bases from the 5′ end of the miRNA may be associated with actual cleavage of the mRNA of the target gene and further degradation, or translational inhibition. This implies putative binding of mtr-miR156a to the MtAVP mRNA target and subsequent cleavage or translational inhibition of the transcript [28,29], leading to possible downregulation of the MtAVP gene.

Table 1.
In silico identification of putative miRNAs targeting AKT1, AVP, SOS3 and alignments of miRNAs and their corresponding mRNAs.
Similarly, a significant match was observed between miR167b and MtAKT1 mRNA in 17 consecutives bases from the 5′ end of the miRNA. A gap has been introduced, indicating that there may be a bulge in that position in the miRNA-target dimer structure. MiR156a and miR167b both belong to well-conserved plant miRNA families and have been implicated in salt responsiveness in a variety of species [30,31,32]. Based on the above observations, we could speculate that if miR156a targets AVP mRNA in M. sativa salt-stressed leaf tissue, the AVP target is downregulated, presumably due to high expression of miR156a. On the other hand, in root tissue miR156a, it is probably downregulated and, consequently, the AVP gene is upregulated (Figure 1). Likewise, if miR167b targets AKT1, a negative correlation between miR167b and AKT1 messenger RNA levels might be expected in salt-stressed leaves and roots in a manner equivalent to the regulation of the miRNA156-AVP putative module. However, these hypotheses await further experimentation and validation. Thus far, miR167 and miR156 have been implicated in the salt response as well as in other abiotic stresses and developmental processes in a variety of species through the targeting and regulation of transcription factor genes, mainly ARF (AUXIN RESPONSE FACTOR) and SQUAMOSA LIKE PROTEIN (SLP), respectively [33,34,35,36,37,38]. Research on miRNA regulation of gene expression has focused on transcription factor target genes, and modules such as miR156-SPL, miR159-MYB, miR164-NAC, miR167-ARF and miR172-AP have been widely investigated. However, to date, little information exists regarding the engagement of other target genes in these regulatory processes through miRNA-mRNA modules, including the genes under study. An ever-increasing number of miRNAs and putative target genes identified in recent years [33,34] will facilitate the study of other miRNA-target gene pairs and their function in salt-tolerance. Our analysis of AKT1, AVP and SOS3 will provide additional information to these studies and contribute towards this goal. Furthermore, candidate miRNAs targeting SOS3 seem to be the miR2605, mir2670f and miR2670g. Nevertheless, there is no information thus far regarding these sequences, and they may represent species-specific miRNAs that need to be studied further.
This work highlights the importance of Na+ compartmentation and the action of the SOS1, SOS3, NHX2, AKT, AVP and HKT1 genes towards salinity tolerance. MiRNAs are also known to contribute to salinity resilience. Expression studies via qPCR experiments for the identified miRNA to validate their significance will possibly enable their future use into breeding programs.
3. Materials and Methods
3.1. Plant Material and Seed Pretreatment
The M. sativa population used in this study was a hybrid of two M. sativa parents [3]. Seeds from both M. sativa and M. arborea were obtained from E. Bingham, Agronomy Department, University of Wisconsin-Madison, Madison, WI, USA. The germplasm of M. arborea is a collection from Greece [3]. The seeds were scarified with absolute sulfuric acid for 10 min and then rinsed thoroughly with sterile distilled water. Bleach solution (3%) was added for 1.5–2 min and then rinsed thoroughly. The scarified seeds were placed on Petri dishes containing 0.5% agar at 4°C overnight, and then transferred to 20°C (dark) for 3–4 days.
3.2. Growth Conditions and Salt Stress Treatments
From each of the Medicago species, 60 seeds were planted, after their germination, in pots of 8.5 cm height and 10 cm in diameter. Each pot contained peat, Kronos N 50–300 mg/L, P2O5 80–300 mg/L, K2O 80–300 mg/L, pH 5–6.5, salinity <1.75 g/L and one seed. The pots were then placed in a growth chamber at a constant temperature of 25 °C, and a photoperiod of 16 h of light and 8 h of darkness, where they were regularly watered and fertilized every week. For each treatment, 20 plants in seedling stage were used for each species in a completely randomized design. The following treatments were applied: (1) salt stress, with initial treatment of 50 mM NaCl and gradual step acclimatization 10 days after, with treatment of 100 mM NaCl, (2) salt shock, with continuous application of 100 mM NaCl concentration and (3) no application of NaCl. Plants were watered every five days after salt treatment initiation. Once per week, Hoagland solution [39] was added to the salt solution. The growth chamber experiment lasted 34 days. After the completion of the experiment, whole shoots, excluding cotyledons and the roots, free of pathogens, were cut and immediately transferred into liquid nitrogen in the middle of the light period. When harvested, all plants were in the vegetative stage, and roots did not show nodules.
3.3. RNA Isolation and cDNA Synthesis
For plant RNA isolation, the TRIzol reagent kit (Sigma-Aldrich, St. Louis, MO, USA) was used and RNA concentration was measured by Nanodrop Spectrophotometer (Thermo Scientific NanoDrop™ 1000 Spectrophotometer). For cDNA synthesis, Superscript II enzyme (Invitrogen, Carlsbad, CA, USA) was used. The quality of the cDNA was evaluated by PCR using an actin gene as the endogenous control reference gene. The primers used in this experiment are based on Sandhu et al., 2017 [10].
3.4. Real-Time PCR Experiments
The StepOnePlus™ Real-Time PCR Systems (Applied Biosystems, Foster City, CA, USA) machine was used to detect relative transcript levels using SYBR Select Master Mix (Applied Biosystems, Foster City, CA, USA). The threshold cycle CT method was used, in which the relative amount of a target gene in the samples can be determined. In our experiment, six target genes were studied (SOS1, SOS3, NHX2, AKT, AVP and HKT1) and the primers that were used were the same as Sandhu et al., 2017. Primer selection was based on Sandhu et al., 2017 [10]. All reactions were performed in triplicates. Melting curves of the reaction products were generated and fluorescence data were collected at a temperature above the melting temperature of non-specific products. Relative expression levels of the studied genes were calculated according to the 2−∆∆Ct method [20] (for calculating ∆∆Ct value, the control sample of each entry was used). Actin-2 was used as an internal control for normalization.
3.5. Statistical Analysis
The effect of variables on gene expression was tested using the one-way analysis of variance (ANOVA) followed by mean comparisons through Tukey’s HSD post hoc test with p-values of p < 0.05. Values are expressed as mean ± SEM (standard error of mean). The statistical analysis was performed using the IBM SPSS Statistics 23 statistical package for Windows (SPSS Inc., Chicago, IL, USA).
3.6. In Silico miRNA Analysis
For the prediction of potential miRNAs that may target and regulate the expression of the genes under study, the “psRNATarget: A Plant Small RNA Target Analysis Server” tool (https://www.zhaolab.org/psRNATarget/-version, 20 December 2022) was employed. The default parameters were used and were as follows: maximum expectation value of 5, penalty for G:U pair of 0.5, penalty for other mismatches of 1, seed region of 2–13 nt, number of mismatches allowed in the seed region of 2.0 and translation inhibition ranges of 10 nt to 11 nt. Under these parameters, and following the analysis of the results, the most significant score which designates the best possibility for miRNA-mRNA pair formation was selected.
Author Contributions
Conceptualization, E.T. and E.M.A.; methodology, E.S.; software, A.K. and E.M.A.; validation, E.S., A.K., M.G., E.M.A., P.J.B., E.T.; formal analysis, E.M.A.; investigation, E.T.; resources, E.T.; data curation E.T., A.K.; writing—original draft preparation, E.S., A.K., M.G., E.M.A., P.J.B., E.T.; writing—review and editing, E.S., A.K., M.G., E.M.A., P.J.B., E.T.; visualization, E.S., A.K., E.M.A., E.T.; supervision, E.T.; project administration, E.T.; funding acquisition, E.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Applicable upon request.
Acknowledgments
We would like to thank E. Bingham for providing us the genetic material used for the study. We would also like to thank Ioanna Kendrick and Joseph Pentheroudakis for English editing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, C.M.; Zhang, J.L.; Liu, X.S.; Li, Z.; Wu, G.Q.; Cai, J.Y.; Flowers, T.J.; Wang, S.M. Puccinellia tenuiflora maintains a low Na+ level under salinity by limiting unidirectional Na+ influx resulting in a high selectivity for K+ over Na+. Plant Cell Environ. 2009, 32, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Aisset Amel, M.Z. Effect of salinity and water stress on the germination of Medicago arborea L. seeds. J. Appl. Biol. Sci. 2016, 6, 113–121. [Google Scholar]
- Tani, E.; Sarri, E.; Goufa, M.; Asimakopoulou, G.; Psychogiou, M.; Bingham, E.; Skaracis, G.N.; Abraham, E.M. Seedling growth and transcriptional responses to salt shock and stress in Medicago sativa L.; Medicago arborea L.; and their hybrid (Alborea). Agronomy 2018, 8, 231. [Google Scholar] [CrossRef]
- Volkov, V.; Amtmann, A. Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, has specific root ion-channel features supporting K+/Na+ homeostasis under salinity stress. Plant J. 2006, 48, 342–353. [Google Scholar] [CrossRef]
- Pandita, D. Role of miRNA technology and miRNAs in abiotic and biotic stress resilience. In Plant Perspectives to Global Climate Changes; Aftab, T., Roychoudhury, A., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 303–330. [Google Scholar]
- Zelm, E.V.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Zhang, L.Q.; Niu, Y.D.; Huridu, H.; Hao, J.F.; Qi, Z.; Hasi, A. Salicornia europaea L. Na+/H+ antiporter gene improves salt tolerance in transgenic alfalfa (Medicago sativa L.). Genet. Mol. Res. 2014, 13, 5350–5360. [Google Scholar] [CrossRef]
- Wu, G.-Q.; Wang, J.-L.; Li, S.-J. Genome-wide identification of Na+/H+ antiporter (NHX) genes in sugar beet (Beta vulgaris L.) and their regulated expression under salt stress. Genes 2019, 10, 401. [Google Scholar] [CrossRef]
- Sandhu, D.; Cornacchione, M.V.; Ferreira, J.F.S.; Suarez, D.L. Variable salinity responses of 12 alfalfa genotypes and comparative expression analyses of salt-response genes. Sci. Rep. 2017, 7, 42958. [Google Scholar] [CrossRef] [PubMed]
- Toranj, S.; Aliabad, K.K.; Abbaspour, H.; Saeedpour, A. Effect of salt stress on the genes expression of the vacuolar H+ -pyrophosphatase and Na+/H+ antiporter in Rubia tinctorum. Mol. Biol. Rep. 2020, 47, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Chinnusamy, V.; Zhu, J.; Zhu, J.K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007, 12, 301–309. [Google Scholar] [CrossRef]
- Xu, T.; Zhang, L.; Yang, Z.; Wei, Y.; Dong, T. Identification and functional characterization of plant miRNA under salt stress shed light on salinity resistance improvement through miRNA manipulation in crops. Front. Plant Sci. 2021, 12, 665439. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Zhang, H.M.; Liu, Z.H.; Li, H.C.; Guo, X.L.; Li, G.L. The wheat NHX antiporter gene TaNHX2 confers salt tolerance in transgenic alfalfa by increasing the retention capacity of intracellular potassium. Plant Mol. Biol. 2015, 87, 317–327. [Google Scholar] [CrossRef]
- Barciszewska-Pacak, M.; Milanowska, K.; Knop, K.; Bielewicz, D.; Nuc, P.; Plewka, P.; Pacak, A.M.; Vazquez, F.; Karlowski, W.; Jarmolowski, A.; et al. Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front. Plant Sci. 2015, 6, 410. [Google Scholar] [CrossRef]
- Cao, X.; Wu, Z.; Jiang, F.; Zhou, R.; Yang, Z. Identification of chilling stress-responsive tomato microRNAs and their target genes by high-throughput sequencing and degradome analysis. BMC Genom. 2014, 15, 1130. [Google Scholar] [CrossRef]
- Shi, R.; Jiao, W.; Yun, L.; Zhang, Z.; Zhang, X.; Wang, Q.; Li, Y.; Mi, F. Utilization of transcriptome, small RNA, and degradome sequencing to provide insights into drought stress and rewatering treatment in Medicago ruthenica. Front. Plant Sci. 2021, 12, 675903. [Google Scholar] [CrossRef]
- Wani, S.H.; Kumar, V.; Khare, T.; Tripathi, P.; Shah, T.; Ramakrishna, C.; Aglawe, S.; Mangrauthia, S.K. miRNA applications for engineering abiotic stress tolerance in plants. Biologia 2020, 75, 1063–1081. [Google Scholar] [CrossRef]
- Sarri, E.; Termentzi, A.; Abraham, E.M.; Papadopoulos, G.K.; Baira, E.; Machera, K.; Loukas, V.; Komaitis, F.; Tani, E. Salinity stress alters the secondary metabolic profile of M. sativa, M. arborea and their hybrid (Alborea). Int. J. Mol. Sci. 2021, 22, 4882. [Google Scholar] [CrossRef] [PubMed]
- Mäser, P.; Eckelman, B.; Vaidyanathan, R.; Horie, T.; Fairbairn, D.J.; Kubo, M.; Yamagami, M.; Yamaguchi, K.; Nishimura, M.; Uozumi, N.; et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002, 531, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Liu, Z.H.; Wen, Z.Y.; Zhang, H.M.; Yang, F.; Guo, X.L. The vacuolar Na+-H+ antiport gene TaNHX2 confers salt tolerance on transgenic alfalfa (Medicago sativa). Funct. Plant Biol. 2012, 39, 708–716. [Google Scholar] [CrossRef]
- Gaxiola, R.A.; Li, J.; Undurraga, S.; Dang, L.M.; Allen, G.J.; Alper, S.L.; Fink, G.R. Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc. Natl. Acad. Sci. USA 2001, 98, 11444–11449. [Google Scholar] [CrossRef]
- Sandhu, D.; Kaundal, A.; Acharya, B.R.; Forest, T.; Pudussery, M.V.; Liu, X.; Ferreira, J.F.S.; Suarez, D.L. Linking diverse salinity responses of 14 almond rootstocks with physiological, biochemical, and genetic determinants. Sci. Rep. 2020, 10, 21087. [Google Scholar] [CrossRef] [PubMed]
- Arif, A.; Zafar, Y.; Arif, M.; Blumwald, E. Improved growth, drought tolerance, and ultrastructural evidence of increased turgidity in tobacco plants overexpressing Arabidopsis vacuolar pyrophosphatase (AVP1). Mol. Biotechnol. 2013, 54, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Knapp, J.; Koirala, P.; Rajagopal, D.; Peer, W.A.; Silbart, L.K.; Murphy, A.; Gaxiola, R.A. Enhanced phosphorus nutrition in monocots and dicots over-expressing a phosphorus-responsive type I H+-pyrophosphatase. Plant Biotechnol. J. 2007, 5, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Bhaskaran, S.; Savithramma, D.L. Co-expression of Pennisetum glaucum vacuolar Na+/H+ antiporter and Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic tomato. J. Exp. Bot. 2011, 62, 5561–5570. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef]
- Mallory, A.C.; Bouché, N. MicroRNA-directed regulation: To cleave or not to cleave. Trends Plant Sci. 2008, 13, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Long, R.; Zhang, T.; Kang, J.; Wang, Z.; Wang, P.; Sun, H.; Yu, J.; Yang, Q. Genome-wide identification of microRNAs in response to salt/alkali stress in Medicago truncatula through high-throughput sequencing. Int. J. Mol. Sci. 2018, 19, 4076. [Google Scholar] [CrossRef]
- Jodder, J.; Das, R.; Sarkar, D.; Bhattacharjee, P.; Kundu, P. Distinct transcriptional and processing regulations control miR167a level in tomato during stress. RNA Biol. 2018, 15, 130–143. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Y.; Li, J. Global identification and analysis of microRNAs involved in salt stress responses in two alfalfa (Medicago sativa ‘Millennium’) lines. Can. J. Plant Sci. 2019, 100, 445–455. [Google Scholar] [CrossRef]
- Fu, R.; Zhang, M.; Zhao, Y.; He, X.; Ding, C.; Wang, S.; Feng, Y.; Song, X.; Li, P.; Wang, B. Identification of salt tolerance-related microRNAs and their targets in maize (Zea mays L.) using high-throughput sequencing and degradome analysis. Front. Plant Sci. 2017, 8, 864. [Google Scholar] [CrossRef] [PubMed]
- Islam, W.; Waheed, A.; Naveed, H.; Zeng, F. MicroRNAs mediated plant responses to salt stress. Cells 2022, 11, 2806. [Google Scholar] [CrossRef]
- Kang, T.; Yu, C.Y.; Liu, Y.; Song, W.M.; Bao, Y.; Guo, X.T.; Li, B.; Zhang, H.X. Subtly manipulated expression of ZmmiR156 in tobacco improves drought and salt tolerance without changing the architecture of transgenic plants. Front. Plant Sci. 2020, 10, 1664. [Google Scholar] [CrossRef] [PubMed]
- Khraiwesh, B.; Zhu, J.K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Ruiz-Manriquez, L.M.; Serrano-Cano, F.I.; Reyes-Pérez, P.R.; Tovar Alfaro, C.K.; Barrón Andrade, Y.E.; Paul, S. Identification of microRNAs and their expression in leaf tissues of guava (Psidium guajava L.) under salinity stress. Agronomy 2020, 10, 1920. [Google Scholar] [CrossRef]
- Song, C.; Zhang, D.; Zheng, L.; Zhang, J.; Zhang, B.; Luo, W.; Li, Y.; Li, G.; Ma, J.; Han, M. miRNA and degradome sequencing reveal miRNA and their target genes that may mediate shoot growth in spur type mutant “Yanfu 6”. Front. Plant Sci. 2017, 8, 441. [Google Scholar] [CrossRef]
- Suh, C. Evaluation of Bioactivity of Phytotoxins from Pathogenic Fungi of Orobanche sp. Ph.D. Thesis, Agricultural University of Athens, Athina, Greece, 2011. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).