Abstract
Despite the major impact of Blastocystis sp. in terms of prevalence in human and animal populations and the risk of zoonotic transmission, no epidemiological survey has yet been conducted in cattle herds in France. The aim of this study was thus to assess the prevalence and molecular diversity of Blastocystis sp. and associated factors in dairy cattle from the north of France. A total of 1581 fecal samples were collected from 1246 animals reared in 20 farms. Molecular detection of the protozoan was performed by real-time PCR and indicated an overall prevalence of Blastocystis sp. reaching 54.8% in the study population. Important inter-herd variation (from 22.2% to 76.5%) of Blastocystis sp. prevalence was also reported. Sequence analysis of 159 positive samples highlighted a very large predominance of ST10 (36/159) and ST14 (64/159), and ST2 was only found in 2 samples. Mixed subtype infections were common, representing 35.8% of sequenced samples (57/159). A putative correlation between Blastocystis sp. colonization and various animal and herd characteristics or feed intake was subsequently investigated. The protozoan was less prevalent in cows that have recently calved but Blastocystis sp. carriage was not significantly related to age. Blastocystis sp. colonization also decreased with high beet pulp and pasture grass consumption and increased with corn silage intake. Finally, the only significant association between Blastocystis sp. STs and animal and herd characteristics was the number of lactations of cows, with a predominance of ST14 in cows that calved once only.
1. Introduction
Blastocystis sp. is a cosmopolitan protozoan that colonizes the gastrointestinal tract of humans and a wide range of animals worldwide [1,2,3]. This anaerobic microorganism is transmitted by the fecal–oral route through direct contact with infected hosts or through the consumption of food or water contaminated with cysts [4]. Human Blastocystis sp. infection can be associated with a variety of digestive symptoms, but the pathogenicity of this protozoan remains controversial in humans since the vast majority of individuals colonized by Blastocystis sp. do not present intestinal manifestations [5,6,7].
In animals, the lack of a reproducible model of Blastocystis sp. infection is the main obstacle to clarifying its pathogenicity [8]. However, it is noteworthy that Blastocystis sp. colonization is not usually associated with disease in either wild or domestic animals [9]. Particularly, cattle have been widely studied for the presence of this protozoan around the world [10], but reports in Europe remain scarce with only three epidemiological surveys conducted so far in the UK [11], Denmark [12], and Spain [13]. A recent synthesis highlighted a 24.4% (95% CI: 16.9–33.9) prevalence for Blastocystis sp. in cattle after reviewing 29 international studies [10]. Five of these 29 studies relied on a microscopy-based method to detect Blastocystis (mean prevalence: 24.4%; range: 1.8–71%). However, this microscopic detection method is well known to be less sensitive than the molecular detection of the protozoan. In the 25 molecular surveys, the prevalence of Blastocystis sp. in domestic dairy/beef cattle varied between 2.9% and 100% (mean: 31.8%), depending on the country. This significant occurrence of Blastocystis sp. in livestock cattle raises the question of the risk of zoonotic transmission, especially to dairy farmers. Indeed, a large genetic diversity has been described within the Blastocystis genus, with at least 28 subtypes (STs) currently considered as valid and colonizing avian and mammalian hosts with various frequencies [14,15,16]. Among these STs, ST1–ST4 account for more than 90% of the human infections reported globally, while ST5–ST8, ST10, ST12, ST14, and ST16 were rarely found in human stool, and are considered to be of animal origin and likely the result of zoonotic transmission [17,18,19,20,21]. Particularly, ST10 and ST14, the two predominant STs identified in bovids [9,22], were only sporadically found in humans in Senegal and Lebanon [19,20].
Despite the significant impact of Blastocystis sp. in bovid in terms of prevalence, molecular data on the epidemiology of this protozoan in cattle are almost non-existent in Europe and to our knowledge, no survey has been performed in France. Therefore, the aim of this study was to obtain the first molecular data regarding the prevalence and ST distribution of Blastocystis sp. in dairy Prim’Holstein cattle from the north of France. In parallel, another purpose of this survey was to determine if the presence of the protozoan was associated with animal-, herd-, or feed intake-related factors.
2. Results and Discussion
Among the 1581 fecal samples collected from 1246 non-diarrheic Prim’Holstein dairy cows raised on 20 farms in Northern France, 866 of them (54.8%) were shown to be positive for Blastocystis sp. by qPCR. This high rate of Blastocystis sp. colonization in France was roughly similar to those observed using molecular methods in bovid from other countries, including Lebanon (63.4%) [22], Japan (54.1%) [23], Thailand (50.0%) [24], Colombia (80.0%) [25], Libya (41.7%) [11], Malaysia (34.5%) [26], Iran (33.3 and 35%) [27,28], and Spain (32.1%) [13]. On the other hand, it was significantly higher than the ones reported in Korea (6.7%) [29], China (9.5%, 10.3% and 27.9%) [30,31,32], Iran (9.6%) [33], Turkey (11.25%) [34], Brazil (21.4%) [35], the UK (22.6%) [11], United Arab Emirates (22.7%) [36], the United States (19.1% and 2.9%) [37,38], and Malaysia (25% and 25.4%) [39,40]. Although the prevalence of Blastocystis sp. in domestic cattle may greatly vary between geographical areas, it is nevertheless common worldwide.
The genetic diversity of Blastocystis sp. was thus explored in 159 randomly selected positive samples collected from 19 herds. Among them, 102 (64.2%) partial SSU rDNA gene sequences obtained from these isolates showed 99 to 100% similarity with homologous Blastocystis sp. sequences available in databases allowing for their subtyping. Interestingly, Blastocystis sp. ST14 was the most prevalent ST identified in this French animal cohort (64/102, 62.7%) followed by ST10 (36/102, 35.3%). ST10 and ST14 were thus largely predominant by accounting for 98% of the subtyped isolates. These two STs also globally represented the most widely distributed for instance, in Denmark [12], Japan [23], Korea [29], and China [30] and were also the only two STs identified in cattle in two studies conducted in the USA [37,41]. The predominance of ST10 and ST14 observed in the present study confirmed that domestic cattle are natural hosts of these two STs that can be considered cattle-adapted STs as previously suggested [22]. Here, we would like to highlight the potential issue of incorrect assignments of some Blastocystis sp. STs in databases, as recently reported [42]. Indeed, by querying the NCBI databases with our own sequences using BLASTN, we noticed that some Blastocystis sp. sequences (from [40]) were erroneously annotated as ST5 although they share >99% sequence similarity with ST14 sequences. The two last SSU rDNA gene sequences obtained in cattle from this study were representative of ST2. This ST was so far only identified in one dairy cattle from a breeding farm in North Lebanon [22]. Consequently, the sporadic prevalence of ST2 in domestic cattle seems to reflect rather an accidental contamination than a real infection. In this sense, a contamination by the breeders of these cattle cannot be excluded since this ST is frequent in the French population [43].
The 57 remaining positive dairy cattle samples (35.8%) corresponded to infections by at least two different Blastocystis sp. STs (mixed infections) according to the resulting sequence chromatograms presenting double traces. A recent study using Sanger sequencing and next-generation sequencing (NGS) to characterize Blastocystis sp. STs in Spanish cows showed that mixed infections are extremely common in cattle [13]. It was also demonstrated for instance in a large cohort of dairy cattle in Lebanon that showed a frequency of mixed infections of about 30% [22]. It would be of great interest in a future study to use NGS that can better discriminate among the STs involved in mixed infections than Sanger sequencing, to sequence the Blastocystis STs encountered in the numerous samples from the present study. For six dairy cattle that were sampled twice at different times, the same Blastocystis sp. ST was found except for two animals where the combinations ST10-then-ST14 and mixed infection-then-ST10 were encountered.
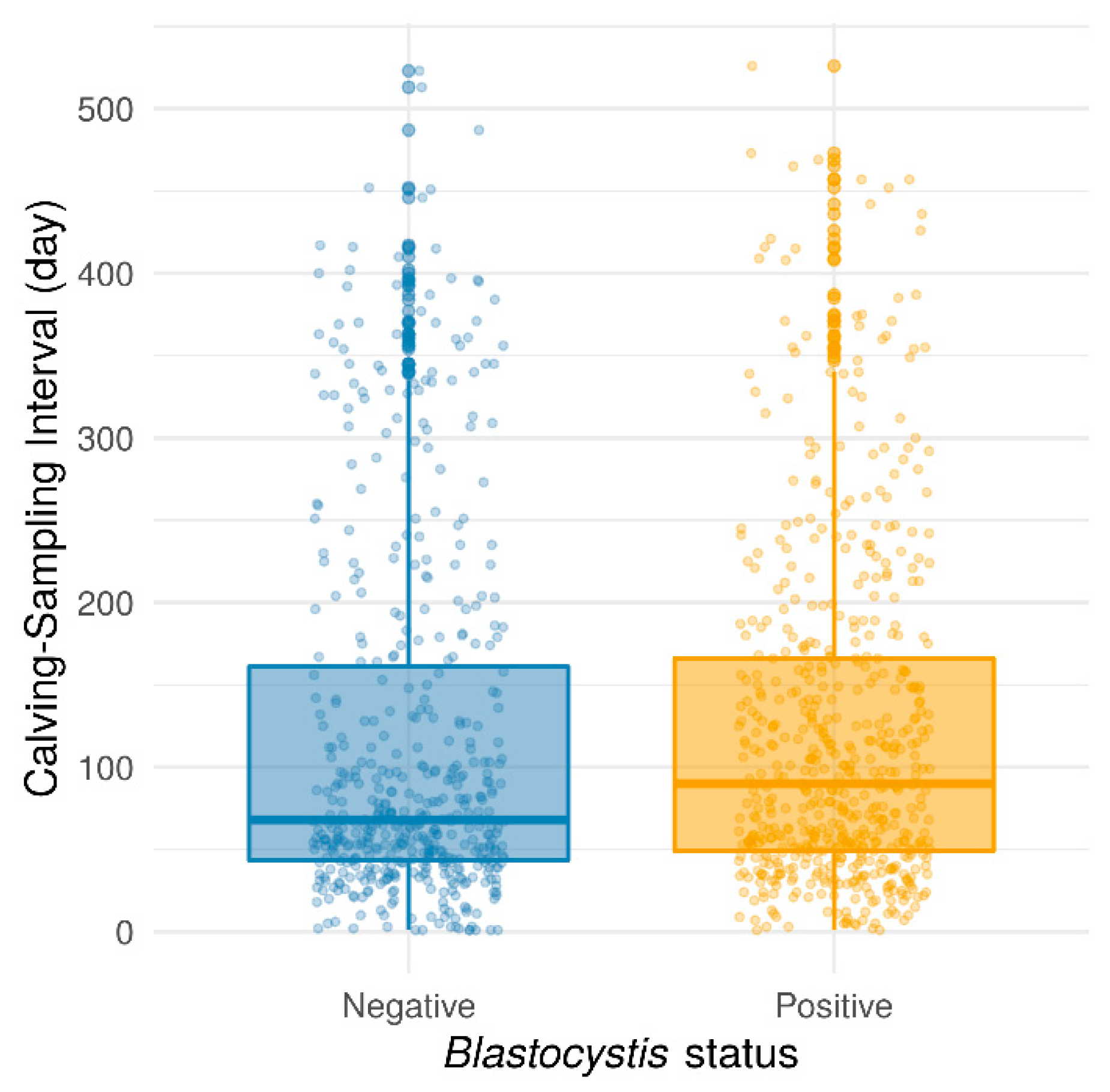
Thereafter, the prevalence of Blastocystis sp. was evaluated according to different variables related to the animals such as age and parity, the herd, or to the breeding practices such as the feeding diet. The age of the animals, with a median of 1025 days (range: 200–4444 days), had no significant effect on Blastocystis sp. prevalence, considering values in days (Wilcoxon rank-sum test with continuity correction, p = 0.9004) or intervals of 500 days (Pearson’s chi-squared test, p = 0.6349). This result conflicted with those obtained in previous reports, in which the highest infection rate was observed in cattle aged from 3 months to 1 year, compared to adults aged over 1 year or calves aged less than 3 months [29,30]. Moreover, no significant association has been found between Blastocystis sp. carriage and the number of parities (Pearson’s chi-squared test, p = 0.0852) even if a trend was emphasized towards a lower prevalence in multiparous cows as compared to nulliparous animals (53.5% vs. 58.0%, respectively) (Pearson’s chi-squared test with Yates’ continuity correction, p = 0.1002). In contrast, the time interval between calving and fecal sampling was found to have a significant effect on Blastocystis sp. carriage. Indeed, the protozoan was less commonly detected in cows that were collected shortly after calving, (Wilcoxon rank-sum test with continuity correction, p = 0.027) (Figure 1). This difference was still significant when considering only cows sampled between 0 and 250 days after calving (Wilcoxon test, p = 5.57 × 10−5, data not shown). The important physiological changes that occur during calving that make them listless (cows do not eat or drink during calving) and which reduce their dry matter intake post-calving could partially explain this result.
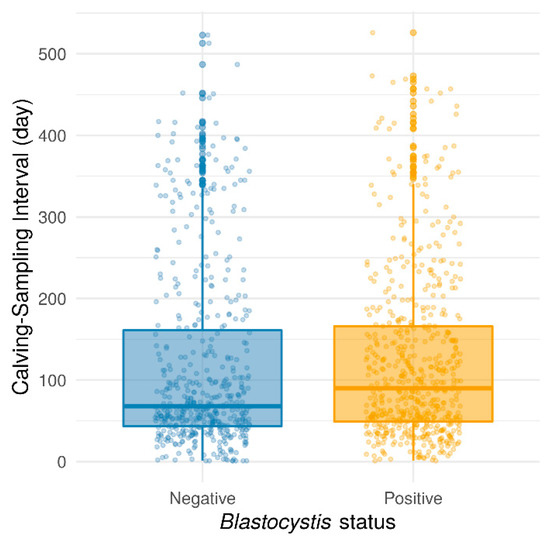
Figure 1.
Box plots representing the time interval (in days) between calving and the fecal sample collection according to the Blastocystis sp. status of the animals. Wilcoxon rank-sum test with continuity correction was used (p = 0.027).
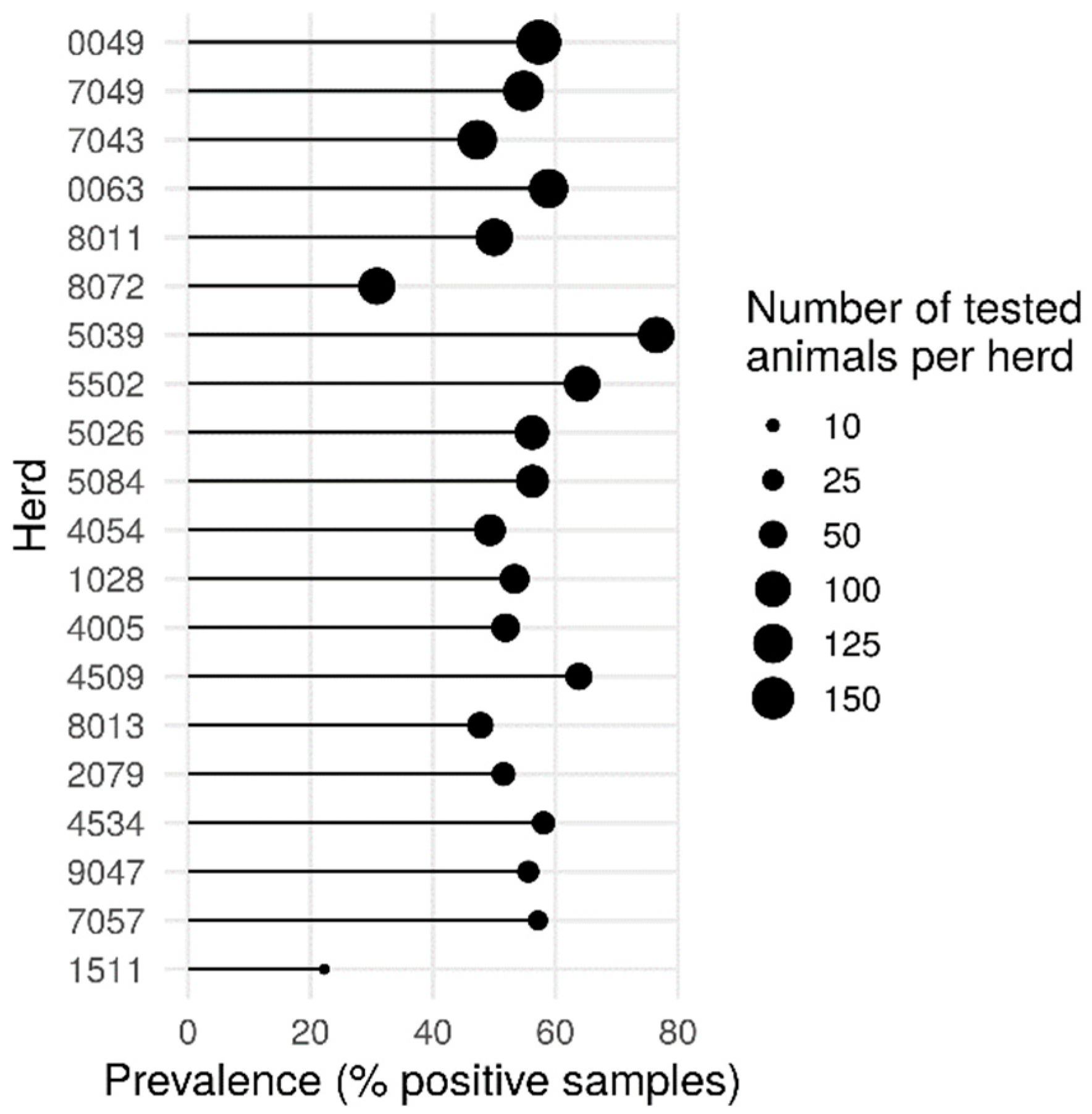
The herd has the most important effect in our study, with an intra-herd prevalence of Blastocystis sp. ranging from 22.2% to 76.5% (Pearson’s chi-squared test, p = 2.868 × 10−6) (Figure 2). These variations could be due to differences in cattle housing and husbandry management practices such as cattle movements on and off the farm (percentage of open grazing or stall-fed over the year), animal housing building, flooring and bedding types, treatment practices, the structure and composition of the herd such as the number of heads, the presence of other animals in the farms, and the sharing of pastures. All these hypotheses deserve further investigation.
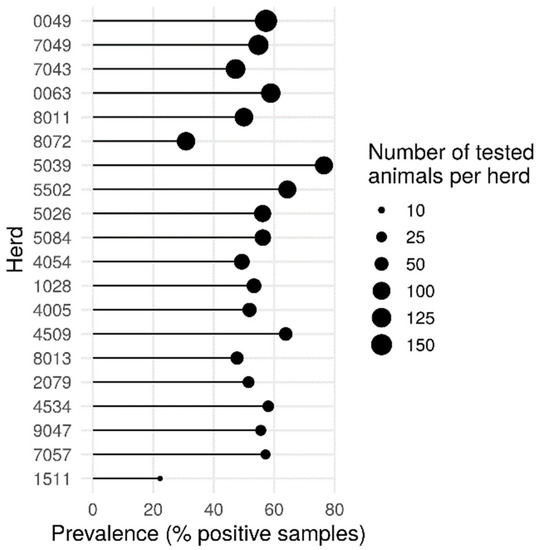
Figure 2.
Percentage of Blastocystis sp.-positive animals within the different herds. The size of the dots is proportional to the number of samples tested in each herd. Pearson’s chi-squared test, (p = 2.868 × 10−6).
Surprisingly, no statistical significance between Blastocystis sp. carriage and water supply (private drilling or public system) was reported (Pearson’s chi-squared test with Yates’ continuity correction, p = 0.1659). However, the transmission of this protozoan is known to occur through ingestion of water (or food) contaminated with Blastocystis sp. cysts. Hence, if all Blastocystis sp. STs are likely to produce resistant cysts in the environment that can be transmitted through contaminated water, it was reasonable to expect a higher prevalence of the protozoan in animals consuming private drilling. It can also be suggested that ST10 and ST14 do not produce resistant cysts and are therefore only transmitted through direct contact between animals. This hypothesis, that remains to be confirmed, could also explain the scarcity of these two STs in the human population.
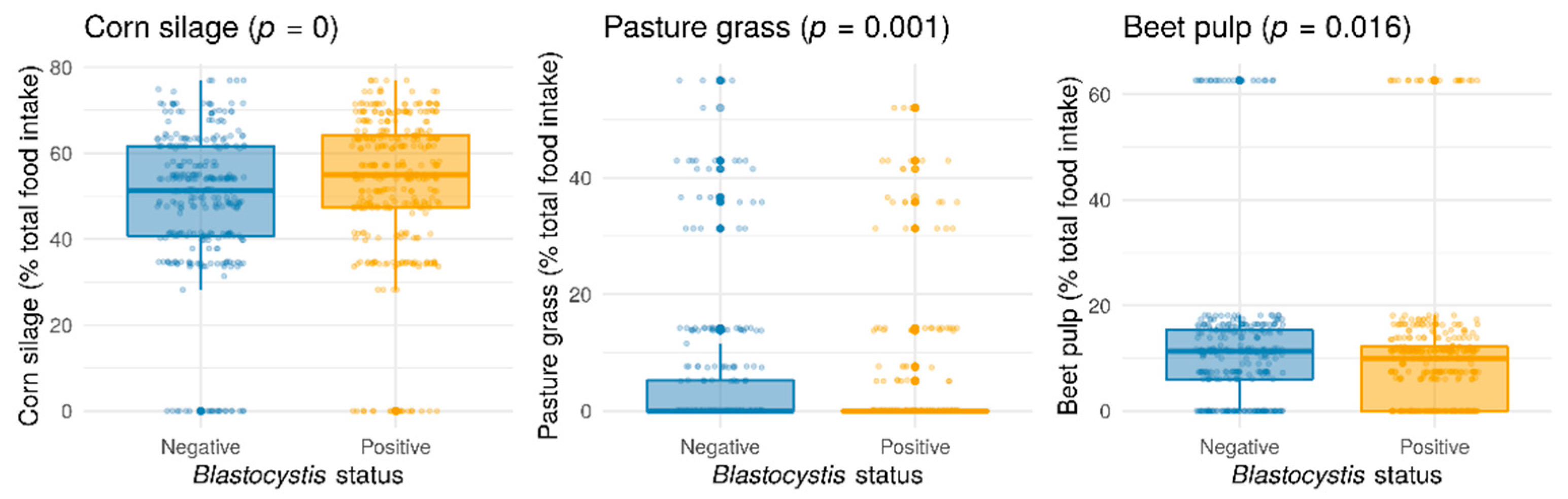
Although the seasonal variations in the occurrence of Blastocystis sp. were difficult to appreciate because of biases induced by unbalanced data over time and herds, animals sampled in winter seemed to be less colonized by the protozoa (Pearson’s chi-squared test with Yates’ continuity correction, p = 0.0003572) (Figure S1). In order to assess the prevalence of Blastocystis sp. as a function of the feeding diet, only samples with the closest interval between the date of diet scoring and the date of fecal sampling, with a maximum difference of 60 days, were selected (728 samples). From the overall analysis, Blastocystis sp. colonization was correlated with three types of feed: corn silage, pasture grass, and to a lesser degree, sugar beet pulp (Figure 3). Unlike usually observed with gastrointestinal nematode parasites, grass grazing was negatively correlated with Blastocystis sp. carriage. Sugar beet pulp diet also appeared to be negatively correlated with the presence of Blastocystis sp., whereas protozoan colonization was positively associated with corn silage consumption (Figure 3). By considering the food effect on Blastocystis sp. prevalence according to seasons, corn silage was still significantly positively correlated with protozoan frequency in winter and in summer. On the other hand, the negative correlation between Blastocystis sp. carriage and beet pulp was only significant in winter, and the negative correlation between protozoan carriage and pasture grass only in summer (Figure S2). These associations could be explained by the nutrient composition of the different types of feed and maybe the interaction with the metabolism of the animals (e.g., negative energy balance during the postpartum period). Moreover, Blastocystis sp. could be vulnerable to some aflatoxins differentially found in beet pulps and corn silage representing two feed whose storage conditions are known to constitute a risk of mold growth and presence of mycotoxins [44]. Other components of the feed rations evaluated in our study, including ensiled grass, alfalfa, nitrogen deficiency corrector, energetic or production concentrates, and sugar beets were not found to be correlated with Blastocystis sp. colonization (data not shown).
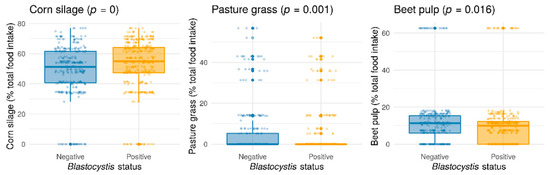
Figure 3.
Statistically significant effects of food items (corn silage (p = 0), pasture grass (p = 0.001), beet pulp (p = 0.016)) on overall Blastocystis sp. prevalence (Wilcoxon test). Each food item value has been transformed into the percentage of total food intake (ingested dry matter).
In addition, since cows are considered to be hypocalcemic during and after parturition (due to a post-calving energy deficit and the high levels of estrogen around calving that inhibit calcium mobilization), and that beet pulps are feeds rich in calcium [45], it can be suggested that Blastocystis sp., less present in animals close to calving or consuming many beet pulps, is sensitive to the calcemia of cows, perhaps not thriving in either hypocalcemic or hypercalcemic conditions.
Finally, no significant associations have been found between the presence of Blastocystis sp. STs (i.e., when classified by ST10, ST14, ST2, or mixed infection) and the herd, sampling, or calving seasons (Pearson’s chi-squared tests, p > 0.05). However, the number of lactations cows have experienced (i.e., lactation rank) seems to be correlated with Blastocystis sp. STs, with a predominance of ST14 in cows that calved only once (Pearson’s chi-squared tests, p = 0.003).
3. Materials and Methods
3.1. Sampling
From September 2017 to December 2018, a total of 1581 fecal samples from 1246 non-diarrheic and non-pregnant Prim’Holstein dairy cattle (nulli-, primi-, or multiparous) were collected by animal reproduction technicians from the Gènes Diffusion company (Douai, France) throughout 20 farms in an area of 110 km around the city of Lille, north of France. The average herd size in these farms during the time frame of the study was 209 (SD, 89) cows (range: 78–439). The sampling strategy aimed at targeting individuals representative of the various breeding conditions and animal characteristics. Particularly, the age of the cows, parity, collection date (season), time interval between calving and sampling, food intake, and drinking water source (personal drilling or public water system) were recorded. For fecal sampling, cows were rectally finger stimulated with sterile-gloved hands to facilitate the collection of a 50 g fecal sample, which was immediately frozen at −20 °C (or stored at 4 °C for 1 to 2 h before freezing). Finally, all frozen samples were centralized in the Research and Development Laboratory of the Gènes Diffusion company (Institut Pasteur of Lille), where the conformity of the samples was checked and the barcodes linked to the samples were scanned.
3.2. Ethical Issues
No approval from the Institutional Animal Care and Use Committee or ethics committee was necessary, as all samples were obtained by duly authorized technicians and no invasive sampling approach was performed. Animals were reared following French guidelines for animal care and use. Sampling was carried out by trained Gènes Diffusion technicians who take part in reproduction monitoring. These approved samplers are holders of the CAFTI (Certificat d’Aptitude aux Fonctions de Technicien d’Insémination-Certificate of Fitness for Insemination Technician Functions), approved by the declaration to the EDE (Departmental Establishments of Breeding) for biological sampling according to animal welfare consideration. The farmers concerned have also signed an agreement document allowing access to their animals’ samples for research purposes.
3.3. Detection of Blastocystis sp. and Subtyping of the Isolates
Total genomic DNA was extracted from approximately 200 mg of animal fecal samples using the NucleoSpin 96 Soil Kit or NucleoSpin Soil Mini Kit (Macherey-Nagel GmbH & Co KG, Düren, Germany) according to the manufacturer’s recommendations. DNA was stored at −20 °C until further processing. Two μL of extracted DNA was subjected to a real-time PCR (qPCR) assay using the Blastocystis-specific primers BL18SPPF1 (5′-AGTAGTCATACGCTCGTCTCAAA-3′) and BL18SR2PP (5′-TCTTCGTTACCCGTTACTGC-3′) targeting the small subunit ribosomal RNA (SSU rRNA) gene as previously described [46]. The qPCR amplicon from each positive sample was purified and used for both strands of Sanger sequencing by Genoscreen (Lille, France). The obtained chromatograms from both forward and reverse strands were visually inspected for the presence of double peaks and overlapping traces. The sequences obtained were compared with all Blastocystis sp. homologous sequences available from the National Center for Biotechnology Information (NCBI) using the Nucleotide Basic Local Alignment Search Tool (BLASTN) program. The STs were identified by determining the exact match or closest similarity to all known Blastocystis STs.
3.4. Statistical Tests
Statistical analyses were conducted using the R software version 4.12 released on 1 November 2021 (R Development Core Team, http://www.R-project.org). Chi-squared and Wilcoxon tests were used to compare the Blastocystis sp. prevalence and Blastocystis sp. STs colonization according to categorical and continuous variables, respectively. The significance level was set at a p-value of 0.05.
4. Conclusions
To the best of our knowledge, this study, including a large number of samples, provides initial data on the prevalence and ST distribution of Blastocystis sp. in domestic cattle in France, together with factors associated with the carriage of this protozoan. Besides the significant occurrence of Blastocystis sp. highlighted herein, this survey confirmed that ST10 and ST14 represent cattle-adapted Blastocystis sp. STs, and that domestic cows in the north of France seem unlikely to be the source of a zoonotic transmission of the protozoan. Overall, age and water supply of cattle livestock were not correlated with Blastocystis sp. colonization. Conversely, some feed and herds were significantly associated with Blastocystis sp. prevalence. Further large-scale studies should be performed to fully understand the husbandry management practices that could explain the inter-herd variability of Blastocystis sp. prevalence.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/parasitologia2010005/s1, Figure S1. Blastocystis sp. prevalence according to the two sampling seasons. Figure S2. Corn silage, pasture grass and beet pulp effects on Blastocystis sp. prevalence in summer or winter.
Author Contributions
Conceptualization, M.C. and C.A.; funding acquisition, M.C. and C.A.; investigation, N.G., S.D., G.E., M.D., E.V., S.M. (Sophie Merlin) and S.M. (Sophie Martel); writing—original draft, M.C.; writing—review and editing, C.A., S.D., E.V. and M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the STIMulE (Soutien aux Travaux Interdisciplinaires, Multi-établissements et Exploratoires) program AGRIMARK STIR-000030 (2020-01182) funded by the Région Hauts-de-France. This study was also supported by the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Institut Pasteur de Lille, the University of Lille and the CHRU of Lille.
Institutional Review Board Statement
Ethical review and approval were waived for this study, as all samples were obtained by duly authorized technicians and no invasive sampling approach was performed.
Informed Consent Statement
Not applicable.
Data Availability Statement
The nucleotide sequence data generated in this study were deposited in GenBank under accession numbers OM522111 to OM522211.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nemati, S.; Zali, M.R.; Johnson, P.; Mirjalali, H.; Karanis, P. Molecular prevalence and subtype distribution of Blastocystis sp. In Asia and in Australia. J. Water Health 2021, 19, 687–704. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, P.A.; Jaimes, J.E.; Ramírez, J.D. A summary of Blastocystis subtypes in North and South America. Parasites Vectors 2019, 12, 376. [Google Scholar] [CrossRef] [PubMed]
- Rauff-Adedotun, A.A.; Termizi, F.H.M.; Shaari, N.; Lee, I.L. The coexistence of Blastocystis spp. in humans, animals and environmental sources from 2010–2021 in Asia. Biology 2021, 10, 990. [Google Scholar] [CrossRef]
- Tan, K.S.W. New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin. Microbiol. Rev. 2008, 21, 639–665. [Google Scholar] [CrossRef] [Green Version]
- Scanlan, P.D.; Stensvold, C.R.; Rajilić-Stojanović, M.; Heilig, H.G.H.J.; De Vos, W.M.; O’Toole, P.W.; Cotter, P.D. The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol. Ecol. 2014, 90, 326–330. [Google Scholar] [CrossRef] [Green Version]
- Andersen, L.O.; Stensvold, C.R. Blastocystis in Health and Disease: Are We Moving from a Clinical to a Public Health Perspective? J. Clin. Microbiol. 2016, 54, 524–528. [Google Scholar] [CrossRef] [Green Version]
- Stensvold, C.R.; Clark, C.G. Current status of Blastocystis: A personal view. Parasitol. Int. 2016, 65, 763–771. [Google Scholar] [CrossRef]
- Ajjampur, S.S.R.; Tan, K.S.W. Pathogenic mechanisms in Blastocystis spp.—Interpreting results from in vitro and in vivo studies. Parasitol. Int. 2016, 65, 772–779. [Google Scholar] [CrossRef]
- Hublin, J.S.Y.; Maloney, J.G.; Santin, M. Blastocystis in domesticated and wild mammals and birds. Res. Vet. Sci. 2021, 135, 260–282. [Google Scholar] [CrossRef]
- Shams, M.; Shamsi, L.; Sadrebazzaz, A.; Asghari, A.; Badali, R.; Omidian, M.; Hassanipour, S. A systematic review and meta-analysis on the global prevalence and subtypes distribution of Blastocystis sp. infection in cattle: A zoonotic concern. Comp. Immunol. Microbiol. Infect. Dis. 2021, 76, 101650. [Google Scholar] [CrossRef]
- Alfellani, M.A.; Taner-Mulla, D.; Jacob, A.S.; Imeede, C.A.; Yoshikawa, H.; Stensvold, C.R.; Clark, C.G. Genetic Diversity of Blastocystis in Livestock and Zoo Animals. Protist 2013, 164, 497–509. [Google Scholar] [CrossRef] [Green Version]
- Stensvold, C.R.; Alfellani, M.A.; Nørskov-Lauritsen, S.; Prip, K.; Victory, E.L.; Maddox, C.; Nielsen, H.V.; Clark, C.G. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int. J. Parasitol. 2009, 39, 473–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abarca, N.; Santín, M.; Ortega, S.; Maloney, J.G.; George, N.S.; Molokin, A.; Cardona, G.A.; Dashti, A.; Köster, P.C.; Bailo, B.; et al. Molecular detection and characterization of Blastocystis sp. And Enterocytozoon bieneusi in cattle in northern spain. Vet. Sci. 2021, 8, 191. [Google Scholar] [CrossRef] [PubMed]
- Stensvold, C.R.; Clark, C.G. Pre-empting Pandora’s Box: Blastocystis Subtypes Revisited. Trends Parasitol. 2020, 36, 229–232. [Google Scholar] [CrossRef]
- Higuera, A.; Herrera, G.; Jimenez, P.; García-Corredor, D.; Pulido-Medellín, M.; Bulla-Castañeda, D.M.; Pinilla, J.C.; Moreno-Pérez, D.A.; Maloney, J.G.; Santín, M.; et al. Identification of Multiple Blastocystis Subtypes in Domestic Animals from Colombia Using Amplicon-Based Next Generation Sequencing. Front. Vet. Sci. 2021, 8, 732129. [Google Scholar] [CrossRef]
- Maloney, J.G.; Jang, Y.; Molokin, A.; George, N.S.; Santin, M. Wide Genetic Diversity of Blastocystis in White-Tailed Deer (Odocoileus virginianus) from Maryland, USA. Microorganisms 2021, 9, 1343. [Google Scholar] [CrossRef]
- Alfellani, M.A.; Stensvold, C.R.; Vidal-Lapiedra, A.; Onuoha, E.S.U.; Fagbenro-Beyioku, A.F.; Clark, C.G. Variable geographic distribution of Blastocystis subtypes and its potential implications. Acta Trop. 2013, 126, 11–18. [Google Scholar] [CrossRef] [Green Version]
- Ramírez, J.D.; Sánchez, A.; Hernández, C.; Flórez, C.; Bernal, M.C.; Giraldo, J.C.; Reyes, P.; López, M.C.; García, L.; Cooper, P.J.; et al. Geographic distribution of human Blastocystis subtypes in South America. Infect. Genet. Evol. 2016, 41, 32–35. [Google Scholar] [CrossRef]
- Khaled, S.; Gantois, N.; Ly, A.T.; Senghor, S.; Even, G.; Dautel, E.; Dejager, R.; Sawant, M.; Baydoun, M.; Benamrouz-Vanneste, S.; et al. Prevalence and subtype distribution of Blastocystis sp. in senegalese school children. Microorganisms 2020, 8, 1408. [Google Scholar] [CrossRef]
- Khaled, S.; Gantois, N.; Ayoubi, A.; Even, G.; Sawant, M.; El Houmayraa, J.; Nabot, M.; Benamrouz-Vanneste, S.; Chabé, M.; Certad, G.; et al. Blastocystis sp. Prevalence and subtypes distribution amongst syrian refugee communities living in North Lebanon. Microorganisms 2021, 9, 184. [Google Scholar] [CrossRef]
- Osorio-Pulgarin, M.I.; Higuera, A.; Beltran-álzate, J.C.; Sánchez-Jiménez, M.; Ramírez, J.D. Epidemiological and molecular characterization of Blastocystis infection in children attending daycare centers in Medellín, Colombia. Biology 2021, 10, 669. [Google Scholar] [CrossRef] [PubMed]
- Greige, S.; El Safadi, D.; Khaled, S.; Gantois, N.; Baydoun, M.; Chemaly, M.; Benamrouz-Vanneste, S.; Chabé, M.; Osman, M.; Certad, G.; et al. First report on the prevalence and subtype distribution of Blastocystis sp. in dairy cattle in Lebanon and assessment of zoonotic transmission. Acta Trop. 2019, 194, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Masuda, A.; Sumiyoshi, T.; Ohtaki, T.; Matsumoto, J. Prevalence and molecular subtyping of Blastocystis from dairy cattle in Kanagawa, Japan. Parasitol. Int. 2018, 67, 702–705. [Google Scholar] [CrossRef] [PubMed]
- Udonsom, R.; Prasertbun, R.; Mahittikorn, A.; Mori, H.; Changbunjong, T.; Komalamisra, C.; Pintong, A.; Sukthana, Y.; Popruk, S. Blastocystis infection and subtype distribution in humans, cattle, goats, and pigs in central and western Thailand. Infect. Genet. Evol. 2018, 65, 107–111. [Google Scholar] [CrossRef]
- Ramírez, J.D.; Sánchez, L.V.; Bautista, D.C.; Corredor, A.F.; Flórez, A.C.; Stensvold, C.R. Blastocystis subtypes detected in humans and animals from Colombia. Infect. Genet. Evol. 2014, 22, 223–228. [Google Scholar] [CrossRef]
- Hemalatha, C.; Chandrawathani, P.; Kumar, S.G.; Premaalatha, B.; Geethamalar, S.; Rozita, L.M.H.; Haziqah, F.M.T.; Sabapathy, D.; Ramlan, M. The diagnosis of Blastocystis sp. from animals—An emerging zoonosis. Malays. J. Vet. Res. 2014, 5, 15–22. [Google Scholar]
- Sharifi, Y.; Abbasi, F.; Shahabi, S.; Zaraei, A.; Mikaeili, F.; Sarkari, B. Comparative genotyping of Blastocystis infecting cattle and human in the south of Iran. Comp. Immunol. Microbiol. Infect. Dis. 2020, 72, 101529. [Google Scholar] [CrossRef]
- Rostami, M.; Fasihi-Harandi, M.; Shafiei, R.; Aspatwar, A.; Derakhshan, F.K.; Raeghi, S. Genetic diversity analysis of Blastocystis subtypes and their distribution among the domestic animals and pigeons in northwest of Iran. Infect. Genet. Evol. 2020, 86, 104591. [Google Scholar] [CrossRef]
- Lee, H.; Lee, S.H.; Seo, M.G.; Kim, H.Y.; Kim, J.W.; Lee, Y.R.; Kim, J.H.; Kwon, O.D.; Kwak, D. Occurrence and genetic diversity of Blastocystis in Korean cattle. Vet. Parasitol. 2018, 258, 70–73. [Google Scholar] [CrossRef]
- Zhu, W.; Tao, W.; Gong, B.; Yang, H.; Li, Y.; Song, M.; Lu, Y.; Li, W. First report of Blastocystis infections in cattle in China. Vet. Parasitol. 2017, 246, 38–42. [Google Scholar] [CrossRef]
- Wang, J.; Gong, B.; Yang, F.; Zhang, W.; Zheng, Y.; Liu, A. Subtype distribution and genetic characterizations of Blastocystis in pigs, cattle, sheep and goats in northeastern China’s Heilongjiang Province. Infect. Genet. Evol. 2018, 57, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Song, J.K.; Yang, F.; Zou, M.; Wang, P.X.; Wang, D.; Zhang, H.J.; Zhao, G.H.; Lin, Q. First genotyping of Blastocystis in yaks from Qinghai Province, northwestern China. Parasites Vectors 2019, 12, 171. [Google Scholar] [CrossRef] [PubMed]
- Badparva, E.; Sadraee, J.; Kheirandish, F. Genetic diversity of Blastocystis isolated from cattle in Khorramabad, Iran. Jundishapur J. Microbiol. 2015, 8, 4–7. [Google Scholar] [CrossRef] [Green Version]
- Aynur, Z.E.; Güçlü, Ö.; Yildiz, I.; Aynur, H.; Hertabaklar, H.; Bozdogan, B.; Ertug, S. Molecular characterization of Blastocystis in cattle in Turkey. Parasitol. Res. 2019, 118, 1055–1059. [Google Scholar] [CrossRef] [PubMed]
- Moura, R.G.F.; de Oliveira-Silva, M.B.; Pedrosa, A.L.; Nascentes, G.A.N.; Cabrine-Santos, M. Occurrence of Blastocystis spp. in domestic animals in triângulo mineiro area of Brazil. Rev. Soc. Bras. Med. Trop. 2018, 51, 240–243. [Google Scholar] [CrossRef] [Green Version]
- AbuOdeh, R.; Ezzedine, S.; Madkour, M.; Stensvold, C.R.; Samie, A.; Nasrallah, G.; AlAbsi, E.; ElBakri, A. Molecular Subtyping of Blastocystis from Diverse Animals in the United Arab Emirates. Protist 2019, 170, 125679. [Google Scholar] [CrossRef]
- Fayer, R.; Santin, M.; MacArisin, D. Detection of concurrent infection of dairy cattle with Blastocystis, Cryptosporidium, Giardia, and Enterocytozoon by molecular and microscopic methods. Parasitol. Res. 2012, 111, 1349–1355. [Google Scholar] [CrossRef]
- Maloney, J.G.; Lombard, J.E.; Urie, N.J.; Shivley, C.B.; Santin, M. Zoonotic and genetically diverse subtypes of Blastocystis in US pre-weaned dairy heifer calves. Parasitol. Res. 2019, 118, 575–582. [Google Scholar] [CrossRef]
- Kamaruddin, S.K.; Mat Yusof, A.; Mohammad, M. Prevalence and subtype distribution of Blastocystis sp. in cattle from Pahang, Malaysia. Trop. Biomed. 2020, 37, 127–141. [Google Scholar]
- Abdullah, D. Molecular Epidemiology, Risk Factors of Zoonotic Enteric Protozoa and Genetic Diversity of Blastocystis Infecting Cattle in Peninsular Malaysia. Ph.D. Thesis, Universiti Putra Malysia, Seri Kembangan, Malaysia, 2017. [Google Scholar]
- Santín, M.; Gómez-Muñoz, M.T.; Solano-Aguilar, G.; Fayer, R. Development of a new PCR protocol to detect and subtype Blastocystis spp. from humans and animals. Parasitol. Res. 2011, 109, 205–212. [Google Scholar] [CrossRef]
- Maloney, J.G.; Santin, M. Mind the gap: New full-length sequences of Blastocystis subtypes generated via oxford nanopore minion sequencing allow for comparisons between full-length and partial sequences of the small subunit of the ribosomal rna gene. Microorganisms 2021, 9, 997. [Google Scholar] [CrossRef] [PubMed]
- El Safadi, D.; Cian, A.; Nourrisson, C.; Pereira, B.; Morelle, C.; Bastien, P.; Bellanger, A.P.; Botterel, F.; Candolfi, E.; Desoubeaux, G.; et al. Prevalence, risk factors for infection and subtype distribution of the intestinal parasite Blastocystis sp. from a large-scale multi-center study in France. BMC Infect. Dis. 2016, 16, 451. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Giuberti, G.; Frisvad, J.C.; Bertuzzi, T.; Nielsen, K.F. Review on mycotoxin issues in ruminants: Occurrence in forages, effects of mycotoxin ingestion on health status and animal performance and practical strategies to counteract their negative effects. Toxins 2015, 7, 3057–3111. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.; Messerschmidt, U. Review: Sugar beets as a substitute for grain for lactating dairy cattle. J. Anim. Sci. Biotechnol. 2017, 8, 25. [Google Scholar] [CrossRef] [Green Version]
- Poirier, P.; Wawrzyniak, I.; Albert, A.; El Alaoui, H.; Delbac, F.; Livrelli, V. Development and evaluation of a real-time PCR assay for detection and quantification of Blastocystis parasites in human stool samples: Prospective study of patients with hematological malignancies. J. Clin. Microbiol. 2011, 49, 975–983. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).