Abstract
Rice is sensitive to salinity at both the seedling and reproductive stages, and it reduces the grain yield from 50 to 100%. In this study, 27 SNP markers significantly associated with 15 QTLs were identified. Three major QTLs were associated with shoot length (Sal_SL 7.1), shoot sodium content (Sal_Na 1.1) and shoot magnesium content (Sal_Mg 2.1). Five QTLs for root length (Sal_RL 1.1, Sal_RL 3.1, Sal_RL 6.1 Sal_RL 8.1 and Sal_RL 12.1), shoot K+/Na+ homeostasis (Sal_K/Na 1.1, Sal_K/Na 4.1, Sal_K/Na 5.1, Sal_K/Na 7.1 and Sal_K/Na 10.1) and single QTLs for shoot potassium (Sal_K 6.1) and calcium content (Sal_Ca 5.1) were also detected. QTL Sal_K/Na 1.1 was found responsible for the ionic ratio associated with the Saltol region, and Sal_K/Na 10.1 was associated with the gene OSCA1;4, which is a hyperosmolality-gated calcium-permeable channel that acts as an osmosensor under salt stress condition. A candidate gene haplotype analysis revealed ten significant genes, LOC_Os06g03940, LOC_Os10g42820, LOC_Os07g36230, LOC_Os02g06410, LOC_Os06g48610, LOC_Os12g12950, LOC_Os03g12050, LOC_Os08g02690, LOC_Os07g47560, and LOC_Os05g08840, responsible for abiotic stress tolerance. The identified potential candidate genes can be used for functional characterization to understand the complex mechanism of salinity tolerance in rice.
1. Introduction
Rice is one of the most important cereal grains, which feed more than half of the world population. However, it is under constant pressure due to biotic and abiotic stresses, which seriously affect its production. Among these stresses, soil salinity is one major constraint, which hinders rice production around the globe. The Global Map of Salt-Affected Soils has estimated more than 833 million hectares of land as salt affected [1]. Salinity affects 50% of the world’s irrigated land, and it is estimated to be rising at a much faster pace [2]. Under saline stress, rice seedlings accumulated high Na+, which interferes with the cellular machinery of a plant cell. Furthermore, it disturbs the water potential of the plant, which leads to low water uptake [3,4]. Seedling stage salinity tolerance is a crucial stage in rice’s life cycle, as it helps the plant to survive the early adverse affects of salinity. Moreover, seedling stage salinity facilitates early plant establishment, which ensures good vegetative growth at the reproductive stage [5]. To identify the genes responsible for salinity tolerance, several QTL mapping studies have been conducted in the past [6,7]. Major Saltol, QTL mapped on chromosome 1, was successfully transferred to a wide array of high yielding genotypes through a series of marker-assisted breeding programs [8,9,10]. In the present study, we employed 27,041 filtered SNPs for identification of novel QTLs for two morphological and five physiological traits using MAGIC rice lines developed from the inter-crossing of eight advanced founder lines.
2. Material and Methods
The plant material consisted of 391 rice MAGIC lines developed from the inter-crossing of eight founder lines at the International Rice Research Institute in the Philippines [11]. These lines were evaluated in hydroponic condition at ICAR-CSSRI, Karnal, using Yoshida nutrient solution. The EC of the solution was raised to ~10 dS/m after 14 DAS, and the anticipated level of salinity was sustained for the next 14 days. Genotyping of the founder and magic lines was done via genotyping by sequencing (GBS) using the Illumina HiSeq method. To identify QTLs for morph-physiological traits, the R-based GAPIT program was employed using the Bayesian-information and Linkage-disequilibrium Iteratively Nested Keyway (BLINK) model. Finally, the marker-associated SNPs with an LOD threshold > 4 were considered to be significant SNPs for traits of interest. Candidate gene haplotype analysis for trait-associated QTLs was conducted using Candi-HAP V2, and violin plots were visualized in ggplot2 program in R.
3. Results and Discussion
QTL mapping for salinity tolerance is an effective way to locate robust genomic regions controlling complex traits. In the present study,15 QTLs were identified for salt tolerance with LOD >4 for 7morph-physiological traits (Table 1).

Table 1.
Associated QTLs with SNP position and probable candidate genes for salinity tolerance in MAGIC population.
A maximum of five QTLs were recorded for root length (Sal_RL 1.1, Sal_RL 3.1, Sal_RL 6.1 Sal_RL 8.1 and Sal_RL 12.1) and shoot K+/Na+ homeostasis (Sal_K/Na 1.1, Sal_K/Na 4.1, Sal_K/Na 5.1, Sal_K/Na 7.1 and Sal_K/Na 10.1). The major QTLs, Sal_Mg 2.1 for Mg2+ content, Sal_Na 1.1 for Na+ content and Sal_SL 7.1 for shoot length, were identified; these explained about 23.74%, 14.14% and 9.43% of the phenotypic variation, respectively. A peak marker at genomic location 11703845 of newly identified QTL Sal_K/Na 1.1was located in previously reported Saltol QTL region [8]. Around 15 genes were found to overlap marker-associated SNPs with diverse functions, such as transcription factor, kinase activity membrane protein with regulatory role in enzyme activity and some other useful functions. The other QTL Sal_Na 1.1 was also found near the Saltol QTL region; the peak of this QTL, lying in the genomic region of LOC_Os01g02750, belongs to the LRR-kinase family protein, and it is a putative candidate gene for cold tolerance [12].Candidate gene haplotype analysis revealed ten significant genes, LOC_Os06g03940, LOC_Os10g42820, LOC_Os07g36230, LOC_Os02g06410, LOC_Os06g48610, LOC_Os12g12950, LOC_Os03g12050, LOC_Os08g02690, LOC_Os07g47560 andLOC_Os05g08840, as having a key role in abiotic stress tolerance. From these, the genes associated with physiological traits were presented in Figure 1. The peak SNP marker of Sal_Mg 2.1 lay in the genomic region of the CBS domain-containing membrane protein (CDCPs) gene (LOC_Os02g06410 (OsCBSX9)) and has a significant role in regulation of the thioredoxin system and stress response/tolerance in rice [13]. The peak marker of Sal_RL 6.1 contains the CCT family protein gene LOC_Os06g48610, which regulates salt tolerance through the abscisic acid-dependent signalling pathway with pleiotropic effects on morphological traits [14]. The peak SNP of Sal_RL_8.1 is associated with the MA3 domain-containing protein coding gene (LOC_Os08g02690). In Arabidopsis, the MA3 domain-containing protein (ECIP1) plays a key role in regulation of salt stress response by interacting with the central membrane protein of ethylene signalling (EIN2) [15].A single SNP peak for the K+ content was associated with the gene LOC_Os06g03940, which encodes for the defence response spastin protein in rice under stress [16]. A group of SNPs associated with the shoot Ca2+ content lay in the genomic region of the Hsp70 domain-containing gene (LOC_Os05g08840) play important role in housekeeping functions under heat stress [17,18]. A significant candidate gene haplotype for LOC_Os10g42820, found to be associated with K+/Na+, belongs to the DUF221 domain-containing gene family [19]. Early-responsiveness to the dehydration protein gene LOC_Os10g42820 (OSCA1;4), responsible for salt and drought tolerance, was detected on QTL Sal_K/Na 10.1. The gene OSCA1;4 is a hyperosmolality-gated calcium-permeable channel that activates an inward current after receiving an osmotic signal exerted by salt stress [20]. Further, a number of uncharacterized genes exhibiting significant haplotypes associated with different traits were also detected in our study. The identified QTLs and respective candidate genes can be further investigated to confirm their role in salt tolerance in rice.
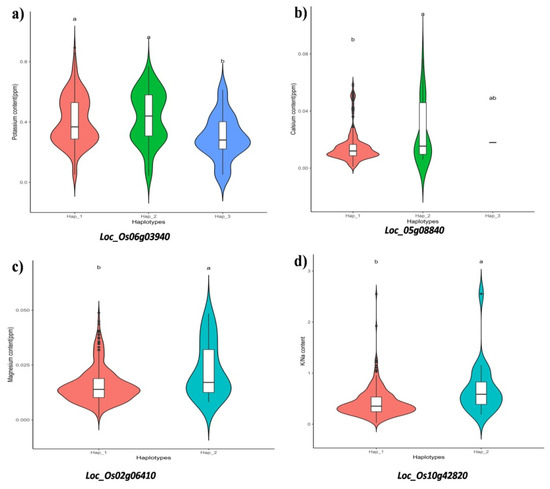
Figure 1.
Violin plots indicating the significant haplotypes of candidate genes for (a) K+ content, (b) Ca2+ content, (c) Mg2+ content and (d) K+/Na+(x-axis=haplotypes; y-axis= trait value). Each box plot with in the violin plot represents minimum, lower quartile, median, upper quartile, and maximum values. Different letters above the violin plots indicate statistically significant differences for the respective haplotypes, at a significance level of p < 0.05 (Duncan test).
4. Conclusions
In the present study, we identify three major QTLs associated with shoot length, shoot sodium content and shoot magnesium content. The QTL for Sal_K/Na 1.1 for the ionic ratio, lying within the Saltol region, and the osmosensor gene OSCA1;4, responsible for hyperosmolality-gated calcium-permeable channel, were detected. In our study, we found significant haplotypes for ten potential candidate genes that play key role in abiotic stress tolerance. The identified SNP markers linked to salt tolerant QTLs and candidate genes with significant allelic variation are potential prospects for marker-assisted breeding for salinity tolerance in rice.
Author Contributions
Conceptualization, R.K.S. and S.L.K.; methodology, S.L.K. and R.K.S.; software, B.M.L. and A.S.W.; validation, S.L.K., P.C.S. and R.K.S.; formal analysis, B.M.L. and A.S.W.; investigation, S.R. and S.Y.; resources, S.L.K. and P.C.S.; data curation, B.M.L., S.Y.; writing—original draft preparation, A.S.W. and S.Y.; writing—review and editing, B.M.L. and S.R.; visualization, B.M.L.; supervision, S.L.K.; project administration, S.L.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by ICAR-New Delhi and IRRI-Phillipines.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are thankful to the Director of ICAR-CSSRI for support to carry out the proposed research work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- FAO Director-General. Global map of salt-affected soils. In Proceedings of the World Map of Salt-Affected Soils, Rome, Italy, 20–22 October 2021. [Google Scholar]
- Singh, R.K.; Kota, S.; Flowers, T.J. Salt tolerance in rice: Seedling and reproductive stage QTL mapping come of age. Theor. Appl. Genet. 2021, 134, 3495–3533. [Google Scholar] [CrossRef] [PubMed]
- Warraich, A.S.; Krishnamurthy, S.L.; Sooch, B.S.; Vinaykumar, N.M.; Dushyanthkumar, B.M.; Bose, J.; Sharma, P.C. Rice GWAS reveals key genomic regions essential for salinity tolerance at reproductive stage. Acta Physiol. Plant. 2020, 42, 134. [Google Scholar] [CrossRef]
- Reddy, N.B.L.; Kim, B.K.; Yoon, I.S.; Kim, K.H.; Kwon, T.R. Salt Tolerance in Rice: Focus on Mechanisms and Approaches. Rice Sci. 2017, 24, 123–144. [Google Scholar] [CrossRef]
- Babu, N.N.; Krishnan, S.G.; Vinod, K.K.; Krishnamurthy, S.L.; Singh, V.K.; Singh, M.P.; Singh, R.; Ellur, R.K.; Rai, V.; Bollinedi, H.; et al. Marker Aided Incorporation of Saltol, a Major QTL Associated with Seedling Stage Salt Tolerance, into Oryza sativa ‘Pusa Basmati 1121’. Front. Plant Sci. 2017, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Prakash, N.R.; Lokeshkumar, B.M.; Rathore, S.; Warraich, A.S.; Yadav, S.; Vinaykumar, N.M.; Dushyanthkumar, B.M.; Krishnamurthy, S.L.; Sharma, P.C. Meta-analysis and validation of genomic loci governing seedling and reproductive stage salinity tolerance in rice. Physiol. Plant. 2022, 174, e13629. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, A.; Rohilla, M.; Bisht, D.; Krishnamurthy, S.L.; Barman, M.; Sarma, R.; Sharma, T.; Mondal, T. Identification and mapping of quantitative trait loci (QTL) and epistatic QTL for salinity tolerance at seedling stage in traditional aromatic short grain rice landrace Kolajoha (Oryza sativa L.) of Assam. India. Euphytica 2020, 216, 75. [Google Scholar] [CrossRef]
- Gregorio, G.B. Tagging Salinity Tolerance Genes in Rice Using Amplified Fragment Length Polymorphism (AFLP). Ph.D. Thesis, University of the Philippines Los Banõs, Laguna, Philippines, 1997. [Google Scholar]
- Krishnamurthy, S.L.; Pundir, P.; Warraich, A.S.; Rathor, S.; Lokeshkumar, B.M.; Singh, N.K.; Sharma, P.C. Introgressed Saltol QTL Lines Improves the Salinity Tolerance in Rice at Seedling Stage. Front. Plant Sci. 2020, 11, 833. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.K.; Kumar, A.; Grover, N.; Ellur, R.K.; Krishnan, S.G.; Bollinedi, H.; Bhowmick, P.K.; Vinod, K.K.; Nagarajan, M.; Krishnamurthy, S.L.; et al. Marker aided introgression of ‘Saltol’, a major QTL for seedling stage salinity tolerance into an elite Basmati rice variety ‘Pusa Basmati 1509’. Sci. Rep. 2020, 10, 13877. [Google Scholar] [CrossRef] [PubMed]
- Bandillo, N.; Raghavan, C.; Muyco, P.A.; Sevilla, M.A.; Lobina, I.T.; Dilla-Ermita, C.J.; Tung, C.W.; McCouch, S.; Thomson, M.; Mauleon, R.; et al. Multi-parent advanced generation inter-cross (MAGIC) populations in rice: Progress and potential for genetics research and breeding. Rice 2013, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Chai, C.; Shankar, R.; Jain, M. Genome-wide discovery of DNA polymorphisms by whole genome sequencing differentiates weedy and cultivated rice. Sci. Rep. 2018, 8, 14218. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Subba, A.; Kaur, C.; Ariyadasa, T.U.; Sharan, A.; Pareek, A.; Soproy, S.K.; Singla-Pareek, S.L. OsCBSCBSPB4 is a two cystathionine-β-synthase domain-containing protein from rice that functions in abiotic stress tolerance. Curr. Genom. 2017, 19, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, M. CCT family genes in cereal crops: A current overview. Crop J. 2017, 5, 449–458. [Google Scholar] [CrossRef]
- Lei, G.; Shen, M.; Li, Z.G.; Zhang, B.; Duan, K.X.; Wang, N.; Cao, Y.R.; Zhang, W.K.; Ma, B.; Ling, H.Q.; et al. EIN2 regulates salt stress response and interacts with a MA3 domain-containing protein ECIP1 in Arabidopsis. Plant Cell Environ. 2011, 34, 1678–1692. [Google Scholar] [CrossRef] [PubMed]
- Fekih, R.; Tamiru, M.; Kanzaki, H.; Abe, A.; Yoshida, K.; Kanzaki, E.; Saitoh, H.; Takagi, H.; Natsume, S.; Undan, J.R.; et al. The rice (Oryza sativa L.) lesion mimic resembling, which encodes an AAA-type ATPase, is implicated in defence response. Mol. Genet. Genom. 2015, 290, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, M.; Gao, P.; Chen, H.; Zheng, Y.; Yang, C.; Yang, Z.; Sun, Q. Expression of heat shock protein (HSP) genes and antioxidant enzyme genes in hybrid rice II YOU 838 during heat stress. Aust. J. Crop Sci. 2021, 4, 38–43. [Google Scholar] [CrossRef]
- Ganie, S.A.; Pani, D.R.; Mondal, T.K. Genome-wide analysis of DUF221 domain-containing gene family in Oryza species and identification of its salinity stress-responsive members in rice. PLoS ONE 2017, 12, e0182469. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Wen, Z.; Han, Y.; Wang, F.; Xi, C.; Liu, J.; Gao, P.; Zhao, H.; Wang, Y.; Wang, Y.; et al. Heterogeneous expression of plasma-membrane-localised OsOSCA1.4 complements osmotic sensing based on hyperosmolality and salt stress in Arabidopsis osca1 mutant. Cell Calcium 2020, 91, 102261. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).