Abstract
Early detection of arrhythmias is very important. Recently, wearable devices are being used to monitor the patient’s heartbeat to detect an arrhythmia. However, there are not satisfactory algorithms for real-time monitoring of arrhythmias in a wearable device. In this work, a novel fast and simple arrhythmia detection algorithm based on YOLO is proposed. The algorithm can detect each heartbeat on long-duration electrocardiogram (ECG) signals without R-peak detection and can classify an arrhythmia simultaneously. The model replaces the 2D Convolutional Neural networks (CNN) with a 1D CNN and the bounding box with a bounding window to utilize raw ECG signals. Results demonstrate that the proposed algorithm has high performance in speed and mean average precisionin detecting an arrhythmia. Furthermore, the bounding window can predict different window lengths on different types of arrhythmia. Therefore, the model can choose an optimal heartbeat window length for arrhythmia classification. Since the proposed model is a compact 1D CNN model based on YOLO, it can be used in a wearable device and embedded system.
1. Introduction
Arrhythmia refers to a group of conditions that cause the heart to beat rapid or irregular. They are detected by an electrocardiogram, which measures the state of the heart’s electrical activity. In general, the process of arrhythmia detection is performed in three steps: beat segmentation, feature extraction and classification. Conventional heartbeat segmentation is the process of R-peak detection in long-duration ECG signals. Differentiation [1] and a digital filter [2] are used in R-peak detection. The R-peak detection algorithm is especially emphasized in bio-signal wearable devices for its computational cost [3].
Feature extraction is the following step in beat segmentation. In machine learning, wavelet transform (WT) [4] and independent component analysis (ICA) [5] are used. Feature extraction reduces the dimensions of beat feature and can greatly improve classification performance. Classification is the process of classifying a beat to each class based on its feature. However, most algorithms are handcrafted features, and such a pipeline processing method involves the problem of loading data into memory in each step and the problem of real-time execution due to operation processing.
A deep learning-based method overcame this problem by end-to-end learning which features extraction and classification fused into one model. Jun [6] used a 2D convolutional neural network (CNN) model which uses ECG images as input. Ji [7] also proposed the Faster R-CNN which converts the ECG signal into an ECG image. They had a 99.21% average accuracy. Kiranyaz [8] used a 1D CNN for classification of five types of arrhythmia. They made a real-time classification model using three consecutive 1D CNN layers. However, deep learning-based methods for classifying arrhythmias still need a beat segmentation process.
In this paper, we proposed a 1D YOLO model which can detect arrhythmia without an R peak in long ECG sequences. The model can detect more than one type of arrhythmia in a long sequence. We modified the YOLO model into a 1D YOLO model and the bounding box was replaced with a bounding window. As a result, the model predicts only one coordinate for the bounding window.
2. Related Work
YOLO (You Only Look Once)
YOLO is an effective real-time object detection deep learning model [10]. The model applies a single neural network to the input image and divides the image into an S × S grid cell. The grid cell approach is faster than using selective search and edge boxes in a region proposal. The Fast R-CNN use selective search and the Faster R-CNN adapts edge boxes for region proposal, but these kinds of region proposal are not good enough to use in real-time systems. Therefore, YOLO is widely used in real-time object detection.
3. Materials and Methods
3.1. MIT-BIH Database
In this study, the MIT-BIH arrhythmia database [9] is used for training and testing. The heartbeats are classified into 5 types: normal, right bundle branch block (RBBB), left bundle branch block (LBBB), ventricular ectopic beat (VEB), and supraventricular ectopic beat (SVEB). Since this is imbalanced, we down-sampled the normal type to match the overall number of other types of arrhythmia.
The MIT-BIH arrhythmia database contains 48 recordings and each recording is about 30 min long. We split 30-min ECG data into 10-s ECG segments. We did not use segments which have only normal-type heartbeats. Therefore, each segment has more than one type of arrhythmia.
3.2. Proposed 1D YOLO Model
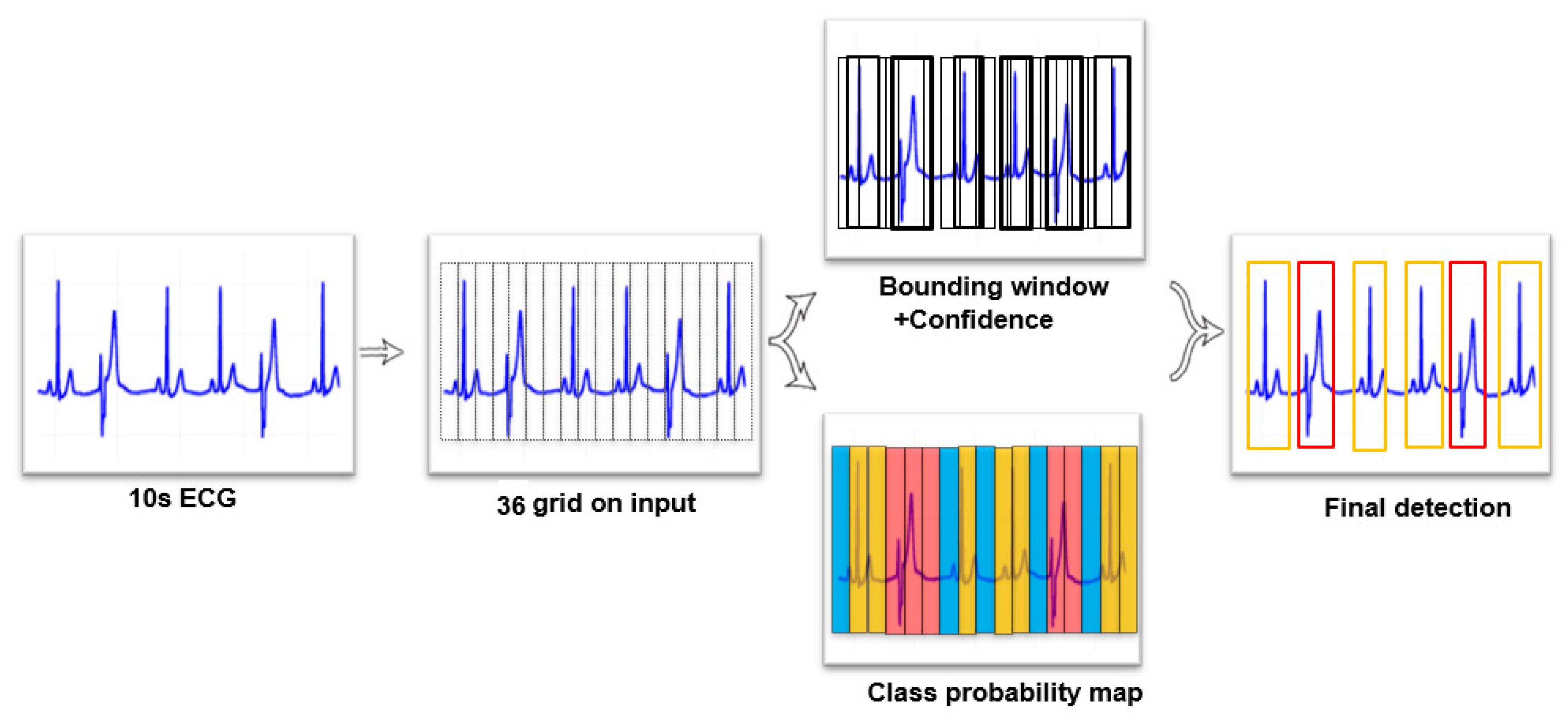
An overview of the proposed 1D YOLO model is shown in Figure 1. We divided 36 grids into ECG segments and each grid predicts 2 bounding windows. Therefore, 72 bounding windows were predicted in each 10-s ECG segment, with a confidence score and position. Most of the bounding windows had a low confidence score, which means that the bounding window was detected as a background. We wanted to focus on the bounding windows which have ECG beats. To remove bounding windows which had low scores, the bounding windows were sorted according to their confidence scores—from high to low. We set a threshold, and windows with a confidence score under this threshold were deleted.
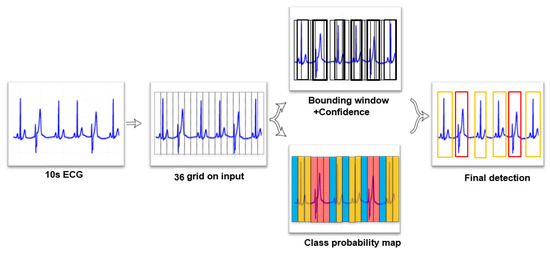
Figure 1.
Proposed YOLO model of detection.
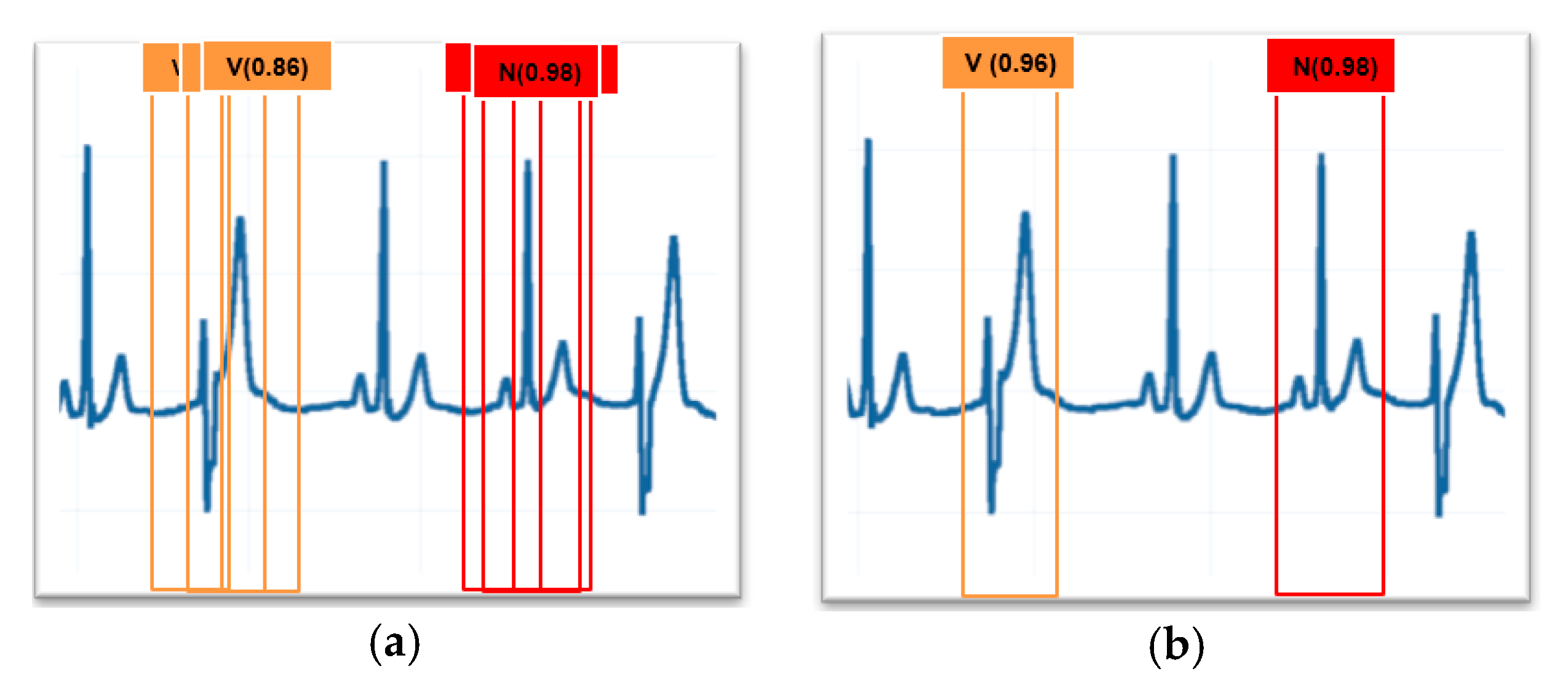
Post-processing was needed to remove several detections of the same heartbeat. Non-maximum suppression was used to remove multiple bounding windows showing the same heartbeat, and the bounding windows which had a high confidence score were left. The results are shown in Figure 2.
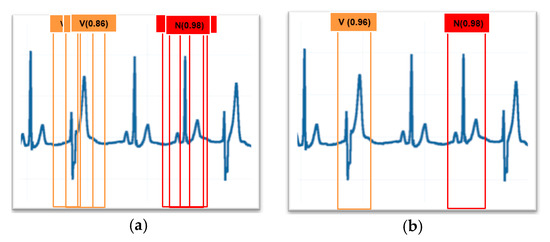
Figure 2.
(a) Before non-maximum suppression; (b) after non-maximum suppression.
4. Result and Discussion
The proposed YOLO model was examined using the MIT-BIH database. We determined five types of arrhythmia and the number of beat in each types of arrhythmia is in Table 1. The precision and recall of each type of arrhythmia is shown in Table 2. Table 3 shows that the proposed model was able to correctly detect the five types of arrhythmia at an average precision of 0.97, an average recall of 0.95 and a 0.96 F1 score.However, SVEB has a low recall value. This may be due to the smaller dataset of SVEB compared to other types of arrhythmia. In addition, SVEB is characterized not only by morphological features but also a beat-to-beat feature. This made the model struggle to detect SVEB. The entire detection time of the proposed 1D YOLO model on 10-s ECG segments is 0.03 s. This makes its real-time application realizable.

Table 1.
Number of beats in each arrhythmia type.

Table 2.
Precision and recall of each arrhythmia type.

Table 3.
Overall performance of proposed model.
5. Conclusions
In this study, we proposed a YOLO-based arrhythmia classification model that can detect each heartbeat and classify it as an arrhythmia in 10-s ECG segments without a beat extraction step. By processing the beat extraction step, feature extraction, and classification step into a single model, the complexity of computation was reduced and presented as a model usable for embedded application. We only classified five types of arrhythmia but the model can be used to detect various types of arrhythmia in the future.
Author Contributions
Conceptualization, W.H.H. and Y.C.J.; methodology, W.H.H.; software, W.H.H., C.H.J. and D.H.H.; validation, W.H.H., C.H.J. and Y.C.J.; formal analysis, W.H.H. and C.H.J.; investigation, W.H.H.; resources, W.H.H.; data curation, W.H.H.; writing—original draft preparation, W.H.H.; writing—review and editing, W.H.H. and Y.C.J.; visualization, W.H.H.; supervision, Y.C.J.; project administration, Y.C.J.; funding acquisition, Y.C.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
This work was supported by the Korean Ministry of Science and ICT and IITP under project contract No. 2020-0-01970-001 (Development of Smart Patch and Multimodal Bio signal Analysis Algorithm for Self-monitoring of Infectious Respiratory Diseases) and partly supported by the Korean Ministry of Trade, Industry, Energy/KEIT under project contract No. 20009959 (Development of stress care device and service technology using tES-based multi-sensory stimulation with clinical effectiveness).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Afonso, V.X.; Tompkins, W.J.; Nguyen, T.Q.; Luo, S. ECG beat detection using filter banks. IEEE Trans. Biomed. Eng. 1999, 46, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Keselbrener, L.; Keselbrener, M.; Akselrod, S. Nonlinear high pass filter for R-wave detection in ECG signal. Med. Eng. Phys. 1997, 19, 481–484. [Google Scholar] [CrossRef]
- Arefin, M.R.; Tavakolian, K.; Fazel-Rezai, R. QRS complex detection in ECG signal for wearable devices. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milano, Italy, 25–29 August 2015; pp. 5940–5943. [Google Scholar]
- Mahmoodabadi, S.Z.; Ahmadian, A.; Abolhasani, M.D. ECG feature extraction using Daubechies wavelets. In Proceedings of the Fifth IASTED International conference on Visualization, Imaging and Image Processing, Benidorm, Spain, 7–9 September 2005; pp. 343–348. [Google Scholar]
- Jiang, X.; Zhang, L.; Zhao, Q.; Albayrak, S. ECG arrhythmias recognition system based on independent component analysis feature extraction. In Proceedings of the TENCON 2006–2006 IEEE Region 10 Conference, Hong Kong, China, 14–17 November 2006; pp. 1–4. [Google Scholar]
- Jun, T.J.; Nguyen, H.M.; Kang, D.; Kim, D.; Kim, D.; Kim, Y.H. ECG arrhythmia classification using a 2-D convolutional neural network. arXiv 2018, arXiv:1804.06812. [Google Scholar]
- Ji, Y.; Zhang, S.; Xiao, W. Electrocardiogram classification based on faster regions with convolutional neural network. Sensors 2019, 19, 2558. [Google Scholar] [CrossRef] [PubMed]
- Kiranyaz, S.; Ince, T.; Gabbouj, M. Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE Trans. Biomed. Eng. 2015, 63, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).