Abstract
A sensitive LC-MS/MS method was developed and validated for simultaneous determination of fungicides from various chemical classes, including strobilurins, triazoles, benzimidazoles, carbamates, and others. Target analytes included azoxystrobin, boscalid, carbendazim, cyazofamid, prochloraz, and tebuconazole. Sample preparation used optimized QuEChERS extraction with d-SPE cleanup to minimize matrix interferences. Chromatographic separation employed a C18 column with gradient elution, while detection used ESI in positive/negative modes with sMRM. Validation (SANTE/11312/2021) showed the deviation of the back-calculated concentrations of the calibration standards from the true concentrations were less than ±20%, recoveries 70–120%, RSD < 20%, and LOQs ≤ 10 µg/kg. The method supports routine monitoring of fungicide residues for regulatory compliance.
1. Introduction
A robust and sensitive multiresidue analytical method based on liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) was developed and validated for the simultaneous determination of fungicides from diverse chemical classes, including strobilurins, triazoles, imidazoles, benzimidazoles, carbamates, dicarboximides, pyrimidines, and anilinopyrimidines. Target analytes included commonly used active substances such as azoxystrobin, boscalid, carbendazim, cyazofamid, prochloraz, and tebuconazole. These fungicides are widely applied in agriculture for fungal disease control but require strict monitoring due to health and environmental concerns.
The QuEChERS procedure has become the standard approach for sample preparation in many laboratories because of productivity improvements [1]. The efficiency of the dSPE clean-up is limited, so high concentrations of matrix-coextractives can remain in the final extract and cause system contamination.
Sample preparation utilized an optimized QuEChERS [2]. Extraction protocol combined with dispersive solid-phase extraction (d-SPE) cleanup to reduce matrix interferences and ensure high analyte recovery. Chromatographic separation was achieved on a reversed-phase C18 column using gradient elution of water and methanol with 0.1% formic acid.
Method validation followed the SANTE/11312/2021 guidelines ([3]; SANTE/11813/2017, Guidance Document on Analytical Quality Control and Method Validation Procedures for Pesticide Residue Analysis in Food and Feed). Matrix effects were evaluated and compensated using matrix-matched calibration.
2. Materials and Methods
2.1. Apparatus and Reagents
The liquid chromatography-tandem mass spectrometry (LC-MS/MS) method was developed using a SCIEX Triple Quadrupole 6500+ system (SCIEX, Framingham, MA, USA) with the Turbo VTM ion Source and an electrospray ionization (ESI) with an integrated ExionLC™ system (SCIEX, Framingham, MA, USA). Data acquisition was carried out using Analyst software (SCIEX, version 1.7.2). Detection was performed using electrospray ionization (ESI) in both positive and negative ion modes with scheduled multiple reaction monitoring (sMRM). For details of LC and MS/MS condition, see Section 3.3, Table 1, Table 2 and Table 3.
Certified standards of all pesticides were purchased from HPC Standards GmbH or Labor Dr. Ehrenstorfer-Schäfers.
Methanol used for chromatography was obtained from VWR (Bratislava, Slovakia). Ultra-pure water was produced using a Milli-Q system (Millipore, Bratislava, Slovakia).
The Nylon membrane filters of pore size 0.22 µm, diameter 25 mm (Albet, Æ 25 mm, lot. 17845000566) were used for the filtration of the final extract.
2.2. Samples and Sample Pretreatment
Homogenized samples (5 g of powdered infant formula or 10 g of liquid ready-to-feed infant formula) were extracted with 10 mL of acetonitrile. Subsequently, 6.5 g of MgSO4:NaCl:Na3Cit·2H2O:Na2HCit·1.5H2O (8:2:2:1) was added. The mixture was shaken, centrifuged, and frozen. Clean-up was performed by dispersive solid-phase extraction (dSPE), in which 1.05 g of MgSO4:PSA (6:1) was added to 6 mL of the extract for non-pigmented samples. Finally, the shaken and centrifuged extract was filtered through nylon filters and acidified with 10 μL of 5% formic acid per 1 mL of extract.
2.3. Preparation of Blank, Matrix-Matched Calibration Standards and Solution for Recovery
Blank samples and samples spiked for recovery determination purposes were prepared in the same way as samples (QuEChERS) and subsequently filtered through a nylon filter. Recoveries were obtained for concentrations corresponding to the limits and multiples of limits (the ratios of analyte concentrations correspond to the MRL ratios—the case of a multi-component residue is also taken into account). Matrix-matched calibration standards were prepared at five levels (see Figure 1).
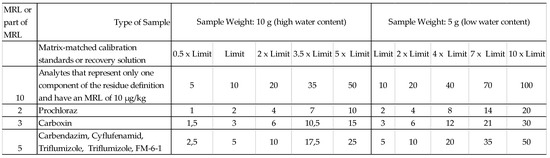
Figure 1.
Concentration (μg/kg) of respective analytes.
3. Results
In this proceeding paper, concern was placed on the determination of fungicides in various matrices such as fruit, vegetables, dairy, meat, cereal, and their combinations in international comparative tests, official control, and European monitoring.
Validation (SANTE/11312/2021) showed that the deviation of the back-calculated concentrations of the calibration standards from the true concentrations was less than ±20%, recoveries 70–120%, RSD < 20%, and LOQs ≤ 10 µg/kg (see Figure 2 and Figure 3).
3.1. LOQ and Analyte Limits
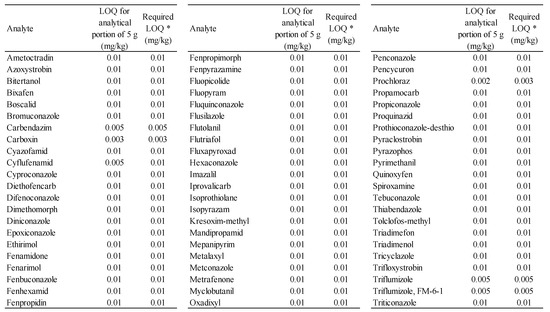
Figure 2.
LOQs and analyte limits. * For an analyte that also forms part of the residue definition (such a pesticide residue is defined as an individual chemical compound), this limit represents the MRL in the original (unreconstituted) matrix. For an analyte that forms part of the residue definition (such a pesticide residue is defined as the sum of n compounds), this limit represents the nth part of the MRL in the original (unreconstituted) matrix.
3.2. Evaluation of Linearity, Selectivity, Specificity, and Accuracy
In Figure 3, the linearity evaluation confirms the LOQ determined within the initial validation or previous verification.
For simplicity, a common LOQ is determined for a given analyte for all matrices from the lowest concentration level that was successfully validated (within the initial validation or previous verification). The LOQ for an analytical dose of 10 g (matrix with a higher water content) is twice the LOQ for an analytical dose of 5 g (matrix with a lower water content). When determining this level, possible co-elution of the pesticide in the blank sample was not taken into account, because the correct selection of the blank sample before preparing the series of measurements eliminates the problem. LOQ corresponds to RL (Reporting Limit) and usually also LCL (Lowest Calibration Level). LOD corresponds to LOQ. The test range (working range) is from LOQ to 1000 times LOQ.
Evaluation of selectivity and specificity (responses in the blank sample; ion ratio in the samples to recovery), linearity (deviations from the calibration line; changes in response at individual calibration levels measured before and after the sample sequence), and accuracy (individual recoveries) is presented in Figure 3.
In each measurement sequence, one blank sample is analyzed. The acceptance criterion is the analyte response in the blank sample being ≤30% of the RL (reporting limit).
In each measurement sequence, the ion ratio (IRa) is evaluated in at least one recovery sample. The acceptance criterion is an IRa within ±30% (relative) of the average of the calibration standards in the sequence.
Each sequence includes the measurement of at least five matrix-matched calibration standard solutions (hereafter referred to as calibration standards). The LCL (lowest calibration level) concentration is lower than or equal to the LOQ. The LOQ is lower than or equal to the MRL (in the case of an analyte that is part of a multi-component residue definition, the LOQ must be lower than or equal to the ratio of the MRL to the number of components in the residue). The maximum possible LCL concentration for an analyte with a single-component residue definition and an MRL (Maximal Residual Limit) of 0.01 mg/kg is: 0.01 µg/mL of undiluted extract for a 10 g sample weight, and 0.005 µg/mL of undiluted extract for a 5 g sample weight. The concentration of the highest calibration point is 50 µg/mL of undiluted extract for such an analyte.
Analysis of the matrix-matched calibration standard solutions is evaluated using external calibration or internal standard method (Chlorpyrifos D10), using weighted linear regression (1/x) with the evaluation software Sciex OS-Q, version 1.7.2.
The acceptance criterion for the maximum absolute deviation from the calibration curve (Δcal) for individual calibration standards is ≤20%. The acceptance criterion for the maximum absolute change in response at individual calibration levels measured before and after the sample sequence (Δresponse) is ≤30%.
A sample with a high concentration of analytes may be diluted up to 100-fold; therefore, the working range is from the LCL up to 100 times the concentration of the highest calibration point.
Recovery is calculated as the ratio of the measured concentration/amount of the substance to the true concentration/amount obtained by the analytical procedure, expressed as a percentage. If no certified reference material is available, recovery is determined by spiking a blank sample.
The analyte recovery is calculated using the following formula:
Recovery = (measured concentration/actual concentration) × 100 (%)
Within each sequence, recovery samples (spiked blank samples) are measured at LOQ and 2 × LOQ levels. The average recovery is calculated for each tested enrichment level. The acceptance criterion is a range of 70–120%. Individual recoveries should be within the range of 60–140%.
The consistency of measurements obtained in the same laboratory under agreed conditions (method, analyte, operator, environment) over an extended period defines the within-laboratory reproducibility. Mathematically, it is expressed as RSDwR of all recoveries obtained for a given analyte within this validation. The acceptance criterion is RSDwR ≤ 20%.
Recoveries and within-laboratory reproducibility are not assessed separately for different matrix categories according to commodity groups but are assessed collectively for all matrices.
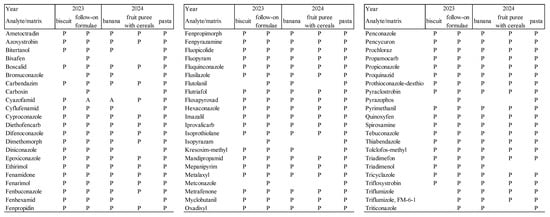
Figure 3.
Evaluation of linearity, selectivity, specificity, and accuracy. P—passes acceptance criteria. A—accepted; empty space—analyte was not analyzed. Accepted: the ratio of the ion ratio in the recovery and the average of the ion ratios in the calibration standards is not ± 30% (IRa), and/or a small deviation occurred in another parameter, while all other parameters are satisfactory.
3.3. Tables for Optimized LC and MS/MS Conditions

Table 1.
Optimized LC conditions.
Table 1.
Optimized LC conditions.
| LC Conditions | |
|---|---|
| Column: | Kinetex® 2.6 μm Polar C18 100 Å, 100 × 2.1 mm |
| Column temperature: | 30 °C |
| Injection volume: | 4 μL |
| Autosampler temperature: | 15 °C |
| Mobile phase A: | 0.1% Formic acid in deionized water |
| Mobile phase B: | 0.1% Formic acid in Methanol |
| Run time: | 22 min |
| Flow rate: | 0.25 mL/min |

Table 2.
LC method, gradient elution.
Table 2.
LC method, gradient elution.
| Time (min) | Mobile Phase A (%) | Mobile Phase B (%) |
|---|---|---|
| 0 | 5 | 95 |
| 2 | 5 | 95 |
| 5.2 | 45 | 55 |
| 10.7 | 95 | 5 |
| 11.7 | 95 | 5 |
| 11.8 | 100 | 0 |
| 15 | 100 | 0 |
| 15.1 | 5 | 95 |
| 22 | 5 | 95 |

Table 3.
Optimized MS/MS conditions.
Table 3.
Optimized MS/MS conditions.
| MS/MS Conditions | |
|---|---|
| Scan type: | Scheduled MRM |
| Target cycle time: | 1 s |
| Ion source: | ESI |
| Curtain gas (CUR): | 40 psi |
| Collision gas (CAD): | Medium |
| Ion Spray Voltage (IS): | 5500 V |
| Temperature (TEM): | 400 °C |
| Ion source Gas 1 (GS1): | 50 psi |
| Ion source Gas 2 (GS2): | 50 psi |
| Entrance Potencial (EP): | 10 V |
4. Discussion
This validated LC-MS/MS method provides a powerful tool for routine high-throughput monitoring of fungicide residues in food and environmental samples, supporting regulatory compliance with maximum residue limits (MRLs) set by the European Union and other authorities.
Analytes that showed significant matrix-dependent response in the initial validation and previous verification are: Carbendazim, Carboxin, Fenpropidin, Fenpropimorph, Propamocarb, and Spiroxamine. In the subsequent milk formula, Carboxin and Pyrimethanil also showed unsatisfactory calibration and/or recovery. In all these cases, problematic calibration and/or recovery must be considered.
Author Contributions
Conceptualization, M.K. and M.D.; methodology, M.D.; software, M.D.; validation, M.D., M.M. and M.K.; formal analysis, M.D.; investigation, M.D.; data curation, M.D.; writing—original draft preparation, M.K.; writing—review and editing, M.D.; visualization, M.K.; supervision, M.D.; project administration, M.D.; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author due to institutional restrictions and the sensitive nature of the raw monitoring data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- QuEChERS. Available online: http://quechers.cvua-stuttgart.de/ (accessed on 30 September 2025).
- Anastassiades, M.; Lehotay, S.J.; Štajnbaher, D.; Schenck, F.J. Fast and Easy Multiresidue Method Employing Acetonitrile Extraction/Partitioning and “Dispersive Solid-Phase Extraction” for the Determination of Pesticide Residues in Produce. J. AOAC Int. 2003, 86, 412–431. [Google Scholar] [CrossRef] [PubMed]
- Maximum Residue Levels. Available online: https://food.ec.europa.eu/plants/pesticides/maximum-residue-levels/guidelines-maximum-residue-levels_en (accessed on 30 September 2025).
Disclaimer/Publisher’s Note: The statements, opinions, and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions, or products referred to in the content. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).