Abstract
A series of novel sulfonylimine derivatives were synthesized via the condensation of aromatic aldehydes with sulfanilamide. Their potential anti-inflammatory activity was evaluated using molecular docking studies against PDE4. The docking protocol was validated through redocking, confirming the accuracy of the computational approach. The docking results showed that the compounds exhibited strong binding affinities, with docking scores ranging from −8.63 to −8.86 kcal/mol, comparable to the reference ligand (−9.20 kcal/mol). The analysis of binding interactions revealed that the sulfonamide moiety played a critical role in PDE4 inhibition through hydrogen bonding, metal coordination, and hydrophobic interactions with key active site residues.
1. Introduction
The sulfonamide group represents a crucial functional moiety in both organic synthesis and pharmaceutical development [1]. Sulfonamides exhibit remarkable chemical stability and unique reactivity, making them valuable as both intermediates and final products in a wide range of chemical transformations. Historically, sulfonamide derivatives gained prominence as the first class of synthetic antibiotics, revolutionizing antimicrobial therapy in the early 20th century [2]. Beyond their well-established antibacterial activity, sulfonamides have been shown to possess a broad spectrum of biological properties, including antidiabetic, antitumor, and anti-inflammatory effects [3].
When sulfonamide derivatives react with aromatic aldehydes or ketones, they form sulfonylimines (also known as Schiff bases). These compounds are characterized by a stable C=N (imine) functional group attached to a sulfonyl moiety. The electron-withdrawing nature of the sulfonyl group (-SO2-) enhances the stability of the imine bond, making sulfonylimines more resistant to hydrolysis than typical imines. Because of their stability, reactivity, and ease of synthesis, sulfonylimines are widely utilized in medicinal chemistry and organic synthesis as versatile intermediates for the development of bioactive molecules, complex molecular architectures, and drug candidates [4]. By combining the structural advantages of both sulfonamides and imines, these compounds play a significant role in drug discovery, chemical manufacturing, and other advanced applications.
From a synthetic perspective, sulfonylimines serve as key intermediates in the preparation of diverse heterocyclic compounds, such as sulfonyl-substituted azoles, oxazolidinones, and thiazoles, which are important scaffolds in modern pharmaceutical development [5]. Their dual reactivity, featuring both nucleophilic and electrophilic centers, allows them to function as versatile building blocks for complex molecular designs. Furthermore, they are widely employed as ligands and chiral auxiliaries in asymmetric synthesis and metal-catalyzed reactions, including hydrogenation, oxidation, and cycloaddition processes [6]. This broad utility extends beyond pharmaceuticals into green chemistry and industrial synthesis, where sulfonylimines are increasingly used in solvent-free and environmentally friendly reactions.
Alongside experimental efforts, computational chemistry has become an essential tool in the rational design and optimization of novel molecules with potential biological or industrial applications. Techniques such as molecular modeling and molecular docking allow for in silico screening of large compound libraries, the identification of promising lead structures, and the optimization of their pharmacokinetic and pharmacodynamic properties prior to synthesis. These approaches significantly reduce both the time and cost associated with drug development while improving the possibility of success.
2. Materials and Methods
The X-ray crystal structure of LEO 29102 in complex with phosphodiesterase 4 (PDE4) (PDB ID: 4WCU) [7], obtained from the RCSB Protein Data Bank, was used in this study. The protein structure was prepared using the Protein Preparation Wizard in the Schrödinger Suite 2021-3 software package to ensure proper geometry, protonation states, and removal of crystallographic artifacts.
The three-dimensional structures of ligands S1, S2, and S3, and the reference ligand (LEO 29102), were generated using Maestro 12.9 and subsequently optimized with LigPrep, employing the OPLS4 force field [8]. Following preparation, the optimized protein and ligand structures were subjected to molecular docking simulations. Docking was performed using Glide software in extra precision (XP) (in the Schrödinger Suite 2023-1) mode [9] to accurately predict binding poses and interactions within the PDE4 active site. The resulting docked complexes were visualized using Chimera X [10], which provided valuable insights into the binding orientation of rutin within the receptor and its interactions with the active site.
3. Results and Discussion
In the present study, a series of sulfonylimine derivatives were synthesized via the condensation of aromatic aldehydes with sulfanilamide, affording the target compounds in good yields (Figure 1).
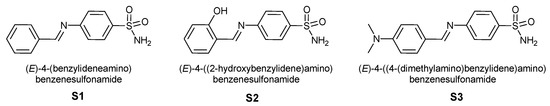
Figure 1.
Structure of synthesized sulfonylimine derivatives (S1, S2, S3).
The potential anti-inflammatory activity of the sulfonylimine derivatives was evaluated through molecular docking studies against phosphodiesterase 4 (PDE4), a key enzyme that regulates inflammatory responses by modulating intracellular cyclic adenosine monophosphate (cAMP) levels. Inhibition of PDE4 leads to elevated cAMP concentrations, which activate protein phosphorylation cascades that ultimately suppress the production of pro-inflammatory cytokines, such as tumor necrosis factor-alpha (TNF-α). This mechanism positions PDE4 as an attractive therapeutic target for the development of novel anti-inflammatory agents [11].
To ensure reliability and predictive accuracy, the docking protocol was validated by redocking of co-crystallized ligand into the active site using the XP docking function. The resulting RMSD between the docked and crystallized ligand poses was 0.40 Å, confirming the accuracy of the docking approach.
The three ligands were then docked into the active site of PDE4. S1 exhibited the strongest binding affinity, with a docking score of −7.86 kcal/mol, followed closely by S2 (−8.79 kcal/mol), and S3 also showed strong binding (−8.63 kcal/mol) (Table 1). These scores are comparable to that of the reference ligand (−9.20 kcal/mol), suggesting that the sulfonylimine derivatives may possess promising anti-inflammatory properties.

Table 1.
Docking score of docked sulfonylimine derivatives and reference ligand.
The analysis of ligand interactions within the active site of the PDE4 enzyme revealed that the sulfonamide group plays a crucial role in enzyme inhibition. Specifically, the amine moiety of the sulfonamide formed strong hydrogen bonds with key catalytic residues Thr271 and Asp318, stabilizing the ligands within the binding pocket. In addition, the oxygen atoms of the sulfone group participated in metal coordination bonds with the catalytic metal ions Mg502 and Zn501 for ligands S1 and S2, while for ligand S3, a single metal coordination bond was observed with Mg502. These interactions are essential, as PDE4 activity is highly dependent on the presence of these divalent metal ions, which are involved in catalysis and substrate stabilization.
For ligands S1 and S2, further stabilization was provided through aromatic interactions. The aromatic ring linked to the sulfonamide group engaged in π-π stacking and π-cation interactions with the residue His160, while a second aromatic ring formed additional π-π stacking interactions with Phe372. These non-covalent interactions play a significant role in anchoring the ligands deep within the hydrophobic regions of the binding pocket, thus enhancing binding affinity and selectivity toward PDE4.
Moreover, hydrophobic contacts were identified with several hydrophobic residues within the active site, including Tyr159, Ile336, Met337, Phe340, Met357, Leu319, and Phe432. These interactions further contribute to the overall stability of the ligands-enzyme complex by minimizing solvent exposure and reinforcing the binding through van der Waals forces.
While S3 exhibited fewer metal coordination interactions compared to S1 and S2, its hydrogen bonding network and hydrophobic interactions still provided a favorable binding conformation (Figure 2).
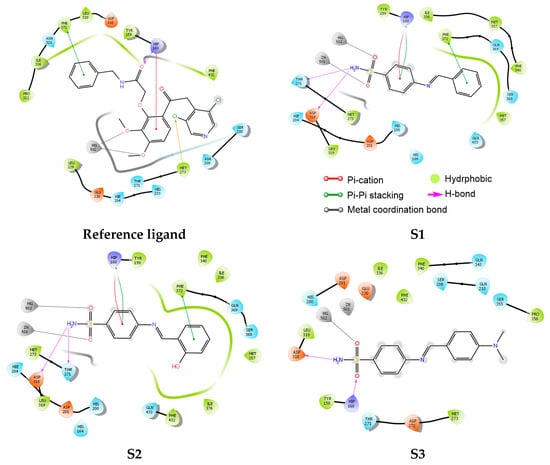
Figure 2.
2D binding interactions of ligands S1, S2, and S3, and the reference ligand, within the active site of the PDE4 enzyme obtained from molecular docking studies.
Taken together, these results highlight the multifaceted role of the sulfonamide moiety in PDE4 inhibition. The hydrogen bonding network, metal coordination, and aromatic interactions collectively ensure strong and specific binding to the enzyme’s active site. The presence of π-π stacking and π-cation interactions indicates that the orientation of the aromatic substituents is critical for activity, while hydrophobic contacts with surrounding residues help to further stabilize the complex. This combination of interactions suggests that structural modifications to the sulfonamide group or its linked aromatic rings could be a promising strategy to optimize binding affinity and selectivity for PDE4 inhibitors.
4. Conclusions
In this work, sulfonylimine derivatives were synthesized and computationally evaluated for their potential anti-inflammatory activity. Molecular docking studies against PDE4 demonstrated that all synthesized compounds displayed favorable binding affinities, closely matching that of the reference inhibitor. Detailed interaction analysis highlighted the crucial role of the sulfonamide group in enzyme inhibition through hydrogen bonding, metal ion coordination, and aromatic interactions, which collectively stabilize the ligand–enzyme complex.
Author Contributions
Conceptualization, A.B.; methodology, R.M. and A.B.; characterization, Y.O.B. and R.M.; writing—original draft preparation, A.B.; writing—review and editing, Y.O.B., A.B. and N.E.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are provided within the article.
Acknowledgments
This work was supported by the General Directorate for Scientific Research and Technological Development (DG-RSDT), Algerian Ministry of Scientific Research, Applied Organic Chemistry Laboratory (FNR 2000).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bouzina, A.; Benzaid, C.; Bentoumi, H.; Bouone, Y.O.; Djendi, M.L.; Mansouri, R.; Sayad, R.; Aouf, N.E. Design, synthesis, and comprehensive in silico and in vitro evaluation of new sulfonamide derivatives with antimicrobial, antioxidant, and enzyme inhibitory properties. J. Mol. Struct. 2025, 1347, 143317. [Google Scholar] [CrossRef]
- Long, P.H.; Bliss, E.A. The clinical use of sulphanilamide and its derivatives in the treatment of infectious diseases. Ann. Intern. Med. 1937, 11, 575–592. [Google Scholar] [CrossRef]
- Bano, S.; Javed, K.; Ahmad, S.; Rathish, I.G.; Singh, S.; Alam, M.S. Synthesis and biological evaluation of some new 2-pyrazolines bearing benzene sulfonamide moiety as potential anti-inflammatory and anti-cancer agents. Eur. J. Med. Chem. 2011, 46, 5763–5768. [Google Scholar] [CrossRef] [PubMed]
- Weinreb, S.M. N-Sulfonyl Imines—Useful Synthons in Stereoselective Organic Synthesis; Stereoselective Heterocyclic Synthesis II; Springer: Berlin/Heidelberg, Germany, 2008; pp. 131–184. [Google Scholar]
- Zhang, H.Z.; Jeyakkumar, P.; Kumar, K.V.; Zhou, C.H. Synthesis of novel sulfonamide azoles via C–N cleavage of sulfonamides by azole ring and relational antimicrobial study. New J. Chem. 2015, 39, 5776–5796. [Google Scholar] [CrossRef]
- Schenkel, L.B.; Ellman, J.A. Novel sulfinyl imine ligands for asymmetric catalysis. Org. Lett. 2003, 5, 545–548. [Google Scholar] [CrossRef] [PubMed]
- Felding, J.; Sørensen, M.D.; Poulsen, T.D.; Larsen, J.; Andersson, C.; Refer, P.; Engell, K.; Ladefoged, L.G.; Thormann, T.; Vinggaard, A.M.; et al. Discovery and early clinical development of 2-{6-[2-(3, 5-dichloro-4-pyridyl) acetyl]-2, 3-dimethoxyphenoxy}-N-propylacetamide (LEO 29102), a soft-drug inhibitor of phosphodiesterase 4 for topical treatment of atopic dermatitis. J. Med. Chem. 2014, 57, 5893–5903. [Google Scholar] [CrossRef] [PubMed]
- Release, S. 2: LigPrep, Schrödinger, LLC, New York. Sci. Rep. 2021, 11, 9510. [Google Scholar]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shaw, D.E.; Shelley, M.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, 4792. [Google Scholar] [CrossRef] [PubMed]
- Perez-Aso, M.; Montesinos, M.C.; Mediero, A.; Wilder, T.; Schafer, P.H.; Cronstein, B. Apremilast, a novel phosphodiesterase 4 (PDE4) inhibitor, regulates inflammation through multiple cAMP downstream effectors. Arthritis Res. Ther. 2015, 17, 249. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).