Abstract
CRISPR-Cas systems have transformed genome engineering with their exceptional precision, programmability, and affordability. Although they originate from microbial defense mechanisms, expanding their use, especially in therapeutics, requires a chemically oriented framework that allows for tunable, reversible, and safe gene editing. This review offers a multidisciplinary look at recent progress in the structural, synthetic, and computational aspects of CRISPR-Cas technologies. Structural analyses examine the domain architectures of Cas enzymes, including the recognition (REC), nuclease (HNH and RuvC), and PAM-interacting domains, emphasizing the catalytic importance of divalent metal ions. Comparative insights into Cas9, Cas12, and Cas13 demonstrate functional diversity across DNA- and RNA-targeting systems, supported by high-resolution structural data on guide RNA pairing and conformational dynamics. The review highlights advances in chemical modulation, such as anti-CRISPR proteins, small-molecule inhibitors, and stimuli-responsive switches, focusing on structure–activity relationships. Additionally, bioorganic delivery systems like lipid nanoparticles, polymers, and cell-penetrating peptides are discussed for their role in improving in vivo delivery through formulation chemistry. Computational chemistry methods—molecular docking, molecular dynamics simulations, and virtual screening—are identified as critical tools for discovering and optimizing modulators. The use of AI-driven tools is proposed as a promising direction for rational CRISPR design. Overall, this chemistry-focused perspective emphasizes the importance of molecular control in developing the next generation of programmable and safe CRISPR-based therapies.
1. Introduction
CRISPR was first described in the 1980s and is recognized as the adaptive immune system together with Cas proteins that defend bacteria and archaea against infectious viruses and foreign DNA [1]. It constitutes Class 1 and Class 2, which are further divided into various types and subtypes, that are the two primary dissimilar classes to which effector protein systems belong. Class I, III, and IV systems utilize multi-protein effector complexes [2]. Class 2 systems utilize single-protein effectors, unlike Class 1. Class 2 systems are subdivided into three categories and several subtypes depending on the effector proteins. They comprise type II CRISPR systems, such as Cas9, type V systems, such as Cas12, and type VI systems, such as Cas13. The most common Cas effector remains the Class 2 type II nuclease Cas9. Genome editing entails accurate alterations at targeted locations within the genome to achieve intended changes to the DNA sequence [3]. CRISPR/Cas9 gene modification is among the most effective genome editing methods. Recent studies have demonstrated that addressing specific factors related to drug resistance with this technique can greatly improve the efficacy of anticancer medications [4]. In bacteria, the CRISPR/Cas9 system functions as an RNA-oriented defense mechanism. This system is capable of editing genes in eukaryotic cells, including those associated with MDR. To achieve this, single-guide RNA (sgRNA) can be crafted to match the intended sequence and be introduced into the target cell together with Cas9, an endonuclease.
The sgRNA directs Cas9 to the target sequence, where Cas9 induces a double-strand break in that sequence. This method allows for the removal or addition of the desired sequence into genes [5]. CRISPR/Cas achieves recognition by base pairing between guide RNA and a target sequence, which is both straightforward and adaptable, with target site selection requiring only adherence to the protospacer-adjacent motif (PAM) specifications of various systems. In contrast to the last two generations of genome editing methods, the CRISPR/Cas system is straightforward, adaptable, reliable, effective, and easily transformed. These attributes have allowed CRISPR/Cas to swiftly supersede ZFN and TALEN as the primary genome editing methods. CRISPR/Cas serves as a protective mechanism, safeguarding bacteria and archaea from invasion by mobile genetic elements and bacteriophages [6].
A chemistry-centric perspective is vital for advancing CRISPR-Cas9 technology because the system’s core functions are inherently chemical in nature. DNA cleavage by Cas9’s HNH and RuvC nuclease domains proceeds through Mg2+-dependent catalysis, where active site residues such as His840 and Lys866 coordinate metal ions to stabilize the transition state—a process elucidated through NMR, molecular dynamics, and QM/MM studies [7]. Understanding these mechanisms enables rational design for improved efficiency. Similarly, off-target effects stem from subtle variations in hydrogen bonding, base stacking, and electrostatic interactions between guide RNA and DNA, which can be mitigated through chemically informed strategies such as truncated guide RNAs (17–18 nt), introduction of mismatched guanines at the 5′ end (“GGX20”), or backbone modifications like 2′-O-methyl, 2′-O-methyl 3′-phosphonoacetate, and phosphorothioate linkages [8]. Functional enhancements also leverage chemical modifications—including 2′-fluoro and locked nucleic acid (LNA) substitutions in guide RNAs or chemical alterations to Cas9 mRNA and donor templates—to improve stability, reduce immune responses, and increase therapeutic viability. Finally, chemistry-driven delivery strategies, inspired by RNAi and oligonucleotide delivery systems, use backbone or sugar modifications and ligand conjugation to enhance cellular uptake, protect CRISPR components from degradation, and enable tissue-specific targeting [9].
In conclusion, enhancement of CRISPR-Cas systems from molecular tools to clinical therapies requires a chemistry perspective. The interface of structural biology, synthetic chemistry, and computation offers next-generation CRISPR applications that are precise, safe, and adaptable.
2. Molecular Mechanisms of Cas Enzymes
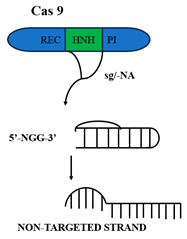
CRISPR-Cas mechanisms operate through the highly regulated and specific activities of CRISPR- associated (Cas) enzymes. The most well-characterized effectors of genome editing are Cas9, Cas12, and Cas13, and they possess distinct structural architectures and catalytic mechanisms. Cas9, Cas12, and Cas13 are three well-characterized CRISPR-associated nucleases with different structures and target specificities. Cas9 (Type II) has a recognition (REC) lobe and a nuclease lobe with HNH and RuvC domains, and a PAM-interacting (PI) domain. An sgRNA binding triggers conformational changes favorable for PAM scanning (e.g., 5′-NGG-3′). After PAM recognition, DNA unwinding makes guide–target base pairing to trigger HNH (target strand) and RuvC (non-target strand) cleavage, with Mg2+/Mn2+ ion dependence [10].
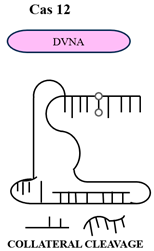
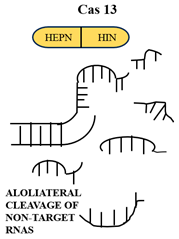
Cas12a (Type V) uses a single RuvC-like domain to cut both DNA strands asymmetrically and generate staggered ends. It targets T-rich PAMs (5′-TTTV-3′) and acts with a single crRNA. Cas12a, after target cleavage, has non-specific ssDNA degradation activity, which is utilized in DETECTR diagnostics. Cas13 (Type VI) is a two-HEPN domain RNA-guided RNase. Binding of the target RNA by crRNA induces structural rearrangements that facilitate cleavage of target and proximal non-target RNAs, facilitating SHERLOCK-based detection [11]. In these mechanisms, as shown in Table 1, guide RNA binding acts as a switch for activation, connecting target recognition and nuclease activation by allosteric mechanisms. Cryo-electron microscopy(Thermo Fisher Scientific, Waltham, MA, USA) and X-ray crystallography (Bruker AXS Inc., Madison, WI, USA) studies revealed multiple conformational states, thus making possible the design of high-fidelity Cas variants with reduced off-target effects.

Table 1.
Molecular Mechanisms of different Cas Enzymes.
3. Synthetic Modulators of CRISPR Activity
Synthetic modulators (Figure 1) signify a growing approach to adjust CRISPR-Cas systems, broadening their use beyond just gene editing. These modulators can be generally classified into protein-based, chemical, and engineered regulatory mechanisms. Anti-CRISPR proteins (Acrs) have developed as natural blockers of CRISPR-Cas systems in response to phage counter-defense strategies. Initially discovered in 2013, more than 120 Acrs spanning almost 100 protein families have since been documented, aiming at various CRISPR subtypes [12]. These proteins are produced early in phage infection to inhibit host CRISPR defense, and their amounts are precisely controlled by anti-CRISPR-associated (Aca) genes. Examples include AcrIIA2 and AcrIIA4 that inhibit SpCas9, along with AcrIIC1/3, which obstructs NmeCas9. Acrs exhibit significant structural and mechanistic variety—spanning from obstructing crRNA-Cas assembly to inhibiting DNA binding or cleavage—and frequently mimic unrelated protein structures, indicating several evolutionary origins. Improvements like AlphaFold2-driven structure prediction are revealing their mechanisms of action. Motivated by these natural proteins, synthetic chemical mimics are being investigated to attain reversible, adjustable regulation of CRISPR activity, thus expanding their possibilities for therapeutic use [13]. Small molecules could serve as a compelling substitute for protein-based anti-CRISPR agents in certain uses. Small molecules have greater permeability through the membrane, are proteolytically stable in vivo, and typically exhibit lower immunogenicity than their protein counterparts; therefore, they are more favorable as agents or pharmaceuticals for regulating Cas9 activity. A small-molecule Cas9 inhibitor was recently discovered through fluorescence polarization-based high-throughput screening (HTS) [14]. Nonetheless, finding novel types of Cas9 inhibitors with varying mechanisms of action may enhance and broaden the possible applications of Cas9 in the future.
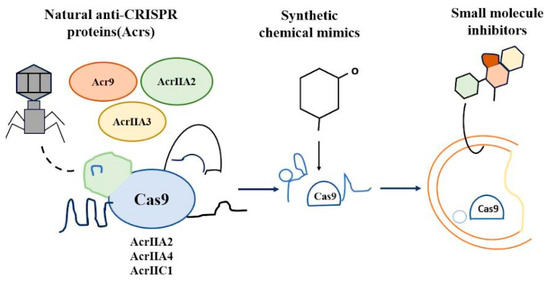
Figure 1.
Synthetic Modulators.
Engineered light- or small-molecule-sensitive switches allow for spatiotemporal control of Cas9 activity. Ligand-gated systems utilize drug-binding sites to switch Cas9 activity on or off, and aptamer-conjugated gRNAs utilize small-molecule-responsive motifs for conditional editing. For example, optogenetic Cas9 variants allow genome editing to be controlled by blue light, with reversible and localized regulation. In addition, rapamycin-inducible dimerization domains have been utilized to control Cas9 nuclease activity in a drug-dependent manner [15]. These strategies add to the toolkit for accurate therapeutic genome editing. SAR-guided design with synthetic analog libraries is being investigated to optimize CRISPR modulators. Systematic alteration of chemical substituents in SAR studies facilitates the identification of structural determinants essential for Cas9 activation or inhibition. For instance, small-molecule screening with SAR optimization resulted in inhibitors that ablate SpCas9 activity with enhanced potency and selectivity [16]. Such rational optimization offers a route to next-generation synthetic regulators of CRISPR systems.
4. Bioorganic Delivery Platforms
Delivery constitutes the primary barrier for CRISPR therapies, particularly in vivo, where stability and targeting present significant challenges. Viral vectors are effective yet pose safety issues, prompting attention to non-viral bioorganic carriers—lipid nanoparticles, peptides, and polymers are safer and adjustable alternatives [17].
Lipid nanoparticles (LNPs) are now the most clinically advanced non-viral delivery platform in the clinic, enabled by FDA-approved RNA-based therapeutics including siRNA (Patisiran) and COVID-19 mRNA vaccines. Translational application of LNPs for CRISPR technology has been demonstrated recently in the first-in-human trial in in vivo gene editing (NCT04601051), where NTLA-2001 (Intellia/Regeneron) was dosed to treat transthyretin (TTR) amyloidosis with cardiomyopathy. In this open-label phase I trial, single intravenous dosing of LNP-encapsulated CRISPR/Cas9 led to a greater than 90% reduction in serum TTR in all dosage groups, with no dose-limiting toxicities and mild infusion-related reactions. This milestone trial is proof of the clinical potential of LNPs for systemic delivery of CRISPR [18]. The salient clinical data are summarized in Table 2, demonstrating the therapeutic efficacy and safety profile of NTLA-2001 as a major advance in systemic CRISPR delivery.

Table 2.
Clinical summary of NTLA-2001, the first in vivo CRISPR/Cas9 trial using LNPs for transthyretin amyloidosis.
Cell-penetrating peptides (CPPs), particularly arginine-rich and amphipathic variants, are now widespread non-viral delivery carriers (Figure 2) for CRISPR/Cas. Traditional sequences like Tat and penetratin promote Cas9 ribonucleoprotein (RNP) uptake by electrostatic interaction and membrane fusion, whereas fusogenic modifications and chemical optimization (e.g., D-amino acid substitution, lipidation) enhance stability and endosomal release. More recently, peptide–nucleic acid (PNA)–CPP conjugates were demonstrated to have higher binding, nuclear targeting, and genome editing efficiency compared to traditional CPPs [19]. While progress has been made, in vivo translation is still hindered by endosomal entrapment and compromised systemic stability.
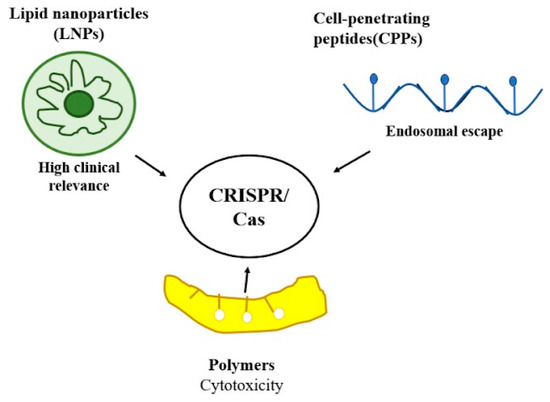
Figure 2.
Overview of non-viral CRISPR delivery systems.
Biodegradable and cationic polymers have been extensively investigated as non-viral nanocarriers for CRISPR/Cas delivery. Poly(lactic-co-glycolic) (PLGA) nanoparticles provide tunability of degradability and biocompatibility, and have been demonstrated to improve the efficiency of homology-directed repair (HDR) in genome editing [20]. Polyethylenimine (PEI), a cationic polymer, provides strong nucleic acid condensation and effective endosomal release, but high-molecular-weight versions have high cytotoxicity; low-molecular-weight or modified PEI derivatives enhance safety. Chitosan, a natural polysaccharide, provides pH-responsive nucleic acid release and excellent biocompatibility. Conjugation chemistries such as disulfide linkages, PEGylation, and incorporation of targeting ligands further enhance polymer-based systems by allowing stability, circulation, and site-specific delivery [21]. Although promising preclinical results have been reported, optimization is still required to optimize delivery efficiency over cytotoxicity.
5. Computational Chemistry and Cheminformatics
Docking is a method for computational modeling. It forecasts the optimal energy-minimized conformation of one molecule when interacting with another molecule to create a stable complex virtually, enhancing the experimental approach. It is fast and economical in comparison to the trial--and-error approaches that involve experimental research. Molecular docking of BRD0539 with SpCas9 was carried out using AutoDock Vina (v1.1.2) [22] through flexible ensemble docking. A 25 × 25 × 25 Å3 grid box was positioned on a recognized binding pocket, considering nearby residue side chains as flexible (≤25 rotatable bonds). The exhaustiveness parameter was configured to 100 (default 8) to enable more extensive conformational sampling at an increased computational expense. Every run kept the highest-ranked binding mode according to Vina’s scoring function. To confirm the results (Table 3), docking was also carried out using GNINA, which incorporates CNN-based scoring. The leading poses from Vina and GNINA coincided within the CTD pocket, demonstrating <2 Å RMSD for ligand heavy atoms, signifying dependable convergence of docking forecasts. Molecular docking and free energy assessments indicate that BRD0539 has the strongest binding affinity for the CTD domain of SpCas9, obstructing the PAM recognition pocket. This stops Cas9 from effectively binding to DNA, thus serving as an allosteric inhibitor of CRISPR- Cas9 function [23].

Table 3.
Free Energies of BRD0539 Binding to SpCas9 Evaluated by Various Methods.
Molecular dynamics (MD) simulations of SpCas9–ligand complexes were performed using AMBER22, initiating with energy minimization, heating, and equilibration, then proceeding with extensive production runs lasting up to 3–4 μs to assess the stability of ligand binding. Binding energies were calculated through the MM-GBSA method to pinpoint essential residues involved in ligand interactions. Additionally, alchemical free energy perturbation (FEP) simulations with suitable restraints were utilized to determine absolute binding free energies, guaranteeing precise handling of ligand positioning throughout decoupling. Free energy landscapes based on principal component analysis (PCA) were subsequently created from backbone movements to reflect key conformational states and their associated stabilities, offering an in-depth perspective on ligand--induced impacts on Cas9 dynamics [24].
Generative protein language models (LMs), trained on extensive protein datasets, can create new CRISPR-Cas proteins by understanding natural evolutionary pressures. By adapting these models to the CRISPR–Cas Atlas (which comprises over 1.2 million operons from microbial genomes, including 389,000 individual effectors like Cas9, Cas12a, and Cas13), researchers increased Cas protein variety beyond natural resources such as UniProt. Utilizing ProGen2-based LM, approximately 4 million novel Cas protein sequences were created, signifying a 4.8-fold rise in sequence diversity. For the Cas9, Cas12a, and Cas13 families, the diversity increased by 4.1×, 6.7×, and 7.1×, respectively. The generated sequences often exhibited 40–60% identity to natural proteins, simulating evolutionary novelty. To verify performance, AlphaFold2 forecasted structures for 2000 generated sequences, with over 81% exhibiting high-confidence folds (pLDDT > 80) akin to natural proteins. This suggests that generative AI can both investigate new Cas variants and direct design towards particular families (e.g., Cas9) with little input, like in Figure 3 [25].
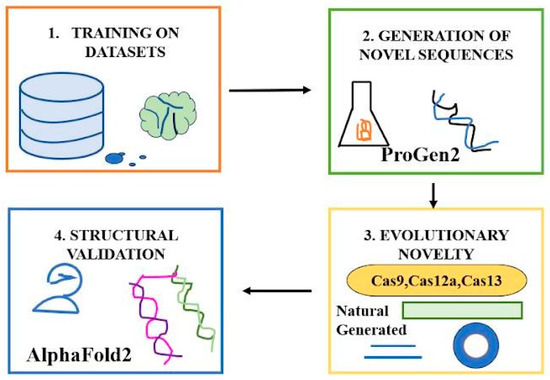
Figure 3.
Generative Protein Language Models to create novel CRISPR-Cas Proteins.
6. Future Scope
The integration of CRISPR with electrochemical biosensors (E-CRISPR) offers rapid, affordable, and portable diagnostics, with potential to expand beyond nucleic acids to proteins and metabolites. Polymer–lipid hybrid nanoparticles provide stable, biocompatible, and targeted delivery platforms for CRISPR, even across barriers like the blood–brain barrier. Exosome-based CRISPR delivery shows promise for precise epigenetic modulation with low immunogenicity, and advancements in cargo loading and imaging could enable combined therapeutic and diagnostic (theranostic) applications.
7. Conclusions
CRISPR Cas systems have evolved from bacterial defense mechanisms into potent and customizable molecular instruments for genome modification. Structural investigations of Cas9, Cas12, and Cas13 have shown how domain arrangements, metal ion dependent catalysis, and conformational shifts regulate their function, offering a framework for systematic engineering. Expanding on this basis, synthetic modulators such as anti-CRISPR proteins, small molecules, and optogenetic or ligand-responsive switches provide accurate and adjustable regulation of gene editing results. At the translational stage, bioorganic delivery systems like lipid nanoparticles, polymers, and cell-penetrating peptides have tackled the primary issue of in vivo administration, with initial clinical trials showing safety and effectiveness. Concurrent progress in computational chemistry, molecular dynamics, and AI-based design is speeding up the identification of new Cas variants and chemical regulators, enhancing the variety and adaptability of CRISPR technologies. Collectively, these advancements position CRISPR as a chemically adjustable system instead of a fixed editing instrument. Its future depends on combining structural, synthetic, delivery, and computational methods to create interventions that are effective, manageable, and clinically safe. With the rise of next-gen smart modulators, hybrid carriers, and theranostic applications, CRISPR technologies are set to revolutionize medicine, agriculture, and biotechnology with unmatched accuracy and flexibility.
Author Contributions
Conceptualization, Y.S. and R.G.; methodology, Y.S.; software, Y.S.; validation, Y.S., A.S. and C.D.; formal analysis, Y.S.; investigation, Y.S.; resources, R.G.; data curation, Y.S.; writing—original draft preparation, Y.S.; writing—review and editing, A.S., C.D. and S.M.; visualization, Y.S.; supervision, R.G.; project administration, R.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable, The study did not involve humans or animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analysed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Garneau, J.E.; Dupuis, M.-È.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadán, A.H.; Moineau, S. The CRISPR/Cas Bacterial Immune System Cleaves Bacteriophage and Plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, Classification and Evolution of CRISPR-Cas Systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, X.; Zhang, Y.; Zhang, Q. CRISPR/Cas Genome Editing Improves Abiotic and Biotic Stress Tolerance of Crops. Front. Genome Ed. 2022, 4, 987817. [Google Scholar] [CrossRef] [PubMed]
- Vaghari-Tabari, M.; Hassanpour, P.; Sadeghsoltani, F.; Malakoti, F.; Alemi, F.; Qujeq, D.; Asemi, Z.; Yousefi, B. CRISPR/Cas9 Gene Editing: A New Approach for Overcoming Drug Resistance in Cancer. Cell Mol. Biol. Lett. 2022, 27, 49. [Google Scholar] [CrossRef]
- Mohammadzadeh, I.; Qujeq, D.; Yousefi, T.; Ferns, G.A.; Maniati, M.; Vaghari-Tabari, M. CRISPR/Cas9 Gene Editing: A New Therapeutic Approach in the Treatment of Infection and Autoimmunity. IUBMB Life 2020, 72, 1603–1621. [Google Scholar] [CrossRef]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef]
- Palermo, G.; Casalino, L.; Jinek, M. Two-Metal Ion Mechanism of DNA Cleavage in CRISPR-Cas9. Biophys. J. 2020, 118, 64a. [Google Scholar] [CrossRef]
- Nierzwicki, Ł.; East, K.W.; Binz, J.M.; Hsu, R.V.; Ahsan, M.; Arantes, P.R.; Skeens, E.; Pacesa, M.; Jinek, M.; Lisi, G.P.; et al. Principles of Target DNA Cleavage and the Role of Mg2+ in the Catalysis of CRISPR–Cas9. Nat. Catal. 2022, 5, 912–922. [Google Scholar] [CrossRef]
- Lennox, K.A.; Behlke, M.A. Chemical Modifications in RNA Interference and CRISPR/Cas Genome Editing Reagents. In RNA Interference and CRISPR Technologies; Sioud, M., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; Volume 2115, pp. 23–55. ISBN 978-1-0716-0289-8. [Google Scholar]
- Gupta, R.; Gupta, D.; Ahmed, K.T.; Dey, D. Modification of Cas9, gRNA and PAM: Key to Further Regulate Genome Editing and Its Applications. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2021; Volume 178, pp. 85–98. ISBN 978-0-12-821590-6. [Google Scholar]
- Antropov, D.N.; Stepanov, G.A. Molecular Mechanisms Underlying CRISPR/Cas-Based Assays for Nucleic Acid Detection. CIMB 2023, 45, 649–662. [Google Scholar] [CrossRef]
- Choudhary, N.; Tandi, D.; Verma, R.K.; Yadav, V.K.; Dhingra, N.; Ghosh, T.; Choudhary, M.; Gaur, R.K.; Abdellatif, M.H.; Gacem, A.; et al. A Comprehensive Appraisal of Mechanism of Anti-CRISPR Proteins: An Advanced Genome Editor to Amend the CRISPR Gene Editing. Front. Plant Sci. 2023, 14, 1164461. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Maji, B.; Gangopadhyay, S.A.; Lee, M.; Shi, M.; Wu, P.; Heler, R.; Mok, B.; Lim, D.; Siriwardena, S.U.; Paul, B.; et al. A High-Throughput Platform to Identify Small-Molecule Inhibitors of CRISPR-Cas9. Cell 2019, 177, 1067–1079.e19. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, A.P.; Fussenegger, M. Synthetic Gene Circuits for Regulation of Next-Generation Cell-Based Therapeutics. Adv. Sci. 2024, 11, 2309088. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, D.; Wan, F.; Chen, B.; Wu, G.; Li, F.; Ren, Y.; Liang, P.; Wan, J.; Songyang, Z. Identification and Analysis of Small Molecule Inhibitors of CRISPR-Cas9 in Human Cells. Cells 2022, 11, 3574. [Google Scholar] [CrossRef]
- Hajebi, S.; Yousefiasl, S.; Rahimmanesh, I.; Dahim, A.; Ahmadi, S.; Kadumudi, F.B.; Rahgozar, N.; Amani, S.; Kumar, A.; Kamrani, E.; et al. Genetically Engineered Viral Vectors and Organic-Based Non-Viral Nanocarriers for Drug Delivery Applications. Adv. Healthc. Mater. 2022, 11, 2201583. [Google Scholar] [CrossRef]
- Kotit, S. Lessons from the First-in-Human in Vivo CRISPR/Cas9 Editing of the TTR Gene by NTLA-2001 Trial in Patients with Transthyretin Amyloidosis with Cardiomyopathy. Glob. Cardiol. Sci. Pract. 2023, 2023, e202304. [Google Scholar] [CrossRef]
- Volpi, S.; Cancelli, U.; Neri, M.; Corradini, R. Multifunctional Delivery Systems for Peptide Nucleic Acids. Pharmaceuticals 2020, 14, 14. [Google Scholar] [CrossRef]
- Lotfi, M.; Morshedi Rad, D.; Mashhadi, S.S.; Ashouri, A.; Mojarrad, M.; Mozaffari-Jovin, S.; Farrokhi, S.; Hashemi, M.; Lotfi, M.; Ebrahimi Warkiani, M.; et al. Recent Advances in CRISPR/Cas9 Delivery Approaches for Therapeutic Gene Editing of Stem Cells. Stem Cell Rev. Rep. 2023, 19, 2576–2596. [Google Scholar] [CrossRef]
- Rodríguez-Rodríguez, R.; Carreón-Álvarez, C.; Cruz-Medina, C.A.; Knauth, P.; López, Z.; Fletes-Vargas, G.; Sahagún, M.R. A Review of pH-Responsive Chitosan-Based Hydrogels for Drug Delivery Applications. Eur. Polym. J. 2025, 237, 114173. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- McNutt, A.T.; Francoeur, P.; Aggarwal, R.; Masuda, T.; Meli, R.; Ragoza, M.; Sunseri, J.; Koes, D.R. GNINA 1.0: Molecular Docking with Deep Learning. J. Cheminform. 2021, 13, 43. [Google Scholar] [CrossRef]
- Zuo, Z.; Liu, J. Structure and Dynamics of Cas9 HNH Domain Catalytic State. Sci. Rep. 2017, 7, 17271. [Google Scholar] [CrossRef]
- Johnson, S.R.; Fu, X.; Viknander, S.; Goldin, C.; Monaco, S.; Zelezniak, A.; Yang, K.K. Computational Scoring and Experimental Evaluation of Enzymes Generated by Neural Networks. Nat. Biotechnol. 2025, 43, 396–405. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).