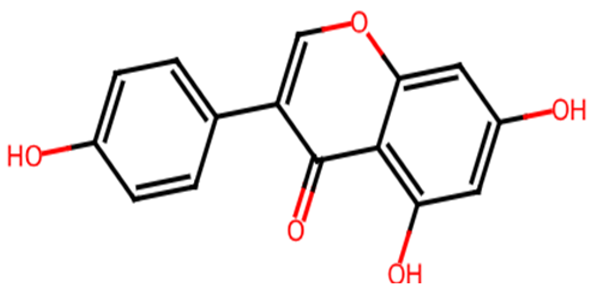
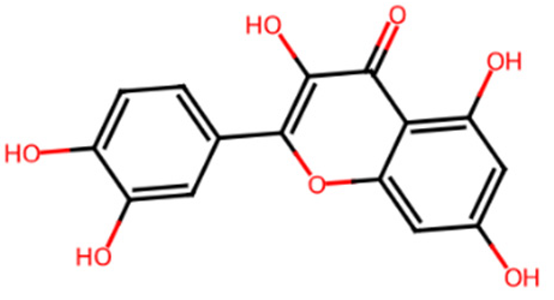
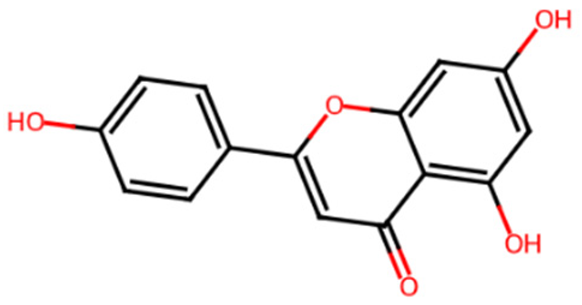
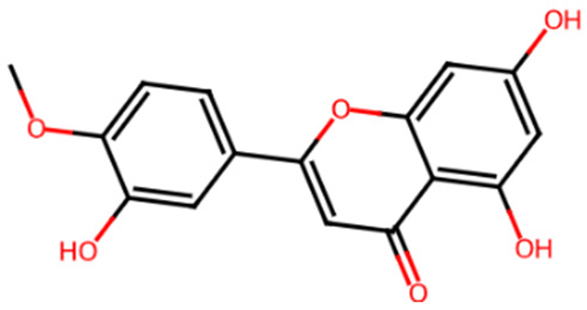
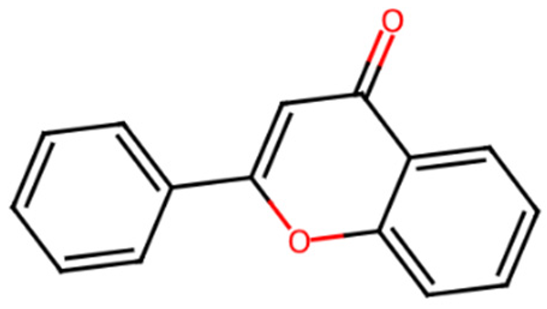
Abstract
Diabetes is a chronic metabolic disorder characterized by persistently high blood glucose levels due to insulin malfunction, defective insulin secretion, or both. Chromen-4-one, known to have diverse biological activity, is a core structure found in many natural products, particularly in the flavonoid and isoflavonoid families. The study aims to explore the potential of Chrome-4-one derivatives as a potential antidiabetic agent through the α-glucosidase inhibition mechanism. The compounds were retrieved from the PubChem database, optimized, and prepared using ChemDraw 12.0, Spartan14, and UCSF Chimera. The post-docking analysis was performed using BIOVIA Discovery Studio. Theoretical oral bioavailability and toxicity predictions were performed using ADMETlab3.0. Molecular docking of the compounds against the α-glucosidase enzyme (PDB ID: 3A4A) was carried out using AutoDock Vina 1.2.5. According to Lipinski’s rule of five (5), all the ligands passed the oral bioavailability and are druggable. The binding score of all the ligands was better than the native ligand (−5.7 Kcal/mol) but slightly lower than that of Acarbose (−9.0 Kcal/mol), except for L7 (Myricetin), which equals the standard drug. The ligands revealed good interaction with the enzyme’s active site residues. The most notable interactions were hydrogen bonding, van der Waals, Pi–anion, Pi–cation, Pi–Pi T-shape, Pi–Sigma, and carbon–hydrogen bonds. The ligands interacted with the key catalytic residues: Asp352, Glu277, Glu411, Trp158, and Arg442, which are responsible for α-glucosidase inhibition. The result of the study suggests that the chrome-4-one derivatives have the potential to be utilized as a lead molecule for orally available α-glucosidase inhibitors.
1. Introduction
Diabetes represents a prevalent condition impacting millions worldwide, distinguished by elevated blood glucose levels resulting from insulin-related complications [1]. Diabetes classification based on the WHO identifies two main types [2]: type 1, caused by the destruction of pancreatic β-cells, resulting in the inability to produce insulin, and type 2, primarily due to insufficient insulin secretion or malfunction [3]. Studies revealed that long-term diabetes has been associated with various health complications, including cardiovascular disease, kidney dysfunction, and neuropathy [4]. The inhibition of α-glucosidase, an enzyme responsible for the hydrolysis of starch into simple sugars, serves as a recognized and effective approach to regulating blood glucose levels in the management of type 2 diabetes [5]. Chromones (4H-chromen-4-ones) represent a class of heterocyclic compounds characterized by a benzo-γ-pyrone framework [6]. The naturally occurring benzopyrones possess diverse biological properties like antiallergic, anti-inflammatory, and antidiabetic [7]. This study aims to explore the potential of Chrome-4-one derivatives as an antidiabetic agent through the α-glucosidase inhibition mechanism.
2. Methods
2.1. Ligand Preparation
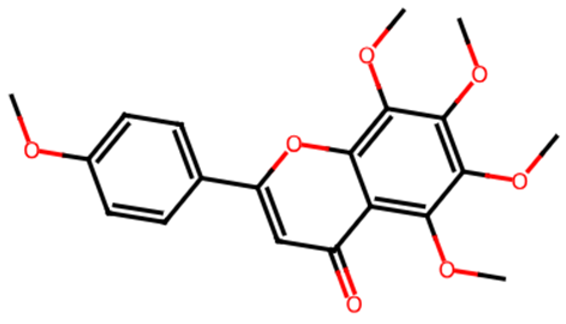
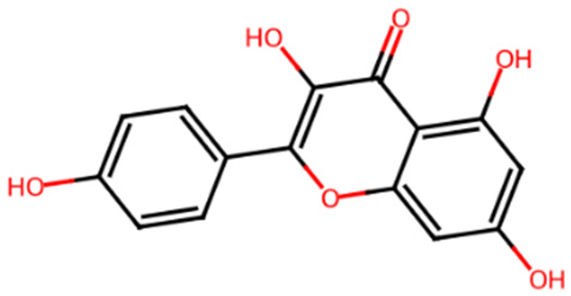
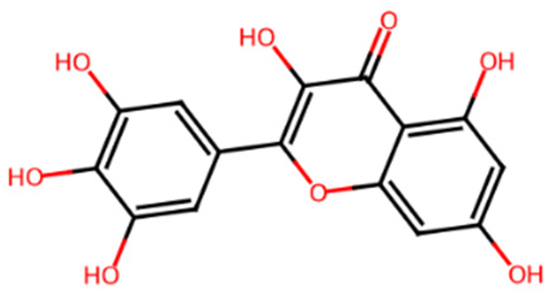
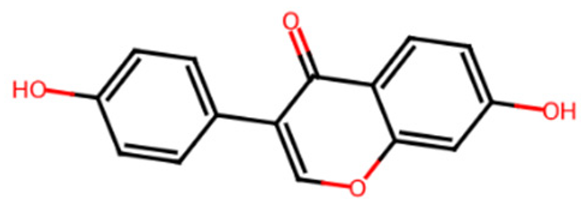
Chromen-4-one derivatives used in this research were queried from the PubChem database. The 2D structures were generated using ChemDraw Ultra version 12.0.2, and Spartan 14v 114 was used to convert the 2D structures to 3D structures. The ligands were optimized and saved as mol2 files.
2.2. Receptor Preparation
The crystal structure of yeast α-glucosidase (PDB: 3A4A) protein was downloaded from the Protein Data Bank (PDB, http://www.rcsb.org accessed on 8 August 2025). The enzyme was prepared by removing all non-residues followed by the addition of hydrogen atoms and Gasteiger charges to the amino acid residues, using UCSF Chimera version 1.17.3 [8].
2.3. Molecular Docking
The prepared 3D ligands and the receptor were converted to pdbqt by utilizing AutoDockTool version 1.5.6 [9]. The molecular docking of the ligands and target enzyme was carried out using Autodock Vina 1.2.5 [10] with the aid of a Cygwin64 terminal. The docking calculations were viewed using UCFS chimera version 1.17.3 [8] and were saved in pdb format. The saved pdb files were viewed using Discovery Studio Visualizer version 20.1.0 for receptor–ligand interactions.
2.4. Theoretical Oral Bioavailability
The oral bioavailability prediction was carried out using an online web server (ADMETlab 3.0).
3. Results and Discussion
Table 1 shows the chemical description of the ligands. The result of theoretical oral bioavailability in Table 2 showed that all the designed ligands passed Lipinski’s rule of five, meaning they are all druggable. All the ligands have a good synthetic accessibility score (<6). The binding score of all the ligands in Figure 1 were better than the native ligand (−5.7 Kcal/mol) but slightly lower than that of Acarbose (−9.0 Kcal/mol), except for L7 (Myricetin), which equals the standard drug. Figure 2 reveals the docking validation of the native ligand. The ligands revealed good interaction with the enzyme’s active site residues. The most significant interactions were hydrogen bonding, van der Waals, Pi–anion, Pi–cation, Pi–Pi T-shape, Pi–Sigma, and carbon–hydrogen bonds. The ligands interacted with the key catalytic residues: Asp352, Glu277, Glu411, Trp158, and Arg442, which are responsible for α-glucosidase inhibition (Figure 3, Figure 4, Figure 5 and Figure 6). The molecular docking studies revealed that chromen-4-one derivatives have a promising inhibitory activity. This is in agreement with the work of Kumara et al. (2025) which reveals that chromones have the potential as α-glucosidase inhibitors with good pharmacokinetic properties [3]. The result in Table 2 correlates with the binding affinity (Figure 1). L7 shows higher lipophilicity (LogP value) in Table 2, which correlates with the binding affinity (related to biological activity) of −9.0 Kcal/mol, which has the best affinity.

Table 1.
Common name, PubChem ID, IUPAC name, chemical structure, and SMILES of the selected chrome-4-one derivatives.

Table 2.
Insilico theoretical oral bioavailability of the designed chromen-4-one derivatives.
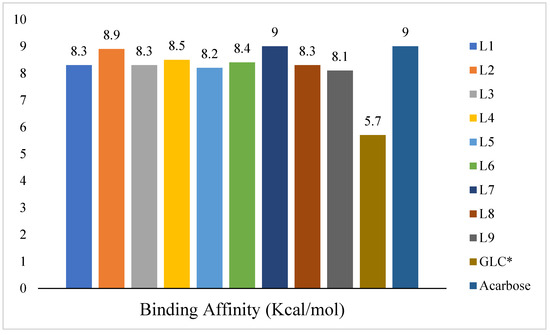
Figure 1.
Binding affinities of the docked ligands (chromen-4-one derivatives), native ligand (GLC*), and Acarbose (standard drug).
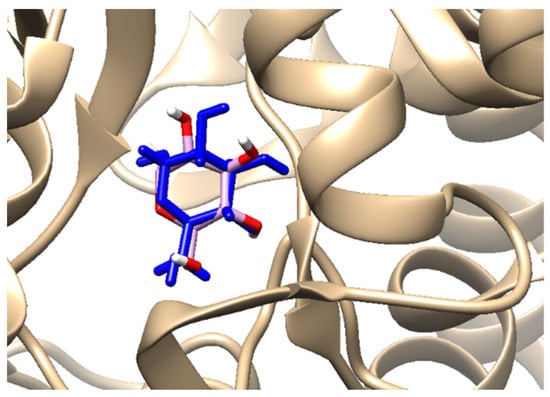
Figure 2.
Crystal structure of α-glucosidase (blue) and re-docked ligand (pink–red edge) superimposed on the crystal structure for validation purposes.
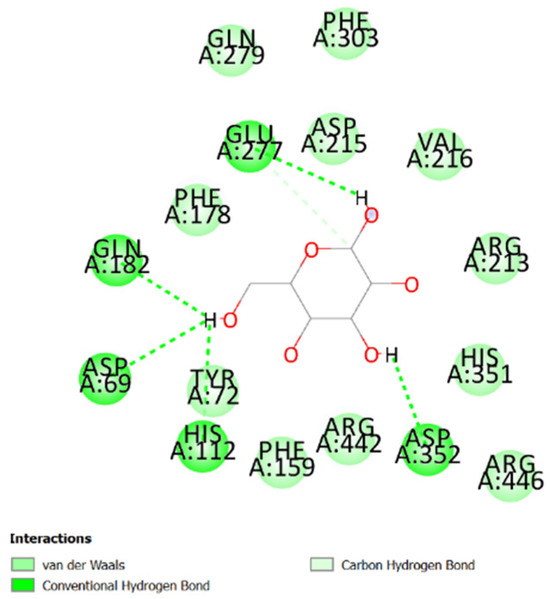
Figure 3.
Two-dimensional pose interaction of native ligand at the active site of α-glucosidase.
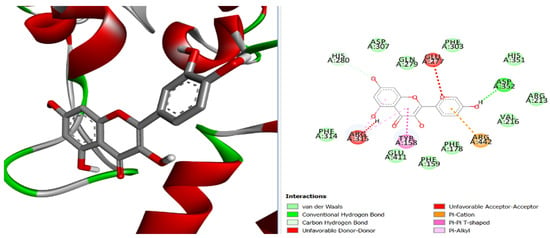
Figure 4.
Three-dimensional and two-dimensional pose interaction of L2 at the active site of α-glucosidase.
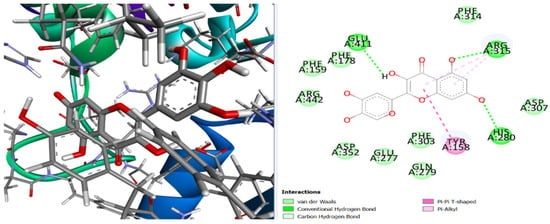
Figure 5.
Three-dimensional and two-dimensional pose interaction of L7 at the active site of α-glucosidase.
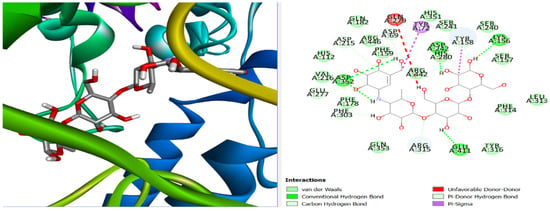
Figure 6.
Three-dimensional and two-dimensional pose interaction of Acarbose at the active site of α-glucosidase.
4. Conclusions
The ligand interacts with amino acid residues at the enzyme’s active site, which is responsible for α-glucosidase inhibition. This suggests that the chromen-4-one derivatives have the potential to be used as α-glucosidase inhibitors.
Author Contributions
Conceptualization, writing—original draft preparation, writing—review and editing, project administration, funding acquisition, I.G.; methodology, software, validation, A.S.B. and M.I.; formal analysis, investigation, resources, data curation, visualization, supervision, Y.A.G. and Y.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available in this manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2009, 37, S62–S67. [Google Scholar]
- Mendes, A.L.; Miot, H.A.; Junior, V.H. Diabetes mellitus and the skin. An Bras. Dermatol. 2017, 92, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Kumara, S.T.; Patrudu, T.B.; Polisetti, V.K.; Chinnachennaiahgari, V.B.; Yatam, S.; Katari, N.K.; Gundla, R. Dihydropyrimidinone-Based Chromones as New α-Glucosidase Inhibitors. ChemistrySelect 2025, 10, e05463. [Google Scholar] [CrossRef]
- Padhi, S.; Kumar, A.; Behera, A. Biomedicine & pharmacotherapy type II diabetes mellitus: A review on recent drug-based therapeutics. Biomed. Pharmacother. 2020, 131, 110708. [Google Scholar] [PubMed]
- Dirir, A.; Daou, M.; Yousef, A.; Yousef, L. A review of alpha-glucosidase inhibitors from plants as potential candidates for the treatment of type-2 diabetes. Phytochem. Rev. 2021, 21, 1049–1079. [Google Scholar] [CrossRef]
- Kuldeep, T.P.; Santosh, S.C.; Manoj Damale Jaiprakash, N.S.; Navanand, B.W.; Gokul, V.S.; Ramesh, S.N.; Sunil, V.G. Design, synthesis, and pharmacological profiling of 2-(Furan-2-yl)- chromen-4-one derivatives: Theoretical and molecular insights. Results Chem. 2025, 17, 102588. [Google Scholar] [CrossRef]
- Benny, A.T.; Arikkatt, S.D.; Vazhappilly, C.G.; Kannadasan, S.; Leelabaiamma, M.S.N.; Thomas, R.; Radha, E.K.; Shanmugam, P. Chromone, A Privileged Scaffold in Drug Discovery: Developments in the Synthesis and Bioactivity. Mini-Rev. Med. Chem. 2022, 22, 1030–1063. [Google Scholar] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Sanner, M.F. Python: A Programming Language for Software Integration and Development. J. Mol. Graph. Mod. 1999, 17, 57–61. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).