Abstract
The Aurora kinase A (Aurora-A), overexpressed in cancer cells, represents a promising anti-cancer therapeutic target due to its role in mitotic progression and chromosome instability. Aurora-A contains a recently described drug pocket within its Targeting Protein for Xklp2 (TPX2) interaction site, offering a promising target for small-molecule disruption and selective inhibition. In this study, 1281 natural products from Argentina’s database (NaturAr), encompassing chemically diverse and structurally rich metabolites, were evaluated using a machine learning model based on molecular fingerprints and variational autoencoders (VAEs) to predict inhibitory activity with high-throughput efficiency. From this initial screening, 624 compounds were classified as active type against Aurora-A, and subsequently subjected to molecular docking using FRED software (v4.3.0.3) against the Aurora-A crystal structure (PDB: 5OSD), focusing on the TPX2-binding interface. Among them, 117 compounds with various scaffolds showed better binding scores than the co-crystallized ligand, highlighting their potential to interact with the druggable target site through stable and specific molecular contacts. This workflow effectively prioritized compounds of natural origin from Argentina for the discovery of new Aurora-A kinase inhibitors, demonstrating the value of integrating AI-driven screening with structure-based modeling. These findings highlight the identification of novel scaffolds with high binding potential, offering promising starting points for the development of selective Aurora-A inhibitors.
1. Introduction
Cancer remains a leading cause of mortality worldwide, with dysregulation of cell cycle progression and mitotic machinery contributing significantly to tumor development and progression [1]. Aurora kinase A (Aurora-A), a serine/threonine kinase overexpressed in various cancers, plays a critical role in mitotic spindle assembly, centrosome maturation, and chromosome segregation, making it an attractive therapeutic target for anti-cancer drug discovery. Inhibition of Aurora-A disrupts mitotic progression, induces apoptosis, and reduces chromosome instability in malignant cells [2]. A recently identified druggable pocket at the Aurora-A–Targeting Protein for Xklp2 (TPX2) interaction site offers a novel opportunity for selective inhibition by small molecules, potentially minimizing off-target effects associated with ATP-competitive inhibitors [3,4].
Natural products from diverse ecosystems, such as those found in Argentina, represent a rich source of chemically diverse scaffolds with potential bioactivity against cancer targets [5]. The NaturAr database, comprising approximately 1281 Argentinian natural products, provides a valuable repository for virtual screening efforts [6]. Computational approaches, including machine learning (ML) models and molecular docking, have revolutionized drug discovery by enabling high-throughput identification of promising candidates while reducing experimental costs and time [7]. ML models based on molecular fingerprints and variational autoencoders (VAE) can efficiently predict inhibitory activity, while structure-based docking refines hits by evaluating binding affinities and interactions within target pockets [7,8].
In this study, we screened the NaturAr database using a concatenated computational protocol. Firstly, we implemented an ML model with a VAE architecture to classify compounds as potential Aurora-A inhibitors. Then we docked the active-predicted molecules against the TPX2-binding interface of Aurora-A (PDB: 5OSD), identifying compounds with superior binding scores compared to the co-crystallized ligand of the target protein. Molecular dynamics simulations further assessed the stability of top hits, revealing favorable RMSD profiles. This integrated workflow highlights the potential of Argentinian natural products as novel Aurora-A inhibitors, paving the way for further experimental validation and development of selective anti-cancer therapies.
2. Materials and Methods
2.1. Compounds Database
The NaturAr database “https://naturar.quimica.unlp.edu.ar/es/ (accessed on 15 May 2025)”, a collaborative, open-source repository cataloging 1281 natural products from Argentinian biodiversity to date, was utilized for this study [9]. Structures were provided in SMILES format.
2.2. Machine Learning Models
The dataset for training the models was obtained from PubChem BioAssay AID 1803719, which documents the inhibition of Aurora A kinase (UniProt ID: O14965). Compounds were classified as “active” (IC50 ≤ 10 μM) or “inactive” based on dose–response curves (accessed on 20 May 2025). Molecular representations needed for model implementation, including Morgan fingerprints (2,048-bit circular patterns) and MACCS keys (166 structural fragments), were computed using RDKit [10]. The VAE architecture, featuring an encoder with two hidden layers (256 → 128 neurons, ReLU activation) that compresses inputs into a 32-dimensional latent space and a symmetric decoder reconstructing the original features, was trained for 100 epochs using mean squared error loss to reduce dimensionality [11]. The VAE’s latent representations then served as optimized inputs for downstream classifiers—RBF-kernel SVM, class-weighted Random Forest, and AUC-optimized XGBoost—further optimized via Bayesian optimization (25 iterations) with stratified 5-fold cross-validation [12,13]. The best model was selected and applied to the NaturAr database, identifying compounds as potential Aurora-A inhibitors (probability > 0.5/best scoring than A9B).
2.3. Protein Preparation and Molecular Docking
The crystal structure of Aurora-A kinase in complex with the co-crystallized ligand A9B at the TPX2-binding interface, PDB ID: 5OSD “https://www.rcsb.org/ (accessed on 22 May 2025)”, was retrieved from the Protein Data Bank [14]. Preparation involved removing water molecules and non-essential ions, followed by protonation using Make Receptor v4.3.0.3 software, OpenEye Scientific. The binding pocket was defined around A9B [15].
SMILES representations of ML-predicted active ligands were converted into 3D structures using OMEGA v5.0.0.3 software in “pose” mode, generating up to 800 conformers per molecule. Molecular docking was performed using FRED software v4.3.0.3 OpenEye Scientific, with the Chemgauss4 scoring function [16]. The protocol was validated by redocking A9B, and a total of 104,949 molecules (up to 800 possible conformers per 624 active-predicted/A9B molecules) were docked.
2.4. Molecular Dynamics Simulations
The top three docked-protein complexes and the 9AB-protein complex underwent 25 ns MD simulations using NAMD2 v2.14 “http://www.ks.uiuc.edu/Research/namd/ (accessed on 25 May 2025)”, with the AMBER ff14SB forcefield (protein), GAFF (ligands), and TIP3P water models. Systems were solvated in a padded cubic box, neutralized with NaCl, and energy-minimized for 1000 steps [17]. Production simulations were carried out in the NPT ensemble at 310 K and 1 atm, using a 1 fs time step and SHAKE constraints, with coordinates saved every 5000 steps (0.005 ns intervals).
RMSD trajectories were calculated and analyzed using VMD v1.9.4 “http://www.ks.uiuc.edu/Research/vmd/ (accessed on 30 May 2025)”, and plots were generated with Python v3.12.3 “https://www.python.org (accessed on 30 May 2025)”, with the pandas v2.2.2 and matplotlib v3.9.2 libraries. The binding free energy between ligands and Aurora A was evaluated using the molecular mechanics Poisson–Boltzmann surface area (MM/PBSA) approach, applied to previously equilibrated molecular dynamics trajectories [18]. Representative sampling intervals from the equilibrated complexes were selected, and interaction energies were calculated using molecular force fields and an implicit solvation model under physiological ionic conditions.
3. Results and Discussion
3.1. Machine Learning Model Performance
Three molecular representations (Morgan fingerprints, MACCS keys, and a hybrid of both) were evaluated with SVM, Random Forest, and XGBoost classifiers after dimensionality reduction using a VAE. The performance metrics are presented in Table 1. The Morgan fingerprints-SVM model emerged as the optimal choice based on rigorous performance evaluation. While XGBoost achieved a marginally higher test AUC (0.7204), the SVM model demonstrated superior generalization with a smaller train-test AUC and precision disparity, indicating greater resilience to overfitting. In contrast, Random Forest (RF) showed near-perfect performance in the training AUCs of all representations, but suffered a substantial drop in the test AUC. This large difference reflects a marked tendency toward overfitting.

Table 1.
Performance metrics of ML models for predicting Aurora-A inhibitory activity.
The poor performance of MACCS-based models, particularly with SVM (AUC < 0.5), reflects the limitations of MACCS fingerprints, as they provide a simpler and more generalized view of molecular structures that capture only a limited set of chemical features. Similarly, hybrid representations did not offer significant improvements over Morgan, suggesting that redundancy between descriptors does not provide additional predictive value. Morgan fingerprints capture structural features at higher resolution than MACCS keys, resulting in more robust and accurate predictions. This trend reflects greater specificity, focusing on a smaller range of actual interactions, which results in higher overall prediction reliability [19,20]. This suggests the superiority of circular fingerprints for encoding local connectivity patterns with higher resolution, making them more suitable for identifying actual interactions between drugs and targets [20].
The Morgan fingerprints-SVM model was selected for screening the NaturAr database, classifying 624 out of 1281 compounds as potential Aurora-A inhibitors (probability > 0.5/best scoring than A9B).
3.2. Molecular Docking Results
The docking protocol was validated by redocking the co-crystallized ligand A9B into the TPX2-binding interface of Aurora-A (PDB: 5OSD), showing a 1.011 Å RMSD value between the docked and crystal poses, indicating good accuracy of the molecular docking protocol [21]. The 624 ML-predicted active compounds were docked, along with A9B as a control (a total of 625 molecules). Of these, 117 compounds displayed superior FRED Chemgauss4 scores compared to A9B, suggesting stronger predicted affinities for the F-pocket. These hits exhibited diverse scaffolds typical of natural products, with molecular weights ranging from approximately 166 to 466 Da, and LogP values between −2.89 and 6.98.
The top five scoring ligands are summarized in Table 2. Docking scores range from −9.93 to −8.10 kcal/mol, which indicates a stronger binding affinity toward Aurora A than A9B. Figure 1 shows the amino acid residues involved in stabilizing the Aurora A–ligand 534 complex. The hydrogen bond formed between Ser155 and ligand 534, which is also conserved in ligand 533, emerges as the primary interaction anchoring the ligands within the binding site, enhancing the stability of the complexes [22]. Hydrophobic contacts, consistently present across all complexes, play a central role in increasing the affinity between the ligands and the surrounding residues in the active site. This finding aligns with previous reports, indicating that Aurora A-ligand binding is largely driven by van der Waals forces and nonpolar (lipophilic) interactions [22,23,24,25].

Table 2.
FRED Chemgauss4 docking scores and interaction profiles with Aurora-A for the top five compounds.
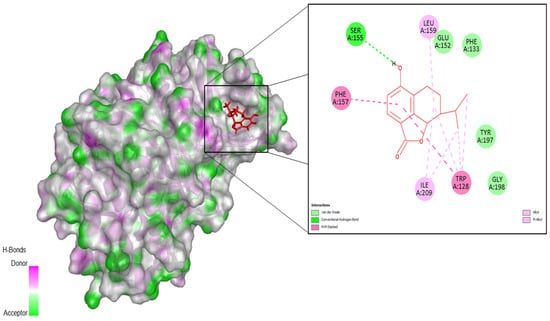
Figure 1.
Interaction of Aurora-A with the docked ligand 534. The binding conformation of ligand 534 within the F pocket of Aurora-A was obtained from molecular docking studies. Key interactions of ligand 534 with protein residues include Ser155, Phe157, Trp128, Leu159, Ile209, Glu152, Phe133, Tyr197, and Gly197. Figure generated using Discovery Studio Visualizer 2024.
The 117 compounds with docking scores superior to A9B comprise naphthalene and cinnamic acid derivatives, flavonoids, terpenic lactones, and prenylated lipids. Representative scaffolds of the ligands exhibiting affinity toward Aurora kinase A are shown in Figure 2, highlighting the structural diversity of the most promising candidates.
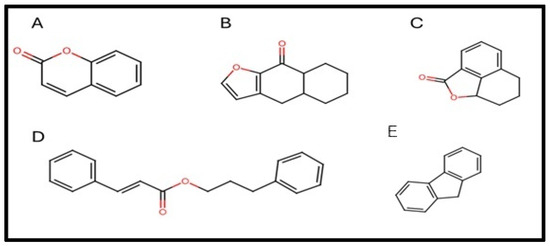
Figure 2.
Scaffolds identified among the active-predicted compounds against Aurora kinase A. The structures correspond to (A) coumarin, (B) benzofuran derivate, (C) isobenzofuranone derivate, (D) Aromatic ester with a stilbene-like framework, and (E) fluorene. Figure generated using Marvin JS 23.8.0.
3.3. Dynamic Stability and Conformational Behavior
To assess binding stability, the top three docked compounds (NaturAr IDs: 534, 1231, and 533), together with A9B as a control, underwent 25 ns MD simulations. Molecular dynamics simulations revealed distinct behaviors regarding the stability and binding of the top-ranked ligands against Aurora-A (Figure 3). Ligands 534 and 533 exhibited relatively low RMSD values (~4–5 Å) (Table 3), indicative of stable complexes throughout the 25 ns trajectory, likely due to their reduced conformational flexibility and rigidity, which fit well into the F pocket. In contrast, ligands A9B and 1231 showed higher fluctuations (~6–7 Å), reflecting increased conformational plasticity within the binding pocket [23]. Despite the higher RMSD, ligand 1231 achieved the most favorable binding free energy (−16.56 kcal/mol), suggesting that adaptive binding and favorable residue-level interactions (π–π stacking and biphenyl/phenolic hydrophobic contacts) can compensate for greater structural fluctuations. Such conformational flexibility may represent induced-fit or conformational selection processes that enhance enthalpic contacts without necessarily diminishing overall affinity.
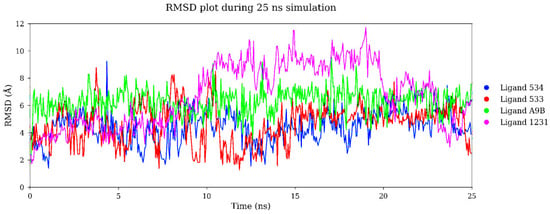
Figure 3.
RMSD values of ligands at the binding pocket of Aurora-A during a 25 ns molecular dynamics simulation.

Table 3.
Molecular dynamics simulation metrics.
4. Conclusions
The identification of novel molecular scaffolds in this study highlights the unique chemical properties of natural products in the search for cancer therapies. The prioritized natural compounds (e.g., coumarins, isochromanones, stilbene-type frameworks, and indenes) exhibited structures that have been little explored in known Aurora-A inhibitors, which are typically based on conventional heterocycles (e.g., pyrrolopyrazoles or quinolines) [22,23,24,25]. This unprecedented structural diversity is relevant as it broadens the chemical space available for anticancer drug design. Resources such as natural product databases (e.g., NaturAr, with more than 1200 metabolites from Argentine biodiversity) are essential for harnessing this chemical diversity in drug innovation. In fact, studies indicate that approximately half of all approved drugs come from natural products or their derivatives [5], highlighting the importance of continuing to explore natural sources using modern computational approaches. Taken together, this study underscores the value of integrating the wealth of natural products with advanced computational methodologies to discover selective Aurora-A inhibitors, providing new starting points for more effective and selective anti-cancer therapies.
Author Contributions
Conceptualization, J.D.G.; methodology, J.D.G.; software, G.C., E.J. and J.D.G.; validation, G.C., E.J. and J.D.G.; formal analysis, J.D.G.; investigation, G.C., E.J. and J.D.G.; data curation, G.C., E.J. and J.D.G.; writing—original draft preparation, G.C. and E.J.; writing—review and editing, J.D.G.; visualization, G.C., E.J. and J.D.G.; supervision, J.D.G.; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Acknowledgments
The authors thank Martin Lavecchia, a member of our research group at CEQUINOR (UNLP-CONICET, CCT La Plata, affiliated with CIC PBA), for his assistance with computational calculations using the OpenEye toolkits through his academic license.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hanahan, D.; Weinberg, R.A. Hallm. Cancer: Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Polverino, F.; Mastrangelo, A.; Guarguaglini, G. Contribution of AurkA/TPX2 Overexpression to Chromosomal Imbalances and Cancer. Cells 2024, 13, 1397. [Google Scholar] [CrossRef]
- McIntyre, P.J.; Collins, P.M.; Vrzal, L.; Birchall, K.; Arnold, L.H.; Mpamhanga, C.; Coombs, P.J.; Burgess, S.G.; Richards, M.W.; Winter, A.; et al. Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches. ACS Chem. Biol. 2017, 12, 2906–2914. [Google Scholar] [CrossRef] [PubMed]
- Janeček, M.; Rossmann, M.; Sharma, P.; Emery, A.; Huggins, D.J.; Stockwell, S.R.; Stokes, J.E.; Tan, Y.S.; Almeida, E.G.; Hardwick, B.; et al. Allosteric Modulation of AURKA Kinase Activity by a Small-Molecule Inhibitor of Its Protein–Protein Interaction with TPX2. Sci. Rep. 2016, 6, 28528. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Martínez Heredia, L.; Quispe, P.A.; Fernández, J.F.; Lavecchia, M.J. NaturAr: A Collaborative, Open-Source Database of Natural Products from Argentinian Biodiversity for Drug Discovery and Bioprospecting. J. Chem. Inf. Model. 2025, 65, 1889–1900. [Google Scholar] [CrossRef]
- Jusoh, A.S.; Remli, M.A.; Mohamad, M.S.; Cazenave, T.; Fong, C.S. How Generative Artificial Intelligence Can Transform Drug Discovery? Eur. J. Med. Chem. 2025, 295, 117825. [Google Scholar] [CrossRef]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and Scoring in Virtual Screening for Drug Discovery: Methods and Applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef]
- NaturAr. Base de Datos de Productos Naturales de Argentina; Universidad Nacional de La Plata: La Plata, Argentina, 2025; Available online: https://naturar.quimica.unlp.edu.ar/es/ (accessed on 15 May 2025).
- RDKit. Open-Source Cheminformatics. 2023. Available online: http://www.rdkit.org (accessed on 20 May 2025).
- Kingma, D.P.; Welling, M. Auto-Encoding Variational Bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar]
- Gómez-Bombarelli, R.; Wei, J.N.; Duvenaud, D.; Hernández-Lobato, J.M.; Sánchez-Lengeling, B.; Sheberla, D.; Aguilera-Iparraguirre, J.; Hirzel, T.D.; Adams, R.P.; Aspuru-Guzik, A. Automatic Chemical Design Using a Data-Driven Continuous Representation of Molecules. ACS Cent. Sci. 2018, 4, 268–276. [Google Scholar] [CrossRef]
- Wu, J.; Chen, Y.; Wu, J.; Zhao, D.; Huang, J.; Lin, M.; Wang, L. Large-Scale Comparison of Machine Learning Methods for Profiling Prediction of Kinase Inhibitors. J. Cheminform. 2024, 16, 13. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- OpenEye Scientific Software. FRED v4.3.0.3: Flexible Docking for Structure-Based Drug Discovery; OpenEye Scientific: Santa Fe, NM, USA, 2020; Available online: https://www.eyesopen.com (accessed on 22 May 2025).
- McGann, M. FRED and HYBRID Docking Performance on Standardized Datasets. J. Comput.-Aided Mol. Des. 2012, 26, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Rogers, D.; Hahn, M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef]
- He, T.; Caba, K.; Ballester, P.J. A Precise Comparison of Molecular Target Prediction Methods. Digit. Discov. 2025, 4, 2548–2558. [Google Scholar] [CrossRef]
- Bell, E.W.; Zhang, Y. DockRMSD: An Open-Source Tool for Atom Mapping and RMSD Calculation of Symmetric Molecules through Graph Isomorphism. J. Cheminform. 2019, 11, 40. [Google Scholar] [CrossRef]
- Tian, Y.-Y.; Tong, J.-B.; Liu, Y.; Tian, Y. QSAR Study, Molecular Docking and Molecular Dynamic Simulation of Aurora Kinase Inhibitors Derived from Imidazo[4,5-b]pyridine Derivatives. Molecules 2024, 29, 1772. [Google Scholar] [CrossRef]
- Bathula, S.; Sankaranarayanan, M.; Malgija, B.; Kaliappan, I.; Bhandare, R.R.; Shaik, A.B. 2-Amino Thiazole Derivatives as Prospective Aurora Kinase Inhibitors against Breast Cancer: QSAR, ADMET Prediction, Molecular Docking, and Molecular Dynamic Simulation Studies. ACS Omega 2023, 8, 44287–44311. [Google Scholar] [CrossRef]
- Siudem, P.; Szeleszczuk, Ł.; Paradowska, K. Searching for Natural Aurora a Kinase Inhibitors from Peppers Using Molecular Docking and Molecular Dynamics. Pharmaceuticals 2023, 16, 1539. [Google Scholar] [CrossRef]
- Beniwal, M.; Jain, N.; Jain, S.; Aggarwal, N. Design, Synthesis, Anticancer Evaluation and Docking Studies of Novel 2-(1-Isonicotinoyl-3-Phenyl-1H-Pyrazol-4-yl)-3-Phenylthiazolidin-4-one Derivatives as Aurora-A Kinase Inhibitors. BMC Chem. 2022, 16, 61. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).