Abstract
Recent advances in computational drug discovery have significantly improved the search for effective treatments for skin cancer, where molecular docking and pharmacokinetics play an important role in identifying new drug-like compounds. This study explores the inhibitory potential of pyrrolopyrazole (4BKY), an enzyme linked to skin cancer progression, using 62 cytotoxic quinoline derivatives. Among these, Ligand 1 and Ligand 4 demonstrated the strongest binding affinities, with docking scores of −8.9519 kcal/mol and −8.7030 kcal/mol, respectively. Their enhanced stability and interaction with key residues GLU 87 and CYS 89 suggest promising inhibitory properties. In addition to docking analysis, these compounds underwent ADMET (Absorption, Distribution, Metabolism, and Excretion) analysis profiling using SWISSADME and pkCSM, to assess their pharmacokinetic Additionally, toxicity assessment was performed using the ProTox-II web server, predicting mutagenicity, carcinogenicity, immunotoxicity, and hepatic toxicity. Findings indicate favorable drug-likeness, efficient synthesis, and compliance with Lipinski’s rule of five, supporting their viability as targeted skin cancer therapeutics. Based on these results, Ligand 1 and Ligand 4 emerge as strong candidates for further research and development in oncology. Future studies will focus on experimental validation and clinical trials to confirm their effectiveness and safety, potentially paving the way for innovative skin cancer treatment strategies involving quinoline-based compounds.
1. Introduction
Skin cancer is a type of cancer that originates in the skin cells and is the most common form of cancer globally. It develops when abnormal skin cells grow uncontrollably, often due to damage caused by ultraviolet (UV) radiation from the sun or tanning beds. There are several types of skin cancer, the most common being basal cell carcinoma, squamous cell carcinoma, and melanoma. Basal cell carcinoma and squamous cell carcinoma are often referred to as non-melanoma skin cancers and are usually less aggressive. Melanoma, however, is more dangerous, as it tends to spread to other parts of the body if not detected and treated early. Risk factors for skin cancer include prolonged exposure to UV radiation, fair skin, a history of sunburns, and a family history of skin cancer. Preventive measures such as using sunscreen, wearing protective clothing, and avoiding excessive sun exposure can significantly reduce the risk of developing skin cancer. Early detection through regular skin examinations is crucial for successful treatment outcomes [1].
The drug discovery process is a complex and multifaceted endeavor that requires a deep understanding of molecular interactions, pharmacokinetic properties, and toxicity profiles to identify potential therapeutic candidates. Quinoline-4-carboxylic acids represent a class of compounds with significant biological activity, including antineoplastic properties. Their versatility and ability to be chemically modified make them attractive for drug discovery endeavors. Advancements in computational chemistry have provided researchers with powerful tools for drug design. MOE (Molecular Operating Environment) [2] is widely used for molecular docking studies, allowing researchers to predict how small molecules, like quinoline-4-carboxylic acids, interact with target proteins. SwissADME [3] offers comprehensive insights into ADME (Absorption, Distribution, Metabolism, Excretion) properties, crucial for understanding the pharmacokinetic profile of potential drug candidates. ProTox-II [4] aids in predicting the toxicity of compounds, helping to identify candidates with the lowest risk of adverse effects. In this study, we designed 65 quinoline-4-carboxylic acid derivatives and evaluated their potential as drug candidates through a series of computational analyses. Our aim was to identify molecules with strong binding affinity, favorable ADME profiles, and low toxicity, thereby streamlining the drug discovery process.
Quinoline-4-carboxylic acids represent a significant class of compounds with a broad range of biological activities. The quinoline ring system, a bicyclic structure comprising a benzene ring fused to a pyridine ring, is known for its versatility in medicinal chemistry. The carboxylic acid group at the 4-position enhances the compound’s ability to form hydrogen bonds, crucial for its interaction with biological targets. These compounds have been extensively studied for their antimicrobial, antiviral, anti-inflammatory, and anticancer properties. Modifications at various positions on the quinoline ring can lead to derivatives with improved potency, selectivity, and pharmacokinetic properties [5,6]. The structure-activity relationship (SAR) of quinoline-4-carboxylic acids indicates that the position and nature of substituents on the quinoline ring significantly influence biological activity. For instance, halogenated derivatives often show enhanced anticancer activity due to increased lipophilicity and improved membrane permeability. Additionally, electron-donating groups can enhance the interaction with nucleophilic sites on target proteins, further increasing the compound’s efficacy.
Vero cells are a line of cells derived from the kidney of an African green monkey and are commonly used in cell culture. These cells are extensively utilized in virology, toxicology, and vaccine production due to their consistent growth and susceptibility to various viruses. In this study, Vero cells were used as a model system to investigate the cytotoxic effects of quinoline-4-carboxylic acid derivatives, providing insights into their potential toxicity and therapeutic efficacy. The application of Vero cells in toxicity testing is crucial for ensuring the safety of new compounds intended for therapeutic use. These cells provide a robust model for studying the mechanisms of drug action and toxicity at the cellular level. In addition to their use in cytotoxicity assays, Vero cells can be employed to study viral replication, vaccine efficacy, and other applications in biomedical research [7].
2. Materials and Methods
2.1. Computational Tools
Throughout this investigation, we used a Dell system equipped with a dual CPU @2.60 GHz, Intel® Core i5-1145G7, and 16 GB RAM. Windows 10 Professional was used as the Computer Operating System. Computational studies, including molecular docking, simulation, and free energy calculations, were conducted using MOE version 17.5 (MOE 2017), while ChemDraw Ultra 16.0 was used for drawing the molecular structures. ADMET predictions were analyzed using SwissADME [3] and ProTox II [4] web servers.
- MOE (Molecular Operating Environment)
MOE is a comprehensive suite for molecular modeling and simulations, offering tools for docking, structure-based design. It facilitates the prediction and analysis of molecular interactions, providing a detailed view of how ligands interact with their protein targets. MOE’s advanced features include flexible receptor docking, which allows for the consideration of protein flexibility during the docking process, enhancing the accuracy of the predictions [8].
- ChemDraw Ultra 16.0
ChemDraw Ultra 16.0 is a powerful and user-friendly software used for creating professional representations of organic, organometallic, polymeric, and biopolymeric materials [9] (https://perkinelmer-chemdraw-professional.software.informer.com/16.0/, accessed on 3 December 2025).
2.1.1. Data Set and Ligand Preparation
A set of 62 cytotoxic quinoline derivative inhibitors against the Vero cell line. The 2D structures of the cytotoxic quinoline derivatives were drawn using ChemDraw Ultra 16.0. was then used to convert the ligands into three-dimensional structures and minimize them. Atomic charges were determined. We used MOE software (Molecular Operating Environment) version 2019.01 [2] for ligand optimization by the AM1 semi-empirical method [10].
2.1.2. Protein Preparation
The crystal structure of unphosphorylated Maternal Embryonic Leucine zipper Kinase (MELK) in complex with a pyrrolopyrazole inhibitor was retrieved from the Protein Data Bank (PDB ID: 4BKY, resolution 1.83 Å) [11]. Prior to docking, the protein was prepared using MOE software: all water molecules, cofactors, and ions were removed; protonation states were adjusted to physiological pH; ligands were treated as flexible, while the enzyme was modeled as a rigid body. This simplification of the enzyme structure is a critical step in molecular modeling, allowing for faster and more efficient calculations. The resulting model provides a reliable framework for evaluating ligand interactions within the active site.
2.2. Molecular Docking
Protein–ligand interactions play a key role in the organization of biological systems. This opens the way for the prediction of their 3D structure, which is a major challenge for the pharmaceutical industry [12]. Molecular docking is one of the most widely used in silico methods. Its purpose is to understand the interactions between a ligand and a receptor to form a protein–ligand complex by studying all the mechanisms and interactions involved in the formation of molecular complexes [13].
Docking essentially comprises two complementary sections:
Docking (selection step): placing the ligand in the active site of the protein and sampling the possible conformations, positions, and orientations, retaining only those that represent the most favorable interaction modes [14].
Scoring (ranking step): estimating the interaction strength and binding affinity between two molecules after docking and assigning a score to the poses obtained [15]. The semiempirical optimization method used in this study follows Stewart’s approach [16].
In this context, we conducted a study of the interactions between the enzyme 4BKY and derivatives of cytotoxic quinolines using MOE software [2]. In our study, we downloaded the protein structure PDB (ID: 4BKY) [11], co-crystallized with the pyrrolopyrazole inhibitor of the formula (3′-{[(4-bromo-1-methyl-1H-pyrrol-2-yl)carbonyl]amino}-N-[(1S)-1-phenyl-2-(pyrrolidin-1-yl)ethyl]-1′,4′-dihydro-5′H-spiro[cyclopropane-1,6′-pyrrolo[3,4-c]pyrazole]-5′-carboxamide: C26H30BrN7O2) using the MOE software [2]. During molecular docking, the inhibitors are oriented towards the active site containing pyrrolopyrazole. The scoring of poses is obtained after 10 search cycles. This result allows for ranking the best poses based on their interaction energies obtained by the “E score”. RMSD is another factor that can be generated from a conformational search to explore the possible conformational space efficiently and exhaustively.
2.3. ADME-T Property Evaluation and Toxicity Assessment
To assess the pharmacokinetic behavior of the selected compounds, their 3D structures were submitted to the SwissADME web tool. The evaluation encompassed key ADME parameters, including gastrointestinal absorption, blood–brain barrier permeability, and potential interactions with cytochrome P450 isoenzymes. These data enabled the prediction of each molecule’s pharmacokinetic profile, facilitating the identification of candidates with favorable drug-like properties for further development.
For toxicity prediction, the molecular structures were analyzed using the ProTox-II platform. This tool provided estimates of acute toxicity levels and highlighted possible adverse effects on major organs and biological systems. Compounds exhibiting the lowest predicted toxicity and acceptable safety margins were prioritized for subsequent investigation.
3. Results and Discussion
3.1. Molecular Docking Studies
Molecular docking analysis was performed to assess the binding affinity of selected quinoline derivatives with the 4BKY protein, a target associated with skin cancer. As presented in Table 1, ligand L_1 exhibited the most favorable binding energy (−8.95 kcal/mol), indicating a strong and stable interaction with the active site residues. Ligands L_3 and L_4 also showed comparable binding energies, suggesting promising inhibitory potential. Notably, L_4 achieved the lowest RMSD_refine value (0.7583 Å), reflecting high spatial accuracy and reliable docking conformation (Figure 1).

Table 1.
Molecular Docking Interactions with 4BKY Protein.
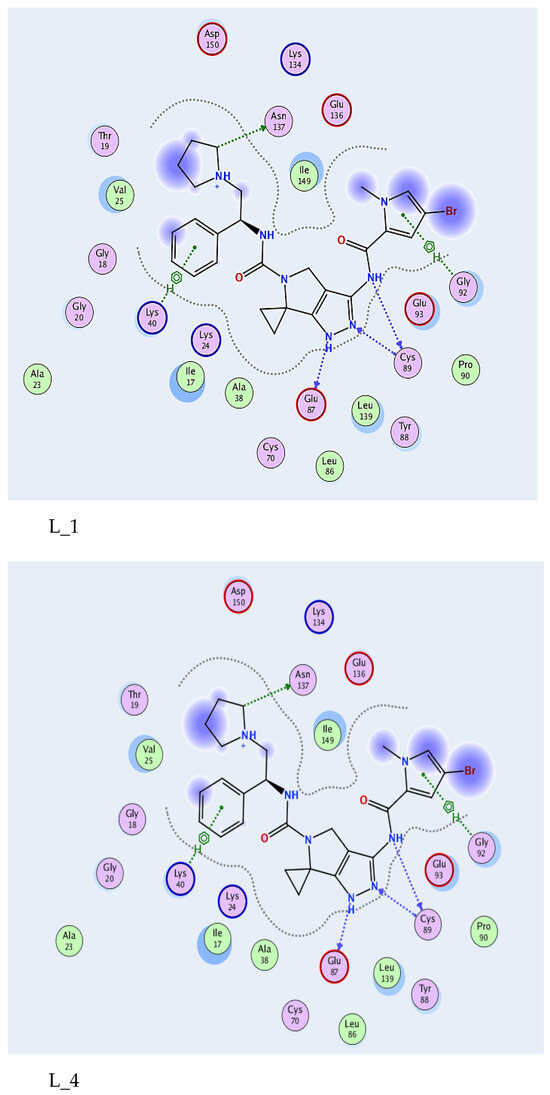
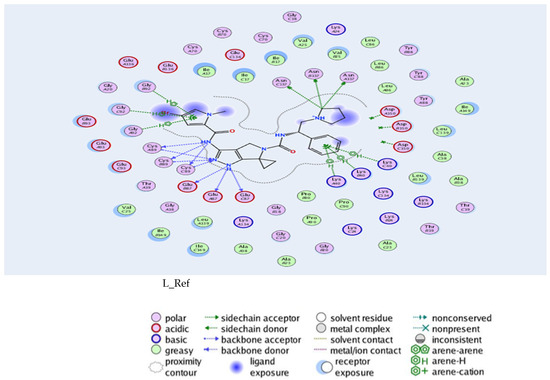
Figure 1.
Two-dimensional interactions and 3D illustration of 4BKY-active site and L_1, L_4.
All tested ligands interacted consistently with key amino acids—GLU87, CYS89, ASN137, LYS40, and GLY92—through hydrogen bonding and π–H interactions, with bond distances ranging from 2.79 to 4.73 Å. The reference compound (L_REF) recorded a binding energy of −8.55 kcal/mol and formed an additional ionic interaction with ASP150, serving as a benchmark for comparison. In contrast, ligand L_26 displayed the weakest binding affinity (−6.74 kcal/mol), which may limit its therapeutic relevance in the context of skin cancer, as detailed in Table 1.
3.2. ADME-Tox Evaluation of Selected Compounds
The ADME–Tox profile of compounds L1, L4, and L_REF (Table 2) reveals that L1 and L4 possess favorable pharmacokinetic properties, despite minor deviations from ideal drug-likeness criteria. Both compounds comply with Lipinski’s Rule of Five and exhibit high gastrointestinal absorption, indicating good oral bioavailability. L4 stands out with the lowest molecular weight (416.47 g/mol) and the best synthetic accessibility score (3.37), suggesting it is easier to synthesize and optimize. However, both L1 and L4 show elevated Log P values, which may affect solubility and distribution. Toxicity alerts such as carcinogenicity, hepatotoxicity, immunotoxicity, and mutagenicity were observed in L1 and L4, requiring further structural refinement. In contrast, L_REF demonstrates a safer toxicity profile but shows blood–brain barrier permeability, which may lead to off-target CNS effects. Overall, L1 and L4 remain promising candidates due to their strong docking performance and acceptable ADME properties, provided that toxicity risks are addressed in future optimization steps.

Table 2.
ADME-T Evaluation of Candidate Ligands Targeting 4BKY Protein.
Author Contributions
Data collection, software, formal analysis, and the first draft of the manuscript were prepared by F.A. and I.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- American Cancer Society. Skin Cancer Facts & Figures. 2023. Available online: https://www.cancer.org/ (accessed on 3 December 2025).
- Chemical Computing Group ULC. Molecular Operating Environment (MOE), 2019.01; Chemical Computing Group ULC: Montreal, QC, Canada, 2019. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xie, X.; Cai, Y.; Yang, X.; Li, J.; Li, Y.; Chen, W.; He, M. Design, Synthesis and Antibacterial Evaluation of Some New 2-Phenyl-quinoline-4-carboxylic Acid Derivatives. Molecules 2016, 21, 340. [Google Scholar] [CrossRef] [PubMed]
- Shoeb, M.; Thakre, P.D.; Dharpure, P.N.; Khan, M. Quinoline Derivatives: A Comprehensive Review of Synthesis, Biological Activities, and Pharmaceutical Applications. IJPPR 2024, 15, 35–51. [Google Scholar] [CrossRef]
- Ammerman, N.C.; Beier-Sexton, M.; Azad, A.F. Growth and maintenance of Vero cell lines. Curr. Protoc. Microbiol. 2008, 11, A.4E.1–A.4E.7. [Google Scholar] [CrossRef] [PubMed]
- PerkinElmer Informatics, Inc. ChemDraw Ultra 16.0. Available online: https://perkinelmer-chemdraw-professional.software.informer.com/16.0/ (accessed on 3 December 2025).
- Vilar, S.; Cozza, G.; Moro, S. Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008, 8, 1555–1572. [Google Scholar] [CrossRef] [PubMed]
- Dewar, M.J.S.; Zoebisch, E.G.; Healy, E.F.; Stewart, J.J.P. Development and use of quantum mechanical molecular models. AM1: A new general purpose quantum mechanical molecular model. J. Am. Chem. Soc. 1985, 107, 3902–3909. [Google Scholar] [CrossRef]
- Protein Data Bank (PDB). Entry: 4BKY. Available online: https://www.rcsb.org/structure/4BKY (accessed on 3 December 2025).
- Lengauer, T.; Rarey, M. Computational methods for biomolecular docking. Curr. Opin. Struct. Biol. 1996, 6, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.Y.; Zhang, H.X.; Mezei, M.; Cui, M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput.-Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.J.P. Optimization of Parameters for Semiempirical Methods I. Method. J. Comput. Chem. 1989, 10, 209–220. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).